Abstract
o-Xylene is one of the most difficult-to-degrade environmental pollutants. We report here Rhodococcus genes mediating oxygenation in the first step of o-xylene degradation. Rhodococcus opacus TKN14, isolated from soil contaminated with o-xylene, was able to utilize o-xylene as the sole carbon source and to metabolize it to o-methylbenzoic acid. A cosmid library from the genome of this strain was constructed in Escherichia coli. A bioconversion analysis revealed that a cosmid clone incorporating a 15-kb NotI fragment had the ability to convert o-xylene into o-methylbenzyl alcohol. The sequence analysis of this 15-kb region indicated the presence of a gene cluster significantly homologous to the naphthalene-inducible dioxygenase gene clusters (nidABCD) that had been isolated from Rhodococcus sp. strain I24. Complementation studies, using E. coli expressing various combinations of individual open reading frames, revealed that a gene (named nidE) for rubredoxin (Rd) and a novel gene (named nidF) encoding an auxiliary protein, which had no overall homology with any other proteins, were indispensable for the methyl oxidation reaction of o-xylene, in addition to the dioxygenase iron-sulfur protein genes (nidAB). Regardless of the presence of NidF, the enzyme composed of NidABE was found to function as a typical naphthalene dioxygenase for converting naphthalene and various (di)methylnaphthalenes into their corresponding cis-dihydrodiols. All the nidABEF genes were transcriptionally induced in R. opacus TKN14 by the addition of o-xylene to a mineral salt medium. It is very likely that these genes are involved in the degradation pathways of a wide range of aromatic hydrocarbons by Rhodococcus species as the first key enzyme.
o-, m- and p-xylenes, which are included in petroleum, are used as solvents in the production of chemicals and drugs, as thinners for paints and enamels, and as intermediates for the synthesis of numerous chemical compounds. They are therefore widespread environmental contaminants in groundwater and soil (4, 11, 27). In m- and p-xylenes, numerous bacteria that are able to utilize them as the sole carbon and energy source have been isolated, and details of their catabolic pathways have been well documented. The most elucidated examples are the catabolism of toluene, m-xylene, and p-xylene with the enzymes encoded on a transmissible TOL plasmid (pWW0) from Pseudomonas putida mt-2 (38, 39). Xylene monooxygenase is the first enzyme on this degradation pathway (14, 34). This enzyme consists of two polypeptide subunits encoded by the xylM and xylA genes. XylA, the NADH acceptor reductase component (34), has been characterized as an electron transport protein transferring reducing equivalents from NADH to XylM, which is the hydroxylase component located in the membrane (18). Toluene and m- and p-xylenes are, respectively, metabolized with XylMA to benzyl alcohol and m- and p-methylbenzyl alcohol. These respective alcohols are further metabolized to benzoate and m- and p-toluates via their corresponding aldehydes before being subjected to meta cleavage. On the other hand, o-xylene has not been accepted as a substrate for XylMA (40), although the reason for this has not been elucidated.
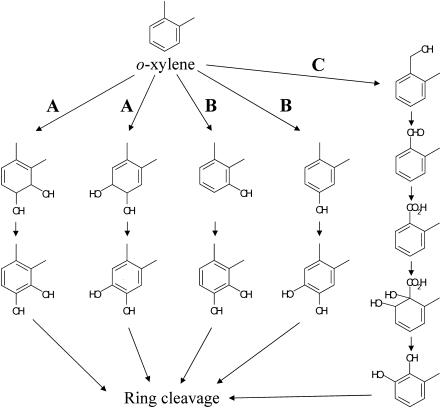
Several studies during the past decade have reported o-xylene as the most difficult-to-degrade aromatic hydrocarbon among the BTEX hydrocarbons (benzene, toluene, ethylbenzene, and o-, m-, and p-xylenes) (31). Only a few bacteria that are able to degrade o-xylene have so far been isolated (4, 7, 10). o-Xylene is oxidatively degraded by way of pathways that fall into the three classes shown in Fig. 1 to produce their corresponding catechols, which are then subjected to aromatic ring cleavage. The first pathway is one initiated by a ring-hydroxylating dioxygenase (Fig. 1, route A). Nocardia sp., Corynebacterium sp. strain C125, and Rhodococcus sp. strain DK17 belong to this class (13, 16, 30). These strains are thought to metabolize o-xylene via 3,4-dimethylcatechol. o-Xylene dioxygenase genes (akbA1a, akbA2a, and akbA3) have recently been isolated from Rhodococcus sp. strain DK17 (16), which can grow on o-xylene but cannot grow on m- or p-xylene (17). The second pathway for o-xylene degradation is one initiated by a ring-hydroxylating monooxygenase (Fig. 1, route B). It has been elucidated with Pseudomonas stutzeri strain OX1 that o-xylene is degraded through two successive monooxygenation reactions of the aromatic ring to form 3,4-dimethylcatechol and 4,5-dimethylcatechol by way of 2,3-dimethylphenol and 3,4-dimethylphenol, respectively (3, 5). The genes in strain OX1 involved in the initial hydroxylation of toluene/o-xylene have been well characterized, and it has been revealed that all of the touABCDEF genes were necessary for full activity of the monooxygenation reaction (6, 8). The third degradation pathway for o-xylene is one initiated by oxidation of the methyl substituent of o-xylene to form 2-methylbenzyl alcohol and subsequent metabolism to form 3-methylcatechol via 2-methylbenzoic acid (Fig. 1, route C). This degradation pathway has been found with Rhodococcus sp. strain B3 (7) and Rhodococcus opacus R7 (10). Another catabolic pathway for o-xylene-mediating aromatic ring hydroxylation has been found in the same bacterial strains. These strains are thus thought to have multioxygenation capability. In Rhodococcus sp. strain B3, o-xylene is simultaneously metabolized by two routes, one via 2-methylbenzoic acid by the methyl oxidation process and the other via 2,3-dimethylphenol and 3,4-dimethylcatechol by successive monooxidation of the aromatic nucleus (7). In R. opacus R7, o-xylene is metabolized by successive monooxygenation in the same way, but 2-methyl-benzyl alcohol that is constitutively generated from o-xylene cannot be further metabolized (10). The genes involved in the hydroxylation of the methyl substituent of o-xylene in both strains have not been isolated or identified.
FIG. 1.
Proposed o-xylene degradation pathway. Dioxygenation and subsequent dehydrogenation of the aromatic ring (A); two successive monooxygenation reactions to the aromatic ring (B); monooxygenation of the methyl group and subsequent oxidations (C).
Rhodococcus opacus TKN14, whose 16S DNA sequence is similar to that of R. opacus DSM 43205T, has been isolated as one of the main bacteria present in soil contaminated with o-xylene (H. Taki, K. Syutsubo, R. G. Mattison, and S. Harayama, unpublished results). This bacterium was able to grow on o-xylene as the sole carbon and energy source and presumed to possess only the above-mentioned third methyl oxidation pathway from the result of a catabolic analysis of o-xylene with this strain (Taki et al., unpublished). In addition to o-xylene, this TKN14 strain was able to grow on p-xylene, ethylbenzene, and toluene as the sole carbon and energy source. We elucidate for the first time in the present work the multicomponent oxygenase system required for hydroxylating the methyl group of o-xylene, which is present in an o-xylene-utilizing bacterium, at the genetic level. We describe here the functional expression of the genes mediating this first oxygenation reaction in Escherichia coli, which have been isolated from R. opacus TKN14, and show that the constructed enzyme system is involved in the degradation pathways of a wide range of aromatic hydrocarbons containing benzene substituents and naphthalene substituents by Rhodococcus species as the first key enzyme.
MATERIALS AND METHODS
Bacterial strains, plasmids, and reagents.
The bacterial strains and plasmids used or constructed in this study are listed in Table 1. Escherichia coli XL1-Blue MR and E. coli JM109 were used as the respective hosts for an in vitro packaging experiment and for the other recombinant DNA and bioconversion experiments. Rhodococcus opacus TKN14 is distributed as MBIC05572 from the Marine Biotechnology Institute Culture Collection (http://www.mbio.jp/mbic/). All chemicals were purchased from Aldrich (Sigma-Aldrich Japan, Tokyo, Japan), Fluka (Sigma-Aldrich Japan), Wako Pure Chemicals (Osaka, Japan), or Tokyo Kasei (Tokyo, Japan).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| Rhodococcus opacus TKN14 | Gram-positive bacterium that grows on o-xylene | MBIC 05572 |
| Escherichia coli XL1-Blue MR | Δ(mcrA) 183 Δ(mcrCB-hsdSMR-mrr) 173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac | Stratagene |
| E. coli JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ(lac-proAB) F′ [traD36 proAB+lacIqlacZ ΔM15] | Takara |
| Plasmids | ||
| SuperCos 1 | Stratagene | |
| pBluescript II SK(+) | Stratagene | |
| pT7Blue T-vector | Novagen | |
| pTrcHisA | Invitrogen | |
| pRTKN700 | Apr; 35-kb Sau3AL-partially digested genome DNA from R. opacus TKN14 in SuperCOS 1 | This study |
| pNot1-9 | Apr; 15-kb NotI fragment from pRTKN700 in pBluescript II SK+ | This study |
| pN54-1-5 | Apr; 7.26-kb PCR fragment from the 15-kb NotI fragment in pT7Blue T-vector | This study |
| pNidABCD | Apr; 4.59-kb EcoRI fragment from the 15-kb NotI fragment in pBluescript II SK+ | This study |
| nidA-disruptant | Apr; pN54-1-5 with disrupted nidA | This study |
| nidB-disruptant | Apr; pN54-1-5 with disrupted nidB | This study |
| ORF1-disruptant | Apr; pN54-1-5 with disrupted ORF1 | This study |
| ORF2-3-disruptant | Apr; pN54-1-5 with disruption in both ORF2 and ORF3 | This study |
| pTrcNidAB | Apr; nidAB (1.94 kb) in pTrcHisA | This study |
| pTrcRd | Apr; Rd (0.28 kb) in pTrcHisA | This study |
| pTrcNidABRd | Apr; nidAB and Rd (2.22 kb) in pTrcHisA | This study |
| pTrcNidABRdORF4 | Apr; nidAB, Rd, and ORF4 (2.82 kb) in pTrcHisA | This study |
| pTrcNidABRdORF5 | Apr; nidAB, Rd, and ORF5 (2.91 kb) in pTrcHisA | This study |
| pTrcNidABRdORF6 | Apr; nidAB, Rd, and ORF6 (2.85 kb) in pTrcHisA | This study |
| pTrcNidABRdORF789 | Apr; nidAB, Rd, ORF7, ORF8; and ORF9 (3.68 kb) in pTrcHisA | This study |
| pTrcNidABRdORF8 | Apr; nidAB, Rd, and ORF8 (2.73 kb) in pTrcHisA | This study |
| pTrcNidABRdORF9 | Apr; nidAB, Rd, and ORF9 (2.94 kb) in pTrcHisA | This study |
| pTrcNidABRdORF78 | Apr; nidAB, Rd, ORF7, and ORF8 (2.93 kb) in pTrcHisA | This study |
| pTrcNidABRdORF89 | Apr; nidAB, Rd, ORF8, and ORF9 (3.45 kb) in pTrcHisA | This study |
| pTrcNidABORF8 | Apr; nidAB and ORF8 (2.45 kb) in pTrcHisA | This study |
Recombinant DNA techniques.
The restriction enzymes and the DNA ligation kit were purchased from New England BioLabs (Beverly, Calif.) and Toyobo (Osaka, Japan), respectively. DNA manipulation was conducted by standard methods (29) or as instructed by the suppliers. Plasmid or cosmid DNA was prepared with the QIAEX II purification kit (QIAGEN, Hilden, Germany) or the PI-200 automatic DNA isolation system (Kurabo, Osaka, Japan). PCR was carried out with an automated thermal cycler (Techne) using LA Taq polymerase (Takara, Kyoto, Japan).
Preparation of genomic DNA and construction of the cosmid library.
Total DNA was extracted from R. opacus TKN14 grown on a 1/10 TSA medium (2.75 g of tryptic soy broth without dextrose [Difco]/liter and 0.25 g of glucose/liter) at 30°C, as previously described (35), partially digested with Sau3AI, and ligated into the BamHI site of SuperCos 1 (Stratagene, La Jolla, Calif.). The ligates were packaged into bacteriophage λ with a Lambda Inn packaging extract (Wako). E. coli XL1-Blue MR was infected with the resulting phages.
Screening of the cosmid library for o-xylene-bioconverting activity through HPLC analysis.
The activity for o-xylene oxygenase was determined by measuring the conversion of o-xylene to o-methylbenzyl alcohol. A total of 1,000 cosmid clones in E. coli XL1-Blue MR were grown overnight at 30°C on an M9 medium (29) containing 0.8% glucose, 0.01% thiamine-HCl, and 150 μg of ampicillin (Ap)/ml until the absorbance at 600 nm reached approximately 1.0. The harvested cells were collected by centrifugation, resuspended in an equal volume of the same broth, and further inoculated with a final concentration of 1 mM isopropyl-1-thio-β-d-galactoside (IPTG) and 0.5 mM o-xylene (from a 50 mM stock solution in dimethyl sulfoxide [DMSO] or ethanol) at 30°C for 3 to 4 days. Samples drawn from the culture were diluted with an equal volume of methanol (MeOH) and then centrifuged for 5 min at 20,000 × g. The supernatant was applied to high-performance liquid chromatography (HPLC) in an XTerra C18 column (4.6 by 150 mm; Waters, Milford, Mass.) with a photodiode array detector (Waters 2996), using a mobile phase of 0.1% phosphoric acid and acetonitrile. Elution was carried out with a decreasing linear gradient of 0.1% phosphoric acid from 75% to 0% acetonitrile over 4 min and then an increasing gradient from 0% to 75% acetonitrile over 5 min at a flow late of 1 ml/min. Metabolites were identified by a comparison of their retention times and UV-visible spectra to those of commercially available standards.
Sequence analysis of an o-xylene oxygenase gene cluster.
The plasmid (pRTKN700) was prepared from the positive clone and digested with various restriction enzymes for further subcloning. Discrete digested DNA fragments were purified from the gel and inserted into the corresponding site of pBluescript II SK(+). A DNA fragment was also amplified by PCR and then cloned into pT7Blue T-vector to generate plasmid pN54-1-5. E. coli JM109 was used as the host. Bioconversion experiments with o-xylene with these subclones were performed as already described.
The nucleotide sequence was determined by using a Taq Dye Deoxy terminator cycle sequencing kit and a model 377 sequencer (Applied Biosystems, Foster City, Calif.). The PCR conditions used were 25 cycles, each consisting of 10 s at 98°C, 5 s at 50°C, and 4 min at 60°C; the PCR solutions contained a final concentration of 5% DMSO. The nucleotide and amino acid sequences were analyzed with the BLASTN or BLASTP program of the National Center for Biotechnology Information.
Construction of several open reading frame (ORF) disruptants.
Oligonucleotides that were used as the primers for PCR amplification are listed in Table 2.
TABLE 2.
Oligonucleotides used for PCR in this study
| Name | Sequence (with restriction site underlined) | Relevant characteristics | Attached restriction site |
|---|---|---|---|
| nidA-AscF | 5′-TTGGCGCGCCTCATGCTCGCGGACACACCGCTGCACGCCGGCA-3′ | Forward primer to construct nidA disruptant | AscI |
| nidA-AscR | 5′-TTGGCGCGCCGCACCTCGGGATCGTTGATGATCGAGGCGGGGA-3′ | Reverse primer to construct nidA disruptant | AscI |
| nidB-AscF | 5′-TTGGCGCGCCGTGCTCCGGCGCGCGGGCGACGGCTTCCGTCT-3′ | Forward primer to construct nidB disruptant | AscI |
| nidB-AscR | 5′-TTGGCGCGCCATTCCCGGTACCTACCGGCGTCGAGCAGCCCGGCCT-3′ | Reverse primer to construct nidB disruptant | AscI |
| ORF1-AscF | 5′-TTGGCGCGCCATCGCGGCGAATGCGCTTGGTGCGTTCGTCGAAG-3′ | Forward primer to construct ORF1 disruptant | AscI |
| ORF1-AscR | 5′-TTGGCGCGCCCCAGCTCGACGAACTGTCCGGCCTCGCCGACGCAG-3′ | Reverse primer to construct ORF1 disruptant | AscI |
| ORF23-AscF | 5′-TTGGCGCGCCCGAACAACGCGACACCCGACCTGGGGGATGATC-3′ | Forward primer to construct ORF2 and -3 disruptant | AscI |
| ORF23-AscR | 5′-TTGGCGCGCCTCGGGCTCAGCCACGCGGCGGCGTTCTTCGCGGAC-3′ | Reverse primer to construct ORF2 and -3 disruptant | AscI |
| nidAB-Bam | 5′-CGCGGATCCATGCTGAGCAACGAACTCCGGCAGACCCTCC-3′ | Forward primer for PCR amplification of nidAB | BamHI |
| nidAB-Pst | 5′-AAAACTGCAGATTCACATGATCAGGGCGAGGTTGTGTGTCATT-3′ | Reverse primer for PCR amplification of nidAB | PstI |
| Rd-Pst | 5′-AAAACTGCAGGAGGAGGGTTCATGAAGATTCTGATCGCG-3′ | Forward primer for PCR amplification of Rd | PstI |
| Rd-Kpn | 5′-GGGGTACCTCATCCCAGCTCTCCCGTTATGAGGTCTTCG-3′ | Reverse primer for PCR amplification of Rd | KpnI |
| ORF4-Kpn | 5′-GGGGTACCAGGAGGCTTCGCATGGGCGGGCTACGGAGACGTGC-3′ | Forward primer for PCR amplification of ORF4 | KpnI |
| ORF4-Hind | 5′-CCCAAGCTTTCATTCCAACCTGAAGCGACGGGCGGGCTGTG-3′ | Reverse primer for PCR amplification of ORF4 | HindIII |
| ORF5-Kpn | 5′-GGGGTACCGAGGAGGAGAGAAGCCATGACACAGCTGT-3′ | Forward primer for PCR amplification of ORF5 | KpnI |
| ORF5-Hind | 5′-CCCAAGCTTTCAGACGGACGCGGCGCGGGGCATACTGGCCG-3′ | Reverse primer for PCR amplification of ORF5 | HindIII |
| ORF6-Kpn | 5′-GGGGTACCAGGTTGGAATGATCAAGGGCCGCAGCG-3′ | Forward primer for PCR amplification of ORF6 | KpnI |
| ORF6-Hind | 5′-CCCAAGCTTTCACCCGAGTGGATGCAGACATACGGCGAG-3′ | Reverse primer for PCR amplification of ORF6 | HindIII |
| ORF7-Kpn | 5′-GGGGTACCGGATCGTTGATGATCGAGGCGGGGACGGTCCA-3′ | Forward primer for PCR amplification of ORF78 or ORF789 | KpnI |
| ORF8-Kpn | 5′-GGGGTACCAGGAGGTCTAACATGTTAGTATCTTTGCTCACATCGGTCAA-3′ | Forward primer for PCR amplification of ORF8 or ORF89 | KpnI |
| ORF8-Hind | 5′-CCCAAGCTTTTACACCGACCAGTCGGTGCCGTCGATCTGACCCA-3′ | Reverse primer for PCR amplification of ORF8 or ORF78 | HindIII |
| ORF9-Hind | 5′-CCCAAGCTTTCAGTGCGCGGGAGGCGCTGAACGACGGGCACCGCAGC-3′ | Reverse primer for PCR amplification of ORF9, ORF89, or ORF789 | HindIII |
| ORF9-Kpn | 5′-GGGGTACCAGGAGGGGTGAAATGTCCGCCCTGCATCGAGCATCCGA-3′ | Forward primer for PCR amplification of ORF9 | KpnI |
| nidAF | 5′-GGACTACCTCGGCGATATGA-3′ | Forward primer for RT-PCR of nidA | |
| nidAR | 5′-TGTGGACGTGCTCTCCATAG-3′ | Reverse primer for RT-PCR of nidA | |
| nidBF | 5′-TCGTCACCAACTTCAAGTC-3′ | Forward primer for RT-PCR of nidB | |
| nidBR | 5′-GGTCTGATCAAGCAGCACAA-3′ | Reverse primer for RT-PCR of nidB | |
| nidEF | 5′-GTCTGCGAGGTCTGTGAAGA-3′ | Forward primer for RT-PCR of nidE (Rd) | |
| NidER | 5′-CTTCGCTCACGTCCTGGTAT-3′ | Reverse primer for RT-PCR of nidE (Rd) | |
| NidFF | 5′-GCCAGGGACTCAAGAAACTG-3′ | Forward primer for RT-PCR of nidF (ORF8) | |
| NidFR | 5′-AGGTGTCCGTATTGGCTGTC-3′ | Reverse primer for RT-PCR of nidF (ORF8) |
Gene disruption experiments were carried out to determine the genes responsible for the conversion of o-xylene. Disruption plasmids were constructed by PCR, using plasmid pN54-1-5 as the template with primers including the AscI site and by cyclization of the subsequent AscI-digested PCR fragments. To construct a nidA disruptant (a disruption was introduced in the middle of the nidA gene), oligonucleotides nidA-AscF and nidA-AscR (Table 2) were used as the forward and reverse primers, respectively. The amplified DNA fragment, which had been digested with AscI, was self ligated and transformed into E. coli JM109. The prepared plasmids were checked by agarose gel electrophoresis of their digested patterns with appropriate restriction enzymes and by their DNA sequence. nidB, ORF1 disruptants, and an ORF2-3 disruptant (a disruption was introduced in both ORF2 and ORF3) were constructed in the same way, using the primers listed in Table 2.
Construction of a variety of plasmids expressing ORFs included in pN54-1-5.
Individual ORFs that may be concerned with the methyl oxidization of o-xylene were amplified by PCR using plasmid pNot1-9 as the template DNA and inserted into expression vector pTrcHisA, and E. coli JM109 was transformed (the position of each amplified gene fragment is shown in Fig. 2). The nidAB genes were amplified with the primer pair nidAB-Bam and nidAB-Pst (Table 2). The amplified 1.94-kb DNA fragment, which had been digested with BamHI and PstI, was inserted into the corresponding site of pTrcHisA to generate pTrcNidAB. The DNA fragment, including the rubredoxin (Rd) gene and its original Shine-Dalgarno sequence, were amplified with the primer pair Rd-Pst and Rd-Kpn (Table 2) and inserted into the PstI-KpnI site of pTrcHisA to generate pTrcRd. The above-mentioned 1.94-kb nidAB fragment (digested with BamHI and PstI) was then inserted into the corresponding site of pTrcRd to generate pTrcNidABRd. To construct the expression plasmids for ORF4, ORF5, ORF6, ORF7, ORF8, ORF9, or their successive ORFs, each DNA region was amplified by PCR with a forward primer having the KpnI site, an original or artificial Shine-Dalgarno sequence, and a reverse primer with the HindIII site (individual primer sequences are shown in Table 2). The individual PCR fragments were digested with KpnI and HindIII and inserted into the KpnI-HindIII site of pTrcRd before the 1.94-kb nidAB fragment (digested with BamHI and PstI) was introduced into the corresponding site to generate pTrcNidABRdORF4, pTrcNidABRdORF5, pTrcNidABRdORF6, pTrcNidABRdORF789, pTrcNidABRdORF8, pTrcNidABRdORF9, pTrcNidABRdORF78, and pTrcNidABRdORF89. Furthermore, a PCR fragment containing ORF8 that had been digested with KpnI and HindIIIwas inserted into the KpnI-HindIII site of pTrcHisA before the 1.94-kb nidABfragment was inserted into the BamHI and PstI sites to generate pTrcNidABORF8.
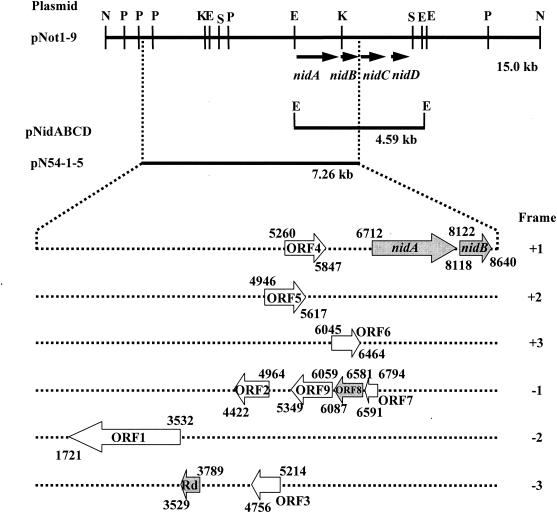
FIG. 2.
Organization of the o-xylene-oxygenase gene clusters isolated from Rhodococcus opacus TKN14. Restriction sites in the 15-kb NotI fragment (pNot1-9) are also shown. Abbreviations: E, EcoRI; K, KpnI; N, NotI; P, PstI; and S, SpeI. Numbers represent nucleotide positions numbered from the 5′ end of the NotI site. The numbers over and under the gene map, respectively, represent the positions of the start and termination of individual genes or ORFs. pNot1-9 and pN54-1-5 allowed the host cells to convert o-xylene into o-methylbenzyl alcohol. pNidABCD was not able to metabolize o-xylene. Individual ORFs and nidAB in the 7.26-kb fragment of pN54-1-5 were functionally analyzed. ORF1 to ORF7 and ORF9 were found to be unnecessary for the methyl oxidation of o-xylene. The genes encoding Rd and ORF8, in addition to nidAB, proved to be necessary for the methyl oxidation reaction and are designated the nidE and nidF genes, respectively.
Bioconversion assays with recombinant E. coli on various monocyclic aromatic hydrocarbons.
E. coli JM109 strains transformed with each plasmid as just described were cultured overnight at 30°C in M9 medium (29) containing 0.2% glucose, 0.01% thiamine-HCl, 100 μM Fe(NH4)2(SO4)2, and 150 μg of Ap/ml and then suspended in the same medium at an optical density at 600 nm (OD600) of 0.1 to 0.2. The cells were further grown to an OD600 of 0.4 to 0.6 and then induced by 1 mM IPTG for 4 to 5 h at 30°C. The cells were harvested, and reactions were carried out with washed cells suspended in fresh M9 medium containing 0.2% glucose, 0.01% thiamine-HCl, 100 μM Fe(NH4)2(SO4)2, 150 μg of Ap/ml, and 1 mM IPTG in screw-cap glass vials at 30°C. Substrates such as toluene; o-, m- and p-xylene; and ethylbenzene were supplied from a 50 mM DMSO or ethanol stock solution to a final concentration of 0.5 mM. After being shaken with the substrate at 30°C for 1 to 2 days, each reaction mixture was analyzed by gas chromatography-mass spectrometry (GC-MS).
GC-MS analysis.
Metabolites from the monocyclic aromatic hydrocarbons were analyzed by GC-MS. After being shaken with the substrate, each reaction mixture was saturated with NaCl and then extracted with ethyl acetate (EtOAc) (the reaction mixture was acidified with 1 N HCl [aqueous phase] to pH 2 before extraction as necessary). The organic phase was analyzed by GC-MS (Shimadzu QP5050A instrument) with a DB-5 column (length, 30 m; diameter, 0.25 mm; J&W Scientific). If necessary, the ethyl acetate extract was evaporated and trimethylsilyl esterified with TMSI-H (hexamethyldisilazane:trimethylchlorosilane:pyridine; GL Science, Tokyo, Japan) before being injected. The following temperature program was applied as follows: from 40°C to 70°C at 15°C/min, from 70°C to 105°C at 5°C/min, and from 105°C to 240°C at 20°C/min. The injector and detector temperatures were both 250°C.
Bioconversion experiments with recombinant E. coli on naphthalene and (di)methylnaphthalenes.
E. coli JM109 harboring pN54-1-5 was grown in LB medium (29) containing Ap at 30°C under reciprocal shaking for 7 to 8 h until the absorbance at OD600 had reached approximately 1. Eight milliliters of this culture was inoculated into 100 ml of M9 medium (29) containing 0.2% glucose, 0.01% thiamine-HCl, 100 μM Fe(NH4)2(SO4)2, 150 μg/ml of Ap, and 1 mM IPTG, as well as 10 mg of the substrate, naphthalene, 1- or 2-methylnaphthalene, or 1,2-, 1,4-, 1,5-, or 2,3-dimethylnaphthalene, in a Sakaguchi flask and cocultivated at 30°C under reciprocal shaking for 2 days.
To extract the converted products as well as the substrates, 100 ml of MeOH was added to the coculture, and the solution was mixed for 30 min. After centrifugation to remove the cells, the liquid phase was checked by HPLC as previously described (15).
Purification and identification of the dihydrodiol products.
The liquid phase (2,000 ml), which had been obtained by the procedure just described, was concentrated in vacuo and extracted with EtOAc (two extractions, 500 ml each). The resulting organic layer was concentrated in vacuo and analyzed by thin-layer chromatography on silica gels (0.25-mm silica gel plates; E. Merck 60F-254). The formed products were purified by column chromatography on Silica Gel 60 (20 by 250 mm2; Merck). The structure of each was identified from its MS data (electric impact [EI]; Jeol DX505W] and nuclear magnetic resonance (NMR) spectral data (400 MHz; Bruker AMX400). A Polari meter (SEPA-300, Horiba) was also used to distinguish the product from naphthalene.
Product converted from naphthalene (product 1).
The crude EtOAc extract (110.8 mg) was subjected to column chromatography (CH2Cl2:MeOH = 30:1) to yield 3.1 mg of product 1. Product 1 was identified as (1R,2S)-cis-1,2-dihydronaphthalene-1,2-diol [(1R,2S)-cis-1,2-dihydro-1,2-naphthalenediol] by comparing its spectral data (MS, NMR, and [α]D) to those of the authentic sample (Fluka).
Product converted from 1-methylnaphthalene (product 2).
The crude EtOAc extract (121.1 mg) was subjected to column chromatography (CH2Cl2:MeOH = 30:1) to yield 1.2 mg of product 2. Product 2 was identified as 8-methyl-cis-1,2-dihydronaphthalene-1,2-diol by a direct comparison of its MS and NMR data to those of an authentic sample which had been prepared as shown previously (15).
Product converted from 2-methylnaphthalene (product 3).
The crude EtOAc extract (52.3 mg) was subjected to column chromatography (CH2Cl2:MeOH = 30:1) to yield 1.8 mg of product 3. Product 3 was identified to be 7-methyl-cis-1,2-dihydronaphthalene-1,2-diol by a direct comparison of its MS and NMR data to those of an authentic sample which had been prepared as shown previously (15).
Product converted from 1,2-dimethylnaphthalene (product 4).
The crude EtOAc extract (90.6 mg) was subjected to column chromatography (CH2Cl2:MeOH = 40:1) to yield 4.2 mg of product 4. The molecular formula of product 4 was determined to be C12H14O2 by high-resolution MS (HRMS) (EI) data. Analyses of 1H detected multiple-quantum coherence (HMQC) and double-quantum-filtered correlation spectroscopy (DQF COSY) spectra showed that this compound was a dihydrodiol derivative of 1,2-dimethylnaphthalene. The region-chemical assignment of the 1,2-diol was confirmed by the long-range carbon-proton connectivity observed from H-10 (δ 2.24) and H-4 (δ 6.31) to C-8a (δ 132.0) in the 1H-detected multiple-bond connectivity (HMBC) spectrum and by the 1H-1H vicinal spin network from H-1 (δ 4.79) to H-4 in the DQF COSY spectrum. Product 4 was thus identified as 7,8-dimethyl-1,2-dihydronaphthalene-1,2-diol.
MS (EI) m/z 190 (M+). HRMS (EI) calculated for C12H14O2 (M+), 190.0994; found 190.0985. 1H NMR (CDCl3) δ: 2.21 (s, 3H), 2.24 (s, 3H), 4.46 (m, 1H), 4.79 (m, 1H), 5.70 (d, 1H, J = 9.7 Hz), 6.31 (dd, 1H, J = 2.7, 9.7 Hx), 6.79 (d, 1H, J = 7.5 Hz), 7.02 (d, 1H, J = 7.5 Hz). 13C NMR (CDCl3) δ: 14.3 (C-10), 20.7 (C-9), 66.6 (C-1), 70.7 (C-2), 124.7 (C-5), 127.4 (C-4), 129.4 (C-3), 129.8 (C-4a), 130.4 (C-6), 132.0 (C-8a), 136.0 (C-8), 137.1 (C-7).
Product converted from 1,4-dimethylnaphthalene (product 5).
The crude EtOAc extract (67.8 mg) was subjected to column chromatography (hexane-EtOAc = 2:1) to yield 1.5 mg of product 5. The molecular formula of product 5 was determined to be C12H14O2 by its HRMS (EI) data. Analyses of HMQC and DQF COSY spectra showed that this compound was a dihydrodiol derivative of 1,4-dimethylnaphthalene. The region-chemical assignment of the 1,2-diol was confirmed by the 1H-1H vicinal spin networks from H-1 (δ 4.71) to H-4 (δ 6.56) and between H-6 (δ 6.97) and H-7 (δ 6.91) observed in the DQF COSY spectrum. Product 5 was thus identified as 5,8-dimethyl-1,2-dihydronaphthalene-1,2-diol.
MS (EI) m/z 190 (M+). HRMS (EI) calculated for C12H14O2 (M+), 190.0994; found 190.0992. 1H NMR (CDCl3) δ: 2.23 (s, 3H), 2.32 (s, 3H), 4.45 (m, 1H), 4.71 (dd, 1H, J = 1.5, 5.4 Hz), 5.76 (m, 1H), 6.56 (dd, 1H, J = 2.7, 9.8 Hz), 6.91 (d, 1H, J = 8.0 Hz), 6.97 (d, 1H, J = 8.0 Hz). 13C NMR δ: 18.3 (C-9), 18.9 (C-10), 66.8 (C-1), 70.4 (C-2), 127.3 (C-4), 130.8 (C-6), 130.9 (C-7), 130.9 (C-3), 130.9 (C-4a), 132.0 (C-5), 132.2 (C-8a), 134.9 (C-8).
Product converted from 1,5-dimethylnaphthalene (product 6).
The crude EtOAc extract (97.3 mg) was subjected to column chromatography (hexane:EtOAc = 2:1) to yield 2.5 mg of product 6. The molecular formula of product 6 was determined to be C12H14O2 by its HRMS (EI) data. Analyses of HMQC and DQF COSY spectra showed that this compound was a dihydrodiol derivative of 1,5-dimethylnaphthalene. The region-chemical assignment of the 1,2-diol was confirmed by the long-range carbon-proton connectivity observed from H-9 (δ 2.01) to C-3 (δ 127.6) in the HMBC spectrum and by the 1H-1H vicinal spin network from H-1 (δ 4.71) to H-3 (δ 5.58) in the DQF COSY spectrum. Product 6 was thus identified as 4,8-dimethyl-1,2-dihydronaphthalene-1,2-diol.
MS (EI) m/z 190 (M+). HRMS (EI) calculated for C12H14O2 (M+), 190.0994; found 190.0999. 1H NMR (CDCl3) δ: 2.01 (s, 3H), 2.38 (s, 3H), 4.44 (m, 1H), 4.71 (d, 1H, J = 4.4 Hz), 5.58 (m, 1H), 7.04 (d, 1H, J = 7.8 Hz), 7.09 (d, 1H, J = 7.7 Hz), 7.19 (dd, 1H, J = 7.7, 7.8 Hz). 13C NMR δ: 18.7 (C-10), 19.3 (C-9), 66.5 (C-1), 70.1 (C-2), 121.9 (C-5), 127.6 (C-3), 128.9 (C-6), 130.1 (C-7), 132.1 (C-4), 132.6 (C-8a), 133.7 (C-4a), 137.2 (C-8).
Product converted from 2,3-dimethylnaphthalene (product 7).
The crude EtOAc extract (110.8 mg) was subjected to column chromatography (CH2Cl2:MeOH = 40:1) to yield 7.5 mg of product 7. The molecular formula of product 7 was determined to be C12H14O2 by its HRMS (EI) data. Analyses of HMQC and DQF COSY spectra showed that this compound was a dihydrodiol derivative of 2,3-dimethylnaphthalene. The region-chemical assignment of the 1,2-diol was confirmed by the 1H-1H vicinal spin network from H-1 (δ 4.54) to H-4 (δ 6.38) observed in the DQF COSY spectrum. Product 7 was thus identified as 6,7-dimethyl-1,2-dihydronaphthalene-1,2-diol dihydronaphthalene.
MS (EI) m/z 190 (M+). HRMS (EI) calculated for C12H14O2 (M+), 190.0994; found 190.1002. 1H NMR (CDCl3) δ: 2.16 (s, 3H), 2.19 (s, 3H), 4.29 (dd, 1H, J = 3.7, 3.8 Hz), 4.54 (d, 1H, J = 3.8 Hz), 5.87 (dd, 1H, J = 3.7, 9.7 Hz), 6.38 (d, 1H, J = 9.7 Hz), 6.81 (s, 1H), 7.19 (s, 1H). 13C NMR δ: 19.4 (C-9), 19.6 (C-10), 68.3 (C-2), 70.5 (C-1), 127.8 (C-3), 128.2 (C-5), 128.7 (C-4), 129.0 (C-8), 129.1 (C-4a), 132.7 (C-8a), 136.8 (C-6), 136.8 (C-7).
Preparation of RNA from R. opacus TKN14 induced with o-xylene and RT-PCR analysis.
R. opacus TKN14 was grown on a 1/10 TSA medium for 3 days at 30°C with reciprocal shaking and washed twice by centrifugation with a mineral salt (MS) medium [0.8 g of KH2PO4/liter, 5.58 g of Na2HPO4/liter, 1.8 g of (NH4)2SO4/liter, 0.12 g of MgSO4 · 7H2O/liter, 0.5 mg of FeSO4 · 5H2O/liter, 1.54 mg of MnSO4 · 5H2O/liter, 2.86 mg of H3BO3/liter, 0.039 mg of CuSO4 · 5H2O/liter, 0.021 mg of ZnCl2/liter, 0.041 mg of CoCl2 · 6H2O/liter, Na2MoO4 · 2H2O, and 11.6 mg of CaCl2 · 2H2O/liter]. The cells were suspended in 20 ml of MS medium, in which the absorbance at OD600 was 1.0; a total of 45 μl of o-xylene was added, and the mixture was cultured at 30°C with reciprocal shaking. The cells were harvested at 4 h and 21 h after the cultivation that started with o-xylene, collected by centrifugation, and washed with saline. Total RNA was prepared with the FastRNA Pro Blue Kit (Qbiogene, Irvine Calif.), as described in the application manual. Contaminated DNA was degraded by RNase-free DNase I (Takara). Three hundred nanograms of total RNA was used for reverse transcription-PCR (RT-PCR) analysis. RT-PCR analysis was conducted with the RNA LA PCR kit (AMV; Takara). Three hundred total RNAs were reversely transcribed with the random 9mers. Half of the resultant cDNA was amplified by using the four sets of primers listed in Table 2. After an initial incubation at 94°C for 2 min, 25 cycles of the following temperature conditions were used, each cycle consisting of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s. All other conditions were as described in the application manual of the supplier.
Nucleotide sequence accession number.
The nucleotide sequence of the 15.0-kb NotI region of pNot1-9 is available in the DDBJ/EMBL/GenBank databases under accession no. AB206671.
RESULTS AND DISCUSSION
Isolation of genes mediating the oxidation of o-xylene from the Rhodococcus opacus TKN14 genomic cosmid library.
Gram-positive bacterium R. opacus TKN14 degrades o-xylene via o-methylbenzoic acid (Taki et al., unpublished), indicating that this strain has the ability to oxidize the methyl substituent in o-xylene, the most recalcitrant isomer of xylenes. To obtain the genes responsible for o-xylene conversion, we screened about 1,000 E. coli cosmid clones by coculturing them with o-xylene and then by analyzing the solvent extracts by HPLC. We found that one cosmid clone, named RTKN700, had the capability to convert o-xylene into o-methylbenzyl alcohol. The identity of 2-methylbenzyl alcohol was confirmed by comparing its UV and HPLC spectral data with those of an authentic sample. No o-tolualdehyde, o-toluic acid, or ring hydroxylation product was present during o-xylene bioconversion by clone RTKN700. A restriction enzyme analysis of a plasmid (pRTKN700) prepared from clone RTKN700 showed this plasmid to have aninsert of approximately 35 kb. Discrete restriction fragments derived from this 35-kb fragment were inserted into the pBluescript II SK(+) vector and introduced into E. coli JM109. The results of o-xylene bioconversion experiments using these subclones showed that E. coli-harboring plasmid pNot1-9 (Fig. 2), which contained a 15-kb NotI fragment derived from pRTKN700, converted o-xylene into o-methylbenzyl alcohol. This result shows that the genes involved in the early stage of o-xylene degradation were located in this 15-kb NotI locus.
Sequence analysis of the gene cluster involved in the methyl oxidation of o-xylene.
The nucleotide sequence of the 15-kb NotI fragment was determined, and its ORF analysis indicated the presence of a gene cluster significantly homologous to that reported as the naphthalene-inducible dioxygenase genes (nidABCD) that convert indene to cis-(1R,2S)-indandiol and other metabolites, which had been isolated from Rhodococcus sp. I24 (36). The amino acid homology (identity) of the nidABCD gene products of strain TKN14 to the corresponding enzymes derived from strain I24 was 98%, 99%, 90%, and 35%, respectively. Thus, we designated these genes from R. opacus TKN14 as nidABCD in the same way (Fig. 2). The TKN14 nidA and nidB genes, respectively, showed high or significant homology to many genes encoding the large (α) and small (β) subunits of a naphthalene-type dioxygenase iron-sulfur protein which appear in the DDBJ/EMBL/GenBank databases. Besides the corresponding enzymes of Rhodococcus sp. I24, the TKN14 NidA and NidB proteins had high amino acid homology to the large and small subunits of naphthalene dioxygenases (NDOs) found in various Rhodococcus species;, i.e., two subspecies (P200 and P400) derived from Rhodococcus rhodochrous NCIMB13064 (DDBJ/EMBL/GenBank accession nos. AY392424 and AY392423) (19, 21), Rhodococcus sp. NCIMB12038 (accession no. AF082663) (1), and Rhodococcus sp. 1BN (accession no. AJ401612) (2, 9). The nidA gene had the known consensus sequence CXHRGX8GNX5CXYHG, which is a Rieske-type iron-sulfur-binding center motif. Downstream of the nidAB genes of TKN14, we found nidC, which could be classified as cis-naphthalene dihydrodiol dehydrogenase, and nidD, which exhibited homology to a putative hydratase-aldolase gene. Hydratase-aldolase was reported to catalyze the reaction from 2′-hydroxybenzalpyruvate to salicylaldehyde and pyruvate in the degradation pathway of naphtha-lene (23). Several uncharacterized genes having homology to those found in other Rhodococcus species were also found downstream of the nidABCD genes (data not shown). Upstream of the nidABCD genes, there were a long 1.81-kb ORF that had moderate homology to the electron transfer flavoprotein family (ORF1 in Fig. 2), Rd, and two ORFs related to regulators, i.e., a putative transcriptional regulator (ORF2) and a putative naphthalene degradation regulator (ORF3).
Functional analysis of the gene cluster involved in the methyl oxidation of o-xylene.
It is important for our knowledge to identify which gene or ORF components in the 15-kb NotI fragment of pNot1-9 are involved in the methyl oxidation of o-xylene in E. coli. We started by subcloning two portions from this 15-kb NotI region. The 7.26-kb region, including nidAB, was amplified by PCR with pNot1-9 as the template DNA and introduced into pT7BlueT-vector by the TA-cloning method to generate pN54-1-5 (Fig. 2). The 4.59-kb EcoRI fragment containing all the nidABCD genes was inserted into the EcoRI site of pBluescript II SK(+) to generate pNidABCD. Bioconversion experiments on o-xylene with E. coli JM109 harboring each plasmid were carried out. E. coli carrying pNidABCD did not metabolize o-xylene. On the other hand, E. coli carrying pN54-1-5 showed the ability to convert o-xylene into o-methyl-benzyl alcohol. It is thus evident that the genes involved in o-xylene oxidation were located in the 7.26-kb DNA region including nidAB.
We next constructed several ORF-disrupting plasmids based on plasmid pN54-1-5, i.e., the nidA, nidB, ORF1, and ORF2-3 disruptants (Table 1). Bioconversion experiments with o-xylene by E. coli carrying these disruption plasmids were carried out, followed by GC-MS analysis. o-Methylbenzyl alcohol was not generated in the extracts from either the nidA-disrupting or nidB-disrupting mutant. In contrast, o-methylbenzyl alcohol was detected in the extracts from both ORF1-disrupting and ORF2-3-disrupting mutants. These results indicated that both nidA and nidB were essential for the methyl oxidation of o-xylene, while ORF1, ORF2, and ORF3 were unnecessary for this. The result of the latter is not surprising, since the three ORFs encode proteins homologous to an electron transfer protein (ORF1) or a transcriptional regulator (ORF2 and ORF3).
Bioconversion experiments with various aromatic hydrocarbons using E. coli harboring pN54-1-5.
We examined the substrate range of E. coli JM109 carrying plasmid pN54-1-5. The bioconversion reactions were conducted by coculturing it with various monocyclic aromatic compounds (benzene substituents) as the substrates. The identity of each product was verified by analyzing the ethyl acetate extract from the coculture with GC-MS, comparing its retention time and mass spectrum to those of an authentic sample. Consequently, the cocultures of E. coli (pN54-1-5) and the substrates generated not only o-methylbenzyl alcohol from o-xylene, but also m-methylbenzyl alcohol from m-xylene, p-methylbenzyl alcohol and a trace amount of 2,5-dimethylphenol from p-xylene, benzyl alcohol from toluene, and 1-phenetylalcohol from ethylbenzene. We also detected the formation of 1-indenol, 1-indanone, and cis-(1,2)-indandiol from indene as the substrate.
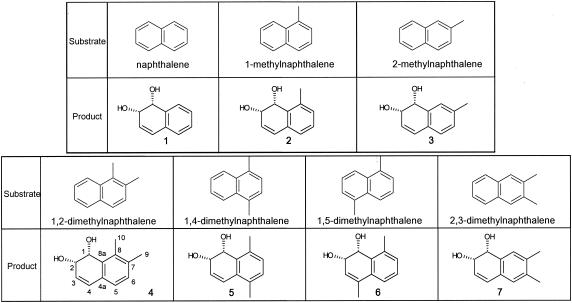
The bioconversion reactions through the cells of E. coli (pN54-1-5) were also carried out by using naphthalene and (di)methylnaphthalenes as the substrates. Each converted product was identified by purifying the ethyl acetate extract of the coculture and by analyzing it with MS and NMR, as shown in Materials and Methods. The results are shown in Fig. 3. The recombinant E. coli converted naphthalene and its methylated or dimethylated substituents into their corresponding cis-dihydrodiol.
FIG. 3.
Structures of products converted from naphthalene and (di)methylated naphthalenes by the cocultures with E. coli JM109 harboring plasmid pN54-1-5. The numbers of products corresponds to those discussed in “Purification and identification of the dihydrodiol products” above, in Materials and Methods. Products 4, 5, and 6 were novel compounds.
Functional identification of the genes needed for the methyl oxidation of o-xylene.
E. coli JM109 expressing only the nidAB genes (plasmid pTrcNidAB) was not able to convert o-xylene into o-methylbenzyl alcohol. We therefore considered that Rd, which is often found accompanying the naphthalene dioxygenase large and small subunit genes in other Rhodococcus species, may be required as an electron transfer protein for the NidAB enzyme. However, E. coli expressing the Rd gene in addition to nidAB (plasmid pTrcNidABRd) was not able to metabolize o-xylene. No homologous gene for reductase such as rubredoxin reductase was included in the 7.26-kb fragment of pN54-1-5. We speculated that an unknown gene(s) indispensable for the methyl oxidation of o-xylene exists between ORF3 and the nidA gene (Fig. 2), since ORF1, ORF2, and ORF3 were not involved in the methyl oxidation reaction of o-xylene.
We constructed various plasmids for expressing the genes for Rd and an ORF(s) between ORF3 and nidA, along with nidAB, using vector pTrcHisA (Table 1). Four E. coli carrying plasmids pTrcNidABRdORF4, pTrcNidABRdORF5, pTrcNidABRdORF6, and pTrcNidABRdORF789 were examined for their o-xylene-converting capability. The results indicated that only E. coli(pTrcNidABRdORF789) had the capability of converting o-xylene into o-methylbenzyl alcohol. It is therefore clear that the ORF789 region comprising ORF7, ORF8, and ORF9 incorporates the indispensable component for o-xylene oxidation. Plasmids incorporating each ORF or a combination of the ORFs were constructed as shown in Table 1. The o-xylene conversion activity was examined by using E. coli-carrying plasmids pTrcNidABRdORF8, pTrcNidABRdORF9,pTrcNidABRdORF78, and pTrcNidABRdORF89. The re- sults revealed that E. coli(pTrcNidABRdORF8), E. coli (pTrcNidABRdORF78), and E. coli(pTrcNidABRdORF89) converted o-xylene into o-methylbenzyl alcohol, indicating that ORF8 was the essential gene for o-xylene conversion. A BLAST analysis of the deduced amino acid sequence of ORF8 showed that this gene product had no overall homology with any other proteins in databases.
We next checked whether Rd was indispensable or not. We expressed only ORF8 and nidAB (plasmid pTrcNidABORF8) (Table 1) in E. coli. A bioconversion experiment on o-xylene with this recombinant E. coli showed that methyl oxidation of o-xylene did not take place by this strain excluding Rd. It was therefore clear that the minimum components necessary for the methyl oxidation of o-xylene were the nidAB genes and the genes for Rd and ORF8. We designated the respective genes for Rd and ORF8 nidE and nidF.
Analysis of the minimum gene set needed to bioconvert several aromatic hydrocarbons.
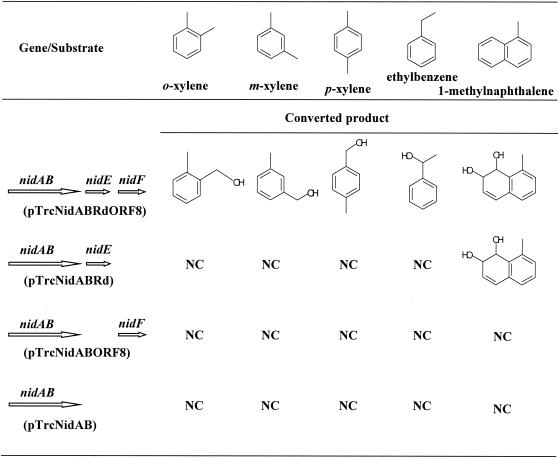
E. coli JM109 cells harboring pTrcNidABRdORF8, pTrcNidABRd, pTrcNidABORF8, or pTrcNidAB were cocultivated to check their bioconversion capability on several aromatic hydrocarbons. o-, m-, and p-xylene, ethylbenzene, and (di)methylnaphthalenes such as 1-methylnaphthalene were used as substrates. The results are shown in Fig. 4. As expected, E. coli(pTrcNidABRdORF8) was able to oxidize the benzylic position of o-, m-, and p-xylene and ethylbenzene to produce the corresponding alcohol, while E. coli harboring pTrcNidABRd, pTrcNidABORF8, and pTrcNidAB could not metabolize these compounds. We also found that the relative reactivity of o- and m-xylene was much higher than that of p-xylene. When the substrate was naphthalene, 1-methylnaphthalene, 2-methylnaphthalene, or 1,2-dimethylnaphthalene, these compounds were converted to the corresponding cis-dihydrodiols (their structure is shown in Fig. 3) not only by E. coli(pTrcNidABRdORF8) but also by E. coli(pTrcNidABRd). No conversion products were detected with E. coli(pTrcNidABORF8) or E. coli(pTrcNidAB). These results show the unique feature of the aromatic hydrocarbon oxygenase system from R. opacus TKN14, i.e., NidF encoded by ORF8 is the auxiliary protein needed for the hydroxylation reaction at the benzylic position of xylenes and ethylbenzene by working with NidAB and NidE (rubredoxin) and is not required for the ring-dihydroxylation reaction of naphthalene and its (di)methyl substituents.
FIG. 4.
Bioconversion tests on monocyclic aromatic hydrocarbons and 1-methylnaphthalene by E. coli JM109 harboring pTrcNidABRdORF8, pTrcNidABRd, pTrcNidABORF8, or pTrcNidAB. NC, not converted.
This R. opacus TKN14 oxygenase system has also shown that rubredoxin, not ferredoxin, is indispensable for the ring dihydroxylation reaction of the naphthalene skeleton, as well as for the methyl group hydroxylation reaction of xylenes. This finding seems very strange, since rubredoxin is considered an essential component of the alkane hydroxylase system, which transfers electrons from rubredoxin reductase to alkane hydroxylase (37). It also seems strange that the oxygenation reaction proceeds in the recombinant E. coli without a foreign reductase gene such as rubredoxin reductase. It is, however, possible that E. coli has an endogenous functional reductase. Several reports have indicated that full or partial dioxygenase activity can be detected without the reductase component (26, 28, 33). In the case of the 2,4-dinitrotoluene dioxygenase (DNT) from Burkholderia sp. strain DNT, which is encoded by the dntAaAbAcAd genes and has structural similarity to NDO from Pseudomonas sp. NCIB 9816-4, deletion of dntAa (the reductase component of DNT) from the gene cluster did not eliminate the oxygenase ability to transform 2,4-dinitrotoluene into 4-methyl-5-nitrocatechol (33). Also, in the case of the diterpenoid dioxygenase from Pseudomonas abietaniphila BKME-9 (26), expression in E. coli of the ditA1A2A3 genes, encoding α and β subunits of oxygenase and ferredoxin without a reductase component, resulted in biocatalytic activity. As for phenanthrene dioxygenase from Nocardioides sp. strain KP7 (28), E. coli cells carrying the phdABC genes, which lack the reductase gene (phnD), exhibited phenanthrene-degrading activity, while this activity was significantly less than that of E. coli (phdABCD). These instances enable us to consider that E. coli cells carrying plasmid pTrcNidABRd or pTrcNidABRdORF8 retain oxygenase activity, probably due to collaboration with an unknown host-derived reductase.
Structural and functional properties of the R. opacus TKN14 oxygenase.
In contrast to the well-studied NDO of the gram-negative bacterium Pseudomonas sp. strain NCIB 9816-4 (32), little information is currently available about the naphthalene dioxygenation system of gram-positive bacteria. Indeed, functional expression of the corresponding genes in E. coli, which are derived from such gram-positive bacteria as the genus Rhodococcus, has been unsuccessful in many cases. In the case of Rhodococcus sp. I24, it was reported that an E. coli clone carrying plasmid pR4-10, which had an insert of about 7.0 kb including the nidAB genes, had indigo formation activity from indole but did not function as naphthalene dioxygenase (36). In the case of Rhodococcus sp. strain NCIMB12038 (22), the iron-sulfur protein of NDO encoded by the narAaAb genes was purified and proved to be necessary for converting naphthalene into naphthalene-cis-dihydrodiol, but an as-yet-uncharacterized protein(s) was necessary for the reconstitution of NDO activity. Kulakov et al. have further reported that the NDO genes from Rhodococcus sp. NCIMB12038 were not functionally expressed in E. coli, presumably because of the assemblage failure of the enzyme (20). Our nidAB genes of R. opacus TKN14 have a high homology to these naphthalene dioxygenases found in Rhodococcus sp. strains I24 and NCIMB12038. No putative ferredoxin or ferredoxin reductase components, which are normally required for transferring electrons from NAD(P)H to a dioxygenase iron-sulfur protein, were found in the nidAB (narAaAb) gene clusters in our TKN14 strain or in the I24 and NCIMB12038 strains, suggesting that rubredoxin (NidE) should work in the Rhodococcus naphthalene dioxygenation system instead of ferredoxin.
The well-known NDO (NahAaAbAcAd) from Pseudomonas sp. NCIB 9816-4 is a multicomponent enzyme system consisting of ferredoxin reductase (NahAa), [2Fe-2S] ferredoxin (NahAb), and large and small subunits (NahAc and NahAd) of an iron-sulfur protein. This NDO is an enzyme with topology different from that of our NidABEF; e.g., the deduced amino acid sequences of NahAc and NahAd derived from strain NCIB 9816-4 have only 31% and 29% identity with NidA and NidB from strain TKN14, respectively, while the substrate range of both NDOs seems very similar. The NCIB 9816-4 NDO can catalyze not only the dioxygenation reaction in the aromatic ring of naphthalene and indene to the corresponding cis-dihydrodiol but also the monooxygenation reaction of isomeric xylenes, including o-xylene, into the corresponding methylbenzyl alcohol (25). On the other hand, the toluene dixoygenase derived from Pseudomonas putida F1 and Pseudomonas sp. strain JS150 metabolizes m- and p-xylene to the corresponding cis-dihydrodiols and does not oxidize o-xylene (12). Lee and Gibson have reported that an NDO from Pseudomonas sp. strain 9816-4 oxidizes styrene to 1-phenyl-1,2-ethanediol (24). We also detected the formation of this product from styrene by E. coli(pTrcNidABRdORF8) (data not shown). However, toluene and styrene dioxygenase oxidize styrene to form styrene cis-2,3-dihydrodiol, and P-450 monooxygenase and cytochrome C peroxidase convert styrene to styrene oxide (24). These results support the notion that NidABEF from R. opacus TKN14 has a substrate range and reaction specificity that are very similar to those of NDO from Pseudomonas sp. NCIB 9816-4, although the two enzymes have different structural properties from each other. These two distinct aromatic hydrocarbon oxygenase systems, which evolved from different ancestors, may acquire the same catalytic function.
Synthesis of nidABEF mRNA in R. opacus TKN14 during the degradation of o-xylene.
R. opacus TKN14 is able to grow on o-xylene as the sole carbon and energy source. We examined whether the individual nidABEF genes were induced or not through mRNA analysis by RT-PCR when this bacterium was cultured with a MS medium including only o-xylene as the carbon source. When culturing was carried out without o-xylene, we were not able to detect the synthesis of the nidA and nidB cDNAs (RT-PCR products), while in the case of nidE and nidF, very faint cDNA bands were detected with the expected sizes, which were 178 bp and 162 bp, respectively, by agarose gel electrophoresis (data not shown). R. opacus TKN14 degraded 90% and 100% of the water-soluble o-xylene in 4 h and 21 h, respectively. Clear RT-PCR products from nidA and nidB were detected in the 4-h sample after o-xylene addition with the expected sizes, which were 233 bp and 191 bp, respectively; the level did not change in the sample at 21 h after o-xylene addition. Clear RT-PCR products from nidE and nidF were also detected in the sample at 4 h and 21 h after o-xylene addition. It has therefore been concluded that the transcriptions of the nidA, nidB, nidE, and nidF genes are induced by the addition of o-xylene to the medium.
It is very likely that the o-xylene monooxygenase-naphthalene dioxygenase genes, nidABEF, are involved in the degradation pathways of a wide range of aromatic hydrocarbons by Rhodococcus species as the first key enzyme.
Acknowledgments
This work was supported by a grant from the New Energy and Industrial Technology Development Organization (NEDO).
REFERENCES
- 1.Allen, C. C., D. R. Boyd, M. J. Larkin, K. A. Reid, N. D. Sharma, and K. Wilson. 1997. Metabolism of naphthalene, 1-naphthol, indene, and indole by Rhodococcus sp. strain NCIMB 12038. Appl. Environ. Microbiol. 63:151-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreoni, V., S. Bernasconi, M. Colombo, J. B. van Beilen, and L. Cavalca. 2000. Detection of genes for alkane and naphthalene catabolism in Rhodococcus sp. strain 1BN. Environ. Microbiol. 2:572-577. [DOI] [PubMed] [Google Scholar]
- 3.Arenghi, F. L., D. Berlanda, E. Galli, G. Sello, and P. Barbieri. 2001. Organization and regulation of meta cleavage pathway genes for toluene and o-xylene derivative degradation in Pseudomonas stutzeri OX1. Appl. Environ. Microbiol. 67:3304-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbieri, P., L. Palladino, P. Di Gennaro, and E. Galli. 1993. Alternative pathways for o-xylene or m-xylene and p-xylene degradation in a Pseudomonas stutzeri strain. Biodegradation 4:71-80. [Google Scholar]
- 5.Bertoni, G., F. Bolognese, E. Galli, and P. Barbieri. 1996. Cloning of the genes for and characterization of the early stages of toluene and o-xylene catabolism in Pseudomonas stutzeri OX1. Appl. Environ. Microbiol. 62:3704-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertoni, G., M. Martino, E. Galli, and P. Barbieri. 1998. Analysis of the gene cluster encoding toluene/o-xylene monooxygenase from Pseudomonas stutzeri OX1. Appl. Environ. Microbiol. 64:3626-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bickerdike, S. R., R. A. Holt, and G. M. Stephens. 1997. Evidence for metabolism of o-xylene by simultaneous ring and methyl group oxidation in a new soil isolate Microbiology 143:2321-2329. [DOI] [PubMed]
- 8.Cafaro, V., R. Scognamiglio, A. Viggiani, V. Izzo, I. Passaro, E. Notomista, F. D. Piaz, A. Amoresano, A. Casbarra, P. Pucci, and A. Di Donato. 2002. Expression and purification of the recombinant subunits of toluene/o-xylene monooxygenase and reconstitution of the active complex. Eur. J. Biochem. 269:5689-5699. [DOI] [PubMed] [Google Scholar]
- 9.Cavalca, L., M. Colombo, S. Larcher, C. Gigliotti, E. Collina, and V. Andreoni. 2002. Survival and naphthalene-degrading activity of Rhodococcus sp. strain 1BN in soil microcosms. J. Appl. Microbiol. 92:1058-1065. [DOI] [PubMed] [Google Scholar]
- 10.Di Gennaro, P., E. Rescalli, E. Galli, G. Sello, and G. Bestetti. 2001. Characterization of Rhodococcus opacus R7, a strain able to degrade naphthalene and o-xylene isolated from a polycyclic aromatic hydrocarbon-contaminated soil. Res. Microbiol. 152:641-651. [DOI] [PubMed] [Google Scholar]
- 11.Fishbein, L. 1985. An overview of environmental and toxicological aspects of aromatic hydrocarbons. III. Xylene. Sci. Total Environ. 43:165-183. [DOI] [PubMed] [Google Scholar]
- 12.Gibson, D. T., V. Mahadevan, and J. F. Davey. 1974. Bacterial metabolism of para- and meta-xylene: oxidation of the aromatic ring. J. Bacteriol. 119:930-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibson, D. T., and V. Subramanian. 1984. Microbial degradation of aromatic hydrocarbons, p. 181-252. In D. T. Gibson (ed.), Microbial degradation of organic compounds, vol. 13. Marcel Dekker, Inc., New York, N.Y. [Google Scholar]
- 14.Harayama, S., M. Rekik, M. Wubbolts, K. Rose, R. A. Leppik, and K. N. Timmis. 1989. Characterization of five genes in the upper-pathway operon of TOL plasmid pWW0 from Pseudomonas putida and identification of the gene products. J. Bacteriol. 171:5048-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasai, Y., K. Shindo, S. Harayama, and N. Misawa. 2003. Molecular characterization and substrate preference of a polycyclic aromatic hydrocarobon dioxygenase from Cycloclasticus sp. strain A5. Appl. Environ. Microbiol. 69:6688-6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, D., J. C. Chae, G. J. Zylstra, Y. S. Kim, S. K. Kim, N. H. Nam, Y. M. Kim, and E. Kim. 2004. Identification of a novel dioxygenase involved in metabolism of o-xylene, toluene, and ethylbenzene by Rhodococcus sp. strain DK17. Appl. Environ. Microbiol. 70:7086-7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, D., Y. S. Kim, S. K. Kim, S. W. Kim, G. J. Zylstra, Y. M. Kim, and E. Kim. 2002. Monocyclic aromatic hydrocarbon degradation by Rhodococcus sp. strain DK17. Appl. Environ. Microbiol. 68:3270-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kok, M., R. Oldenhuis, M. P. van der Linden, P. Raatjes, J. Kingma, P. H. van Lelyveld, and B. Witholt. 1989. The Pseudomonas oleovorans alkane hydroxylase gene. Sequence and expression. J. Biol. Chem. 264:5435-5441. [PubMed] [Google Scholar]
- 19.Kulakov, L. A., M. J. Larkin, and A. N. Kulakova. 1997. Cryptic plasmid pKA22 isolated from the naphthalene degrading derivative of Rhodococcus rhodochrous NCIMB13064. Plasmid 38:61-69. [DOI] [PubMed] [Google Scholar]
- 20.Kulakov, L. A., C. C. Allen, D. A. Lipscomb, and M. J. Larkin. 2000. Cloning and characterization of a novel cis-naphthalene didihydrodiol dehydrogenase gene (narB) from Rhodococcus sp. NCIMB12038. FEMS Microbiol. Lett. 182:327-331. [DOI] [PubMed] [Google Scholar]
- 21.Kulakova, A. N., K. A. Reid, M. J. Larkin, C. C. Allen, and L. A. Kulakov. 1996. Isolation of Rhodococcus rhodochrous NCIMB13064 derivatives with new biodegradative abilities. FEMS Microbiol. Lett. 145:227-231. [DOI] [PubMed] [Google Scholar]
- 22.Larkin, M. J., C. C. Allen, L. A. Kulakov, and D. A. Lipscomb. 1999. Purification and characterization of a novel naphthalene dioxygenase from Rhodococcus sp. strain NCIMB12038. J. Bacteriol. 181:6200-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laurie, A. D., and G. Lloyd-Jones. 1999. The phn genes of Burkholderia sp. strain RP007 constitute a divergent gene cluster for polycyclic aromatic hydrocarbon catabolism. J. Bacteriol. 181:531-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, K., and D. T. Gibson. 1996. Stereospecific dihydroxylation of the styrene vinyl group by purified naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816-4. J. Bacteriol. 178:3353-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, K., and D. T. Gibson. 1996. Toluene and ethylbenzene oxidation by purified naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816-4. Appl. Environ. Microbiol. 62:3101-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin, V. J., and W. W. Mohn. 1999. A novel aromatic-ring-hydroxylating dioxygenase from the diterpenoid-degrading bacterium Pseudomonas abietaniphila BKME-9. J. Bacteriol. 181:2675-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patty, F. A. 1963. Industrial hygiene and toxicology, vol. 2. Toxicology. Interscience Publishers, New York, N.Y.
- 28.Saito, A., T. Iwabuchi, and S. Harayama. 2000. A novel phenanthrene dioxygenase from Nocardioides sp. strain KP7: expression in Escherichia coli. J. Bacteriol. 182:2134-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Schraa, G., B., M. Bethe, A. R. van Neerven, W. J. Van den Tweel, E. Van der Wende, and A. J. Zehnder. 1987. Degradation of 1,2-dimethylbenzene by Corynebacterium strain C125. Antonie Leeuwenhoek 53:159-170. [DOI] [PubMed] [Google Scholar]
- 31.Solano-Serena, F., R. Marchal, J. M. Lebeault, and J. P. Vandecasteele. 2000. Selection of microbial populations degrading recalcitrant hydrocarbons of gasoline by monitoring of culture-headspace composition. Lett. Appl. Microbiol. 30:19-22. [DOI] [PubMed] [Google Scholar]
- 32.Suen, W. C., and D. T. Gibson. 1993. Isolation and preliminary characterization of the subunits of the terminal component of naphthalene dioxygenase from Pseudomonas putida NCIB 9816-4. J. Bacteriol. 175:5877-5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suen, W. C., B. E. Haigler, and J. C. Spain. 1996. 2,4-Dinitrotoluene dioxygenase from Burkholderia sp. strain DNT: similarity to naphthalene dioxygenase. J. Bacteriol. 178:4926-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki, M., T. Hayakawa, J. P. Shaw, M. Rekik, and S. Harayama. 1988. Primary structure of xylene monooxygenase: similarities to and differences from the alkane hydroxylation system. J. Bacteriol. 173:1690-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teramoto, M., S. Takaichi, Y. Inomata, H. Ikenaga, and N. Misawa. 2003. Structural and functional analysis of a lycopene β-monocyclase gene isolated from a unique marine bacterium that produces myxol. FEBS Lett. 545:120-126. [DOI] [PubMed] [Google Scholar]
- 36.Treadway, S. L., K. S. Yanagimachi, E. Lankenau, P. A. Lessard, G. Stephanopoulos, and A. J. Sinskey. 1999. Isolation and characterization of indene bioconversion genes from Rhodococcus strain I24. Appl. Microbiol. Biotechnol. 51:786-793. [DOI] [PubMed] [Google Scholar]
- 37.van Beilen, J. B., M. Neuenschwander, T. H. Smits, C. Roth, S. B. Balada, and B. Witholt. 2002. Rubredoxins involved in alkane oxidation. J. Bacteriol. 184:1722-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams, P. A., and K. Murray. 1974. Metabolism of benzoate and the methylbenzoates by Pseudomonas putida (arvilla) mt-2: evidence for the existence of a TOL plasmid. J. Bacteriol. 120:416-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Worsey, M. J., and P. A. Williams. 1975. Metabolism of toluene and xylenes by Pseudomonas putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J. Bacteriol. 124:7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wubbolts, M. G., P. Reuvekamp, and B. Witholt. 1994. TOL plasmid-specified xylene oxygenase is a wide substrate range monooxygenase capable of olefin epoxidation. Enzyme Microb. Technol. 16:608-615. [DOI] [PubMed] [Google Scholar]