Abstract
The currently accepted culture techniques for the detection of Legionella spp. in water samples (AS/NZS 3896:1998 and ISO 11731 standard methods) are slow and laborious, requiring from 7 to 14 days for a result. We describe a fully validated rapid confirmation technique that uses real-time PCR incorporating the intercalating dye SYTO9 for the direct identification of primary cultures, significantly decreasing turnaround time and allowing faster remedial action to be taken by the industry.
Legionellae are ubiquitous organisms and are commonly found in lakes and rivers, usually at low levels (17). These organisms infiltrate water distribution systems and multiply in a variety of man-made constructions, including cooling towers, spa pools, misters, fountains, showers, and ice machines (4). The conditions in these environments make human infection possible via inhalation (or microaspiration in the case of ice machines [9]) of contaminated aerosols. The majority of human infections are caused by Legionella pneumophila serogroup 1, although in Australia and New Zealand, numerous cases have been attributed to Legionella longbeachae found in potting mixes, supposedly due to the use of pine and eucalypt products (11, 16).
There is still no consensus regarding the infectious dose and environmental levels of Legionella that are necessary for the spread of disease. Data of Legionella counts from cooling towers implicated in outbreaks are not readily available, but counts between 1,000 CFU/ml (3) and 100,000 CFU/ml (6) have been found in suspected sources, whereas counts found in potable water supplies in nosocomial settings have been very low (18).
Regardless of outbreak source or infective dose, there is a need for a faster culture and confirmation technique for Legionella. Legionella detection methods adopted in most testing laboratories are based upon the ISO 11731 (2) or the AS/NZS 3896:1998 (3) method in Australia and New Zealand, which are considered the “gold standards.” These culture methods are similar and require traditional confirmation of Legionella-like isolates by subculture on media that primarily challenge the cysteine requirement of the microorganism. This approach in itself presents interpretational challenges, particularly for slow-growing and unusual species of bacteria. For example, the ISO 11731 method requires subculture onto buffered charcoal-yeast extract (BCYE) agar minus cysteine, but Legionella oakridgensis will grow on this medium (13). Several other methodologies exist, each with advantages and disadvantages. The direct fluorescent-antibody (DFA) assay is tedious, lacks sensitivity, can be cross-reactive with non-Legionella isolates, and cannot discriminate between culturable and nonculturable cells (5). Numerous direct PCR methodologies have been reported, and despite obvious speed advantages, they cannot discriminate between culturable and nonculturable cells. Previously, a commercially available PCR kit, the EnviroAmp Legionella kit (Perkin Elmer), was used for Legionella detection in water samples and was adapted for direct colony confirmation using PCR (14) but was subsequently withdrawn from the market due to specificity concerns. Another limitation of alternative methodologies is that the interpretation of the significance of the results is difficult because action levels for the detection of Legionella are based on counts derived using the standard culture techniques. This is particularly the case for individuals who now employ the AS/NZS 3666.3 part 3 method (1) as part of performance-based maintenance procedures which dictate remedial actions based on reported colony counts.
We describe here a colony-based confirmatory assay for the rapid identification of Legionella pneumophila and Legionella spp. using real-time PCR and a double-stranded-DNA-intercalating dye, SYTO9, recently described by Monis et al. (12). In total, 148 isolates from 144 samples (potable waters, evaporative tower water, and cooling tower water) were included in this evaluation. This assay delivered cost and time savings and also allowed the culture, confirmation, and serogrouping of L. pneumophila in as few as 3 days.
Isolation of Legionella from water samples and latex agglutination assays.
Water samples (500 ml) were examined without preconcentration in accordance with the AS/NZS 3896:1998 method (3). In brief, 0.1 ml was inoculated onto buffered charcoal-yeast extract agar base (code CM0655; Oxoid, Basingstoke, Hampshire, United Kingdom) with MWY selective supplement (code SR0118; Oxoid), and 0.01 ml was inoculated onto BCYE agar with BMPA selective supplement (code SR0111; Oxoid). An aliquot was heat treated at 50°C for 30 min, and 0.1 and 0.01 ml were inoculated onto MWY agar. Additionally, 1 ml of sample was acid treated in 9 ml of HCl-KCl acid buffer (pH 2.2) for 5 min, and 0.1 ml was inoculated onto BMPA agar. All plates were incubated at 35°C. Our laboratory protocol was to examine plates for Legionella-like organisms on days 3, 5, and 7, and the suspect isolates were subcultured onto BCYE and horse blood agar (Medvet, Adelaide, South Australia) and incubated for a further 3 days. For this study, isolates were also subcultured on either MWY or BMPA, depending on their origins during primary isolation. Gram-negative organisms that grew on BCYE (and on MWY/BMPA) and not on horse blood agar were reported to be Legionella. Serogrouping with the Legionella latex kit (code DR0800; Oxoid) was performed according to the manufacturer's instructions with isolates subcultured onto BCYE and onto MWY or BMPA. An alternate isolate confirmation method (real-time PCR) was run concurrently with selective media using the methods described below.
Control organisms and DNA template preparation.
Experiments to optimize and validate PCR were performed using the bacterial strains listed in Table 1. Single colonies of control strains or environmental isolates were used to prepare colony suspensions with a McFarland standard of 0.5 (in phosphate-buffered saline), and 5-μl volumes of the suspensions were added directly to each PCR.
TABLE 1.
ATCC strains used for testing the specificities of the mip and 16S rRNA gene assays
| Organism | ATCC accession no. | 16S rRNA PCR result |
mip PCR
|
|
|---|---|---|---|---|
| Take offa | Result | |||
| Legionella pneumophila serogroup 1 | ATCC 43111 | + | <25 | + |
| Legionella pneumophila serogroup 13 | ATCC 43736 | + | <25 | + |
| Legionella pneumophila serogroup 9 | ATCC 35298 | + | <25 | + |
| Legionella pneumophila serogroup 1 | ATCC 33152 | + | <25 | + |
| Legionella pneumophila serogroup 6 | ATCC 33215 | + | <25 | + |
| Legionella pneumophila serogroup 1 | ATCC 33153 | + | <25 | + |
| Legionella anisa | ATCC 35292 | + | >25 | − |
| Legionella micdadei | ATCC 33218 | + | >25 | − |
| Legionella longbeachae serogroup 1 | ATCC 33462 | + | >25 | − |
| Legionella longbeachae serogroup 2 | ATCC 33484 | + | >25 | − |
| Legionella cincinnatiensis | ATCC 43753 | + | >25 | − |
| Legionella sainthelensi | ATCC 35248 | + | >25 | − |
| Legionella santicrucis | ATCC 43119 | + | >25 | − |
| Legionella oakridgensis | ATCC 33761 | + | >25 | − |
| Legionella bozemanii | ATCC 33217 | + | >25 | − |
| Legionella birminghamensis | ATCC 43702 | + | >25 | − |
| Legionella bozemanii serogroup 1 | ATCC 33217 | + | >25 | − |
| Legionella bozemanii serogroup 2 | ATCC 35545 | + | >25 | − |
| Legionella brunensis | ATCC 43878 | + | >25 | − |
| Legionella cherrii | ATCC 35252 | + | >25 | − |
| Legionella dumoffii | ATCC 33279 | + | >25 | − |
| Legionella erythra | ATCC 35303 | + | >25 | − |
| Legionella fairfieldensis | ATCC 49588 | + | >25 | − |
| Legionella feeleii serogroup 1 | ATCC 35489 | + | >25 | − |
| Legionella feeleii serogroup 2 | ATCC 358449 | + | >25 | − |
| Legionella gormanii | ATCC 33297 | + | >25 | − |
| Legionella gratiana | ATCC 49413 | + | >25 | − |
| Legionella hackeliae | ATCC 35999 | + | >25 | − |
| Legionella israelensis | ATCC 43119 | + | >25 | − |
| Legionella jamestownensis | ATCC 35298 | + | >25 | − |
| Legionella jordanis | ATCC 33623 | + | >25 | − |
| Legionella lansingensis | ATCC 49751 | + | >25 | − |
| Legionella maceachernii | ATCC 35300 | + | >25 | − |
| Legionella moravica | ATCC 43877 | + | >25 | − |
| Legionella parisiensis | ATCC 35299 | + | >25 | − |
| Legionella quinlivanii | ATCC 43830 | + | >25 | − |
| Legionella rubrilucens | ATCC 35304 | + | >25 | − |
| Legionella spiritensis | ATCC 35249 | + | >25 | − |
| Legionella steigerwaltii | ATCC 35302 | + | >25 | − |
| Legionella tucsonensis | ATCC 49180 | + | >25 | − |
| Legionella wadsworthii | ATCC 33877 | + | >25 | − |
| Legionella worsliensis | ATCC 49508 | + | >25 | − |
| Aeromonas hydrophila | ATCC 7966 | − | >25 | − |
| Enterobacter aerogenes | ATCC 13048 | − | >25 | − |
| Escherichia coli | ATCC 1175 | − | >25 | − |
| Citrobacter freundii | ATCC 8090 | − | >25 | − |
| Bacillus subtilis | ATCC 6333 | − | >25 | − |
| Klebsiella pneumoniae | ATCC 13883 | − | >25 | − |
| Pseudomonas aeruginosa | ATCC 10145 | − | >25 | − |
| Pseudomonas fluorescens | ATCC 13523 | − | >25 | − |
“Take off” is defined as the cycle at which exponential amplification starts.
Confirmation of isolates using real-time PCR and melting curve analysis.
Legionella 16S rRNA gene PCR was performed as described previously (7, 10, 19) except that AmpliTaq Gold was used as the DNA polymerase, the reaction volume was 25 μl, and the cycling conditions were changed to an initial hold at 95°C for 10 min, followed by 40 cycles consisting of 94°C for 20 s, 60°C for 20 s, and 72°C for 25 s. L. pneumophila-specific PCR was performed using primers mip 99F (5′ TGTCTTATAGCATTGGTGCC 3′) and mip 213R (5′ CAATTGAGCGCCACTCATAG 3′) (8) under the same cycling conditions. For both mip and 16S rRNA assays, 5 μl of template DNA was used in a 25-μl reaction mixture that included 1× PCR buffer II (Applied Biosystems, New Jersey), 2.5 mM MgCl2, 200 μM deoxynucleoside triphosphate mix (Promega Corporation, Madison, WI), 0.5 μM each of the forward and reverse primers, 3.34 μM SYTO9 (Molecular Probes, OR), and 1 U AmpliTaq Gold (Applied Biosystems, New Jersey). All reactions were carried out in a RotorGene 3000 (Corbett Research, Sydney, Australia) with data acquisition at 72°C on the 6-carboxyfluorescein channel (excitation at 470 nm, detection at 510 nm) at a gain of 5. Amplification takeoff (defined as the cycle at which exponential amplification starts) was determined using the comparative quantitation feature of the RotorGene software for the amplification data acquired at a gain of 5. Following amplification, melting curve data were acquired on the 6-carboxyfluorescein channel (at gains of 2 and 5) using a ramping rate of 1°C/60 s from 75°C to 95°C. The differentiated data were analyzed using RotorGene software with the digital filter set as “none.” When required, samples were analyzed by 1% agarose gel electrophoresis with the addition of Gelstar nucleic acid stain (Cambrex Bio Science, Rockland, Inc.) using standard methods (15).
Results and discussion.
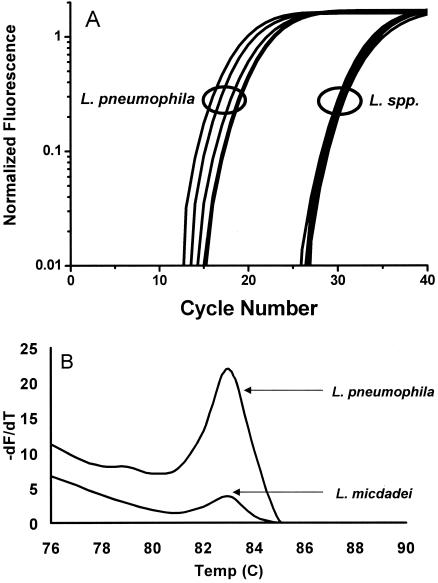
The specificity of the real-time PCR was determined by challenging the assays using the organisms listed in Table 1. The specificity of the 16S rRNA gene assay has been described previously (7, 10, 19) and was confirmed by melting curve analysis, producing a characteristic melting temperature (Tm value) of 88 ± 1°C that corresponded to the detection of a 386-bp fragment by gel electrophoresis (data not shown). PCR using the mip primers amplified a 114-bp product with a Tm value of 82.5 ± 1°C from L. pneumophila and also from some non-pneumophila Legionella species such as L. longbeachae serogroups 1 and 2, L. anisa, L. micdadei, L. cincinnatiensis, L. sainthelensi, and L. santicrucis, but there was a notable difference in the cycles at which amplification started for pneumophila and non-pneumophila species (based on cycle takeoff value). As shown in Fig. 1A, amplification of DNA from L. pneumophila was detected within 12 to 21 cycles for the isolates tested. In comparison, amplification of DNA from other Legionella species occurred after 25 cycles. This difference was supported by melting curve analysis, which found that L. pneumophila samples had relative peak heights threefold greater than those of non-pneumophila Legionella species (Fig. 1B). These observations for the mip reactions can be attributed to differences in primer binding efficiency. The primers are exact matches for L. pneumophila, whereas there are between 3 and 7 base mismatches between the mip 99F or mip 213R primer and the corresponding regions of non-pneumophila Legionella species, including mismatches at the 3′ end of each primer. These mismatches cause poor priming from the genomic DNA of non-pneumophila Legionella species, resulting in an increase in the cycle number at which amplification is detected. Therefore, based on the differential takeoff values, any amplification of mip that is detected after cycle 25 is classed as negative for L. pneumophila.
FIG. 1.
A. Raw cycling data for mip PCR using SYTO9. B. Typical mip melting curve analysis of L. pneumophila and some Legionella spp. (such as L. micdadei) that may produce Tm values similar to those produced by L. pneumophila after 40 cycles. The relative peak height difference is used for differentiation in conjunction with the take-off value.
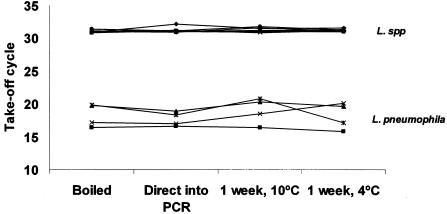
Direct addition of isolate suspensions into the PCR was not significantly different to extraction by boiling at 100°C for 10 min, suggesting that the initial denaturation step of 95°C for 10 min used in the PCR is sufficient to effectively lyse the cells and release template DNA (Fig. 2). Additionally, storage of boiled isolate suspensions at 4°C and −10°C for 1 week did not affect the performance of the assay. The inherent variations seen with the preparation of a suspension with a McFarland standard of 0.5 did not seem to affect the performance of this assay, eliminating the need to quantify DNA inoculums by other methods. It is important, however, to visually compare the isolate suspension to those in 0.5 McFarland standard comparator tubes that are commercially available in order to ensure standardization.
FIG. 2.
Comparison of mip take-off cycles for template DNA preparation and storage.
A total of 144 environmental samples yielding 148 isolates were analyzed by the rapid real-time PCR confirmation method and the traditional confirmation method. Of the 148 isolates tested, the standard method classed 57 as Legionella sp., 36 as L. pneumophila, and 55 as non-Legionella organisms. The rapid assay described here showed complete correlation with the standard method, with no disparities observed (Table 2). All PCR-negative samples were subjected to a repeat PCR (under similar conditions, except approximately 200 copies of L. pneumophila serogroup 1 DNA were incorporated into the PCR master mix) to monitor for PCR inhibition. In all cases the spiked DNA master mix plus the previously negative sample returned a positive result, demonstrating the absence of PCR inhibitors in the original isolate suspensions (data not shown).
TABLE 2.
Comparison of Legionella identifications of 16S rRNA and mip PCR with those of the AS/NZS 3896:1998 method
| Species detected | No. of isolates identified (n = 148)
|
||
|---|---|---|---|
| AS/NZS 3896:1998 method | Real-time PCR
|
||
| 16S rRNA gene | mip | ||
| Legionella species | 57 | 57 | 0 |
| L. pneumophila | 36 | 36 | 36 |
| Non-Legionella spp. | 55 | 0 | 0 |
Fifty Legionella isolates were included in a comparative study of latex agglutination assays from selective (MWY and BMPA) and nonselective (BCYE) media. Twenty-seven L. pneumophila serogroup 1, 9 L. pneumophila serogroup 2-14, and 14 latex agglutination-positive Legionella species were included in this comparison. There was no difference in performance between the latex agglutination assays when isolates were picked from selective and nonselective media. These results therefore indicate that L. pneumophila strains can be serogrouped directly from the primary isolation medium once confirmed by positive reactions for 16S rRNA and mip by real-time PCR.
The combined method of standard culture and real-time PCR confirmation is therefore capable of significantly decreasing turnaround times for Legionella identification and quantitation. For example, the current standard method may yield visible L. pneumophila colonies after 3 to 4 days and will require an additional 2 to 4 days to confirm by standard methods. A confirmed result in this case is available in 5 to 8 days. With the method we describe, the same colony can be used for real-time PCR confirmation and subsequent serogrouping, with a confirmed result available in 3 to 4 days; i.e., isolates can be identified as Legionella or L. pneumophila on the same day that they are visible. Similarly, slower-growing species such as L. anisa or L. micdadei, which typically appear after 4 to 6 days of incubation, can be confirmed and quantitated on the day that colonies are visible, compared to a further 2 to 4 days for traditional confirmation.
The incorporation of SYTO9 instead of the conventionally used dye SYBR green I required minimal optimization and did not lead to any interpretative difficulties. SYTO9 is an exciting alternative to SYBR green I in the diagnostic setting since the assays employing this dye seem more robust and insensitive to changes in DNA concentration, which is in direct contrast to SYBR green I for selected amplicons as described by Monis et al. (12). Monis et al. compared the performance of SYTO9 to that of SYBR green I in a number of PCR targets in both prokaryotic and eukaryotic systems, including the 16S rRNA gene and mip gene described in this assay, and concluded that the use of SYTO9 in real-time PCR melting curve analysis is superior to the use of SYBR green I. The assay described here follows from the work of Monis et al. and is the first reported use of SYTO9 in a diagnostic setting that has been extensively validated with field samples and isolates, and it seems that the use of SYTO9 may lend itself to real-time PCR users wishing to fast track optimization and implementation of real-time PCR assays.
In conclusion, we have described a rapid assay for Legionella that complements the current culture-based standard methods. This rapid method is easy to employ and could be implemented by most water testing laboratories. The results from this assay can be used in the same context as data generated using the current methods, therefore allowing rapid response to a confirmed quantitative count. The need to respond to counts is highlighted by prescriptive standards (e.g., AS/NZS 3666.3) where remedial action is based upon a confirmed colony count. Direct PCR from samples, therefore, continues to be of little use, as little information aside from the presence or absence (of viable, nonviable, or mixed cells) is obtained. As real-time PCR technology becomes more accessible and adopted in routine testing laboratories, rapid-PCR methods such as that described here will eventually supersede traditional methods for confirmation of bacterial identification. In a time when swift responses are not only required but demanded, the uptake of such a method will facilitate the administration of remedial action in a much more timely fashion.
Acknowledgments
We acknowledge the financial support from the Australian Water Quality Centre and the South Australian Water Corporation.
Technical assistance from the Microbiology unit and N. Peart are duly acknowledged.
REFERENCES
- 1.Anonymous. 2000. Air-handling and water systems of buildings—microbial control. Part 3. Performance-based maintenance of cooling water systems. Standards Australia, North Sydney, NSW, Australia.
- 2.Anonymous. 1998. Water quality—detection and enumeration of Legionella. ISO 11731:1998. International Organisation for Standardisation, Geneva, Switzerland.
- 3.Anonymous. 1998. Waters—examination for Legionellae. AS/NZS 3896:1998. Standards Australia, North Sydney, NSW, Australia.
- 4.Atlas, R. M. 1999. Legionella: from environmental habitats to disease pathology, detection and control. Environ Microbiol. 1:283-293. [DOI] [PubMed] [Google Scholar]
- 5.Bartie, C., S. N. Venter, and L. H. Nel. 2003. Identification methods for Legionella from environmental samples. Water Res. 37:1362-1370. [DOI] [PubMed] [Google Scholar]
- 6.Broadbent, C. 1996. Guidance for the control of Legionella. National Environmental Health Forum Monographs, Water Series no. 1. National Environmental Health Forum, Adelaide, South Australia, Australia.
- 7.Cloud, J. L., K. C. Carroll, P. Pixton, M. Erali, and D. R. Hillyard. 2000. Detection of Legionella species in respiratory specimens using PCR with sequencing confirmation. J. Clin. Microbiol. 38:1709-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giglio, S., P. T. Monis, and C. P. Saint. 2003. Demonstration of preferential binding of SYBR Green I to specific DNA fragments in real-time multiplex PCR. Nucleic Acids Res. 31:e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graman, P. S., G. A. Quinlan, and J. A. Rank. 1997. Nosocomial legionellosis traced to a contaminated ice machine. Infect. Control Hosp. Epidemiol. 18:637-640. [DOI] [PubMed] [Google Scholar]
- 10.Jonas, D., A. Rosenbaum, S. Weyrich, and S. Bhakdi. 1995. Enzyme-linked immunoassay for detection of PCR-amplified DNA of legionellae in bronchoalveolar fluid. J. Clin. Microbiol. 33:1247-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koide, M., N. Arakaki, and A. Saito. 2001. Distribution of Legionella longbeachae and other legionellae in Japanese potting soils. J. Infect. Chemother. 7:224-227. [DOI] [PubMed] [Google Scholar]
- 12.Monis, P. T., S. Giglio, and C. P. Saint. 2005. Comparison of SYTO9 and SYBR Green I for real-time polymerase chain reaction and investigation of the effect of dye concentration on amplification and DNA melting curve analysis. Anal. Biochem. 340:24-34. [DOI] [PubMed] [Google Scholar]
- 13.Orrison, L. H., W. B. Cherry, R. L. Tyndall, C. B. Fliermans, S. B. Gough, M. A. Lambert, L. K. McDougal, W. F. Bibb, and D. J. Brenner. 1983. Legionella oakridgensis: unusual new species isolated from cooling tower water. Appl. Environ. Microbiol. 45:536-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saint, C. P. 1998. A colony based confirmation assay for Legionella and Legionella pneumophila employing the EnviroAmp Legionella system and seroagglutination. Lett. Appl. Microbiol. 26:377-381. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Steele, T. W., C. V. Moore, and N. Sangster. 1990. Distribution of Legionella longbeachae serogroup 1 and other legionellae in potting soils in Australia. Appl. Environ. Microbiol. 56:2984-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steinert, M., U. Hentschel, and J. Hacker. 2002. Legionella pneumophila: an aquatic microbe goes astray. FEMS Microbiol. Rev. 26:149-162. [DOI] [PubMed] [Google Scholar]
- 18.Torii, K., Y. Iinuma, M. Ichikawa, K. Kato, M. Koide, H. Baba, R. Suzuki, and M. Ohta. 2003. A case of nosocomial Legionella pneumophila pneumonia. Jpn. J. Infect. Dis. 56:101-102. [PubMed] [Google Scholar]
- 19.Wellinghausen, N., C. Frost, and R. Marre. 2001. Detection of legionellae in hospital water samples by quantitative real-time LightCycler PCR. Appl. Environ Microbiol. 67:3985-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]