Abstract
So far, there is only fragmentary and unconfirmed information on bacteriophages infecting the genus Bifidobacterium. In this report we analyzed three prophage-like elements that are present in the genomes of Bifidobacterium breve UCC 2003, Bifidobacterium longum NCC 2705, and Bifidobacterium longum DJO10A, designated Bbr-1, Bl-1, and Blj-1, respectively. These prophagelike elements exhibit homology with genes of double-stranded DNA bacteriophages spanning a broad phylogenetic range of host bacteria and are surprisingly closely related to bacteriophages infecting low-G+C bacteria. All three prophage-like elements are integrated in a tRNAMet gene, which appears to be reconstructed following phage integration. Analysis of the distribution of this integration site in many bifidobacterial species revealed that the attB sites are well conserved. The Blj-1 prophage is 36.9 kb long and was induced when a B. longum DJO10A culture was exposed to mitomycin C or hydrogen peroxide. The Bbr-1 prophage-like element appears to consist of a noninducible 28.5-kb chimeric DNA fragment composed of a composite mobile element inserted into prophage-like sequences, which do not appear to be widely distributed among B. breve strains. Northern blot analysis of the Bbr-1 prophage-like element showed that large parts of its genome are transcriptionally silent. Interestingly, a gene predicted to encode an extracellular beta-glucosidase carried within the Bbr-1 prophage-like element was shown to be transcribed.
Bacteriophages are widely distributed among eubacteria, where they influence the genomic evolution and adaptive capabilities of their hosts (9, 10). Microbial genomics has revealed that a substantial proportion of many bacterial chromosomes is represented by prophage sequences. Prophages may confer a diverse array of phenotypic traits on their hosts, including those that govern the course and the pathobiology of bacterial infections (3, 9). In fact, based on evolutionary reasoning, prophages are postulated to contribute genes that increase the fitness of lysogenic bacteria in their specific ecological niche (3). This realization has changed our understanding of phage-bacterium interactions from being a simple parasite-host relationship to a mutually beneficial genomic coevolution.
The increasing number of available bacterial genome sequences has compounded this understanding of prophage genome distribution and evolution. The mosaic pattern and localized diversity of many different prophage genomes are obvious from comparative analyses of prophage genome content and organization, as well as similarities of orthologous gene products encoded by these elements (18). Phage lytic functions and particle structure have been considered the main criteria for phage taxonomy (1), although in the absence of such information phage classification may rely on phylogenetic trees that are based on phage genome comparisons (proteomic tree) (31). Such proteomic tree analysis may thus be a useful means to infer phage evolution despite the fact that it is not a molecular taxonomical tool that is fully accepted yet by the International Committee on the Taxonomy of Viruses.
So far, there is only fragmentary information on bacteriophages infecting the genus Bifidobacterium (bifidobacteriophages) (34) although many phage and prophage sequences have been described among the phylogenetically related high-G+C gram-positive bacteria, e.g., the genera Mycobacterium, Corynebacterium, and Streptomyces (9, 10, 25). Few of these prophages appear to be complete and capable of spontaneous excision from bacterial cells. Interestingly, the fully sequenced Streptomyces coelicolor A3, as well as Mycobacterium leprae and Mycobacterium bovis do not appear to contain prophage sequences, whereas Mycobacterium tuberculosis is predicted to possess two small prophage remnants, φRv1 and φRv2 (16). These M. tuberculosis prophages appear to contain an incomplete structural gene module, and it has been proposed that they could act as satellite phages in a manner reminiscent of coliphage P4 (20), through the mobilization of the structural components encoded by another phage to carry their genomes (16). The mycobacteriophage genomes are clearly mosaic in nature, with regions of obvious sequence similarity interspersed with segments that appear to be unrelated, suggesting that extensive horizontal genetic exchange among bacteriophages is common (16). Interestingly, the genome sequences of the Streptomyces phages provide a clear evolutionary connection between mycobacteriophages and λ-like phages (16).
Bifidobacterium breve UCC 2003, a human commensal isolated from an infant stool sample, is phylogenetically closely related to the sequenced Bifidobacterium longum NCC 2705 and Bifidobacterium longum DJO10A strains (33; unpublished results). Analysis of the complete genome sequence of B. longumNCC 2705 predicted a vast repertoire of mobile elements, including multiple copies of different insertion elements, in addition to an integrated plasmid (33, 42).
In this study, comparative and experimental evidence is presented which shows that (i) all three sequenced bifidobacteria are predicted to contain a single but different prophage-like element; (ii) all three prophage-like elements are integrated in the same homologous DNA sequence; and (iii) the B. longum DJO10A prophage-like element can be chemically induced to excise from its host genome. Comparison of these prophage sequences with those available in the public databases indicates that the bifidobacterial prophages possess a typical mosaic genome structure.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All Bifidobacterium strains were grown anaerobically in MRS (Difco, Detroit, MI, USA) supplemented with 0.05% l-cysteine-HCl and incubated at 37°C for 16 h.
RNA isolation and Northern blot analysis.
Total RNA was isolated from 10 ml of B. breve UCC 2003, collected at an optical density at 600 nm of 0.6, 0.8, and 1.1 using macaloid acid (41), and treated with DNase (Roche, Sussex, United Kingdom). Fifteen μg RNA was separated by electrophoresis on a 1.5% agarose-3% formaldehyde denaturing gel, transferred to a Zeta-Probe blotting membrane (Bio-Rad, Hemel Hempstead, United Kingdom) according to Sambrook et al. (32), and fixed by UV cross-linking using a Stratalinker 1800 (Stratagene, USA). PCR amplicons obtained by primer combinations that were designed to target specific genes of the Bbr-1 genome were radiolabeled with α-32P (32) using the random primed DNA labeling system (Roche, East Sussex, United Kingdom) in accordance with the manufacturer's instruction. Prehybridization and hybridization were carried out at 65°C in 0.5 M NaHPO4 (pH 7.2), 1.0 mM EDTA, and 7.0% sodium dodecyl sulfate (SDS). Following 18 h of hybridization, the membrane was rinsed twice for 30 min at 65°C in 0.1 M NaHPO4 (pH 7.2), 1.0 mM EDTA, 1% SDS, twice for 30 min at 65°C in 0.1 mM NaHPO4 (pH 7.2), 1.0 mM EDTA, 0.1% SDS, and exposed to X-OMAT autoradiography film (Eastman Kodak).
DNA amplification of attB sites.
A 600-bp PCR fragment corresponding to the attB region of prophage-like element Bbr-1 was generated using primer combination B1 (5′-GTTCAGAATCACCAGTCTG-3′) and B2 (5′-GAATCTGAGGAGCAGCTG-3′), while a 750-bp PCR fragment corresponding to the attB region of prophage-like element Bl-1 and Blj-1 was generated using primers L1 (5′-CATTCCTATAACGGCCATTTATG-3′) and L2 (5′-GTGGCGATGTGTCGCTTGCCTC-3′), and both fragments were subsequently sequenced on both strands.
The sequences homologous to the presumed Bbr-1 attachment site in Bifidobacterium animalis subsp. lactis LMG 18906 and Bifidobacterium animalis subsp. animalis DSM 25527 were amplified using primers Bal−1 (5′-GTCACCATTAACAGAAATCAATG-3′) and Bal-2 (5′-GTGACGTAGATTCGATTGGTG-3′).
Pulsed-field gel electrophoresis and Southern blots.
Agarose-embedded bacterial cells were prepared as described by Ventura et al. (36). Briefly, bifidobacteria were grown to an A600 of 0.5 in MRS medium. Chloramphenicol (100 μg/ml) was added and incubation was continued for 2 h. Cells were harvested by centrifugation, embedded in agarose and subsequently incubated overnight at 37°C in a lysis buffer (100 mM EDTA, 6 mM Tris, 1 M NaCl) containing 1 mg/ml of lysozyme (Sigma, United Kingdom) and 20 U/ml of mutanolysin (Sigma, United Kingdom). Proteinase K (1 mg/ml) treatment was performed in 100 mM EDTA, 1% Sarcosyl for 18 h at 50°C. Prior to restriction enzyme digestion the agarose blocks were washed twice in 1x Tris-EDTA for 1 h each.
For digestion with restriction endonuclease, bacterial cells embedded in agarose blocks were treated with 50 units of SpeI (Roche, East Sussex, United Kingdom) as described by the manufacturer. Digestion was stopped by washing the blocks for 20 min in Tris-EDTA buffer. Pulsed-field gel electrophoresis was performed by a contour-clamped homogenous electric field mode in a CHEF-DRII apparatus (Bio-Rad). DNA fragments were separated using 1% agarose gels in 0.5 × Tris-borate-EDTA (TBE) buffer, cooled to 14°C. The pulse time was 1 to 6 seconds, with a voltage of 6 V/cm at 14°C for 22 h. The agarose gels were stained with ethidium bromide (0.5 μg/ml) and visualized under UV light at 260 nm.
Southern blots of agarose gels were performed on Hybond N+ membranes (Amersham, United Kingdom) and developed according to Sambrook et al. (32) using radiolabeled PCR generated probes described in the text.
Sequence analysis.
Open reading frames were predicted using the ORF Finder (NCBI) accepting ATG, TTG, and GTG as possible start codons and requiring a minimum size of 50 amino acids. Identified open reading frames (ORFs) were subsequently manually checked for validity. Nucleotide and predicted amino acid sequences were compared with sequences in public databases (GenBank, EMBL, PIR-Protein, SWISS-PROT, and PROSITE), using BLAST (2), PSI-BLAST, and FASTA (21). A scan for tRNA genes was performed using the tRNAscan-SE program (23). Motif searches were achieved by interrogation of the Pfam database (4). Likely transmembrane domains were determined using the HMMTOP server (www.enzim.hu/hmmtop) and the DAS (Dense Alignment Surface) transmembrane prediction server (www.sbc.su.se/∼miklos/DAS/maindas.html).
Excision of Bbr-1 prophage-like element following addition of mitomycin C.
B. breve UCC2003 was grown until it reached the mid-exponential growth phase at which point mitomycin C (Sigma, United Kingdom) was added to a final concentration of 0 (control), 1, 2, 3, or 5 μg/ml. Furthermore, the inducibility of the Blj-1 prophage-like element was also assayed by adding hydrogen peroxide to a final concentration of 2 mM. Growth was allowed to continue for 12 h at 37°C after which cells were collected by centrifugation at 8,000 × g for 15 min. DNA was extracted as described previously (43, 44). Possible excision of Bbr-1 and Blj-1 prophage-like elements was monitored by PCR with reverse Bbr-1 int primer (5′-CTGTGAGCTGGCATCGTC-3′) and Bbr-1-ORF1429 primer (5′-GTCGTGGTCTGGGTCTTG-3′) or with reverse Blj-1 int primer (5′-CAGGAACTCGCTCGGATC-3′) and forward Blj-1 cI primer (5′-CAACGACAACCTGAATATGTTC-3′) specific for circularized Bbr-1 phage or Blj-1 phage to generate a 350-bp amplicon or 777 bp, respectively.
In the same PCR the 1451-1 (5′-GTAGGCGATCTGGTTGCTG-3′) and 1449-2 (5′-GACGACGTCTACAGGTTCGAC-3′) or 26-1 (5′-GTCGGTTCCATCCGAAGAG-3′) and 26-2 (5′-CAGCAATGGAGGAAAGCCAAC-3′) primers were added as positive controls. These primer pairs target a DNA fragment that encompasses parts of ORFs 1451 to 1449 or ORF 26, and should generate a 2,000-bp or 610-bp amplicon, respectively. Amplifications were performed with a Perkin-Elmer thermocycler (Cetus 9700, Perkin Elmer) with the following temperature profiles: one cycle of 95°C for 10 min and 35 cycles of 95°C for 30 seconds, 54°C for 30 seconds, and 72°C for 1 min.
Assay for viable bacteriophages.
Tests for bacteriophage were performed by spotting 50 μl of induced culture on a soft agar medium seeded individually with four B. breve and 10 B. longum strains as indicator strains. Plates were incubated anaerobically for 24 h at 37°C and then observed for plaque-forming ability as described previously (14, 34).
Proteomic tree analysis.
The phylogenetic analysis was performed as described previously (31). Every ORF-derived amino acid sequence in Bbr-1, Bl-1, and Blj-1 prophage-like sequences was compared to all ORF-derived proteins deposited in the complete phage and prophage genome database. The database includes deduced proteins from 375 bacteriophage genome from NCBI and from the prophages sequences as described previously (10).
Nucleotide sequence accession numbers.
The GenBank accession number forthe B. breve Bbr-1 prophage sequence is AY840979, B. longum Bl-1 pro-phage sequences is NC_004307, and B. longum Blj-1 prophage sequences is NZ_AABM02000005.
RESULTS AND DISCUSSION
Localization of prophage sequences.
Integrases are useful markers for identifying mobile DNA elements such as pro-phages in bacterial genomes (9, 10, 38, 40). The B. breve strain UCC 2003 contains 11 homologues of integrase genes in its 2.4-Mb genome (data not shown), while the B. longum NCC 2705 strain harbors nine predicted integrase paralogues in its 2.2-Mb genome. To precisely map likely prophage sequences, we searched for predicted phage-associated genes in the vicinity of these integrase genes. The genes located adjacent to int1452 of B. breve revealed many phage gene matches, whereas no evidence for a prophage association was obtained for any of the other ten identified integrase homologues.
One integrase in NCC 2705 belongs to a 17,462-bp phage (designated Bl-1), whereas the other eight integrases were not linked to any prophage sequences (data not shown).
The genome sequence of B. longum DJO10A revealed 17 ORFs that shared similarity with integrase genes and two ORFs that shared similarity with the host recombinases XerC and XerD, which have been described to mediate phage or other foreign gene integration into genomes (17). Of these, only ORF 1032 showed phage-like genes in its vicinity (data not shown).
Genome analysis of prophage-like element Bbr-1.
The predicted prophage-like element Bbr-1 in B. breve UCC 2003 extends from ORF 1452 (integrase gene) to ORF 1429 (lysin gene) (Fig. 1a). These ORFs are flanked by a 38-bp repeat indicating the existence of putative attL and attR sites. Moreover, PCR primers (B1 and B2) placed in the flanking bacterial genes, encoding a MetA protein and a hypothetical protein, generated a 600-bp amplicon with genomic DNA from B. breve NCTC 11815, B. breve JCM 7017, B. breve JCM 7020, and B. breve NCDO 2257, a size which concurs with a chromosome lacking prophage sequence at this site. Sequencing of this PCR product identified the presence of a single copy of a 38-bp repeat region suggesting an attB site (Fig. 2a). Phage integration complements the 3′ end of a tRNA carrying the anticodon specific for Met.
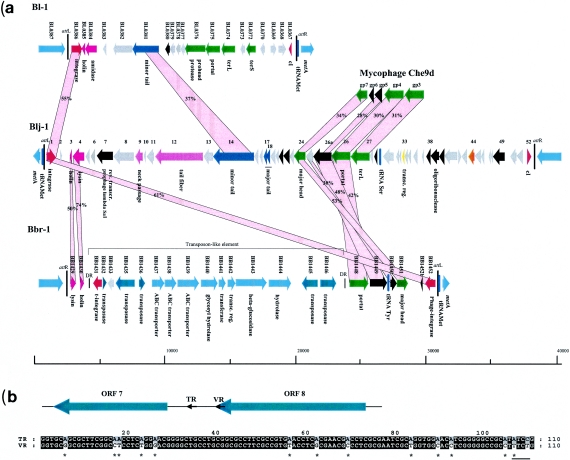
FIG. 1.
Panel a, comparative genome maps of the prophage-like elements Bbr-1, Bl-1, and Blj-1 and alignment of the partial genetic map of mycobacteriophage Che9d with Bbr-1 and Blj-1 prophage-like elements. Genes sharing similarity are linked by Probable functions of encoded proteins identified by bioinformatics analysis are indicated. The modular structure is color coded: red, lysogeny; green, DNA packaging and head; blue, tail; mauve, tail fiber; violet, lysis module; yellow, transcriptional regulator; orange, DNA replication; gray, unknown genes; black, genes similar to other functionally unknown bacteriophage genes. Vertical blue lines: tRNA genes. Panel b, schematic representation of the putative diversity generating retroelement and nucleotide sequence alignments of TR and VR segments from Blj-1.Adenine substitutions are indicated by asterisks, the TTCT motif is underlined.
FIG.2.
Integration sites of prophage-like elements Bbr-1 (panel a) and Bl-1 (panel b). The predicted host genes are depicted by black arrows and annotated according to their predicted functions. The prophage lysin and integrase genes at the prophage extremities are indicated by gray arrows. A gray vertical bar represents the deduced attachment site attL and attR; their nucleotide sequences are provided as an enlarged insert in which the core sequence of the attL and attR sites are underlined. The corresponding sequence of the attB site from a B. breve Bbr-1 prophage free strain or B. longum Bl-1 prophage free strain is provided in the bottom part of panels a and b, respectively. The boxed sequence indicates part of the tRNA gene. B1-B2 and L1-L2 provide the locations of the primers for PCR. Panel c: PCR amplifications of the attB site for Bbr-1 from the indicated B. breve strains. Lane MK, 1-kb DNA ladder (Gibco BRL). Panel d: PCR of the attB site from different B. longum strains.
Database matches (see Table 1) suggested that the Bbr-1 prophage genome could be subdivided into functional modules (Fig. 1a). In this prophage-like element the lysogenic module is limited to a phage integrase gene (ORF1452), whose deduced protein product shares significant amino acid (amino acids) similarity with a tyrosine recombinase of Xanthomonas campestris and with phage integrases of Mycobacterium and Corynebacterium species. The integrase gene is followed by what appears to be a partially deleted gene, designated ORF 1452b, which exhibits 51% amino acids identity to a transcriptional regulator of high-G+C bacteria and 29% amino acid identity to an anonymous gene from mycobacteriophage Che9c. The next ORFs (ORF 1451 to 1448) constitute the likely head morphogenesis module. More specifically, ORF1451 displays 32% and 28% amino acid identity to a major head protein of the mycobacteriophage Che9d and the lactococcal bacteriophage φLC3, respectively. Also, the protein encoded by ORF 1448 shares similarity to the predicted portal proteins of the same phages. In the intergenic region within the head morphogenesis module a tRNA gene is located, which is specific for Tyr (GTA codon). Interestingly, this is a prominent codon both in the Bbr-1 prophage-like element and in the bacterial host. This finding corroborates the frequent presence of tRNA genes within prophage genomes (39, 40), suggesting that prophage tRNAs might be of selective value to the lysogenic cells (lysogenic conversion role).
TABLE 1.
Database matches for Bbr-1
| ORFa | Representative similarity to protein in database | e value | % identity (% similarity) |
|---|---|---|---|
| 1452 | Tyrosine recombinase, Xanthomonas campestris | 4e-11 | 27 (43) |
| 1452b | gp30 mycobacteriophage Che9c | 1e-04 | 29 (42) |
| 1451 | Major head protein gp7 mycobacteriophage Che9d | 1e-25 | 32 (46) |
| 1449 | Putative phage protein, Corynebacterium diphtheriae | 7e-06 | 30 (46) |
| 1448 | Structural protein of Lactococcus bacteriophage rlt | 6e-34 | 30 (47) |
| 1446 | Putative transposase, Mycobacterium bovis | 1e-138 | 61 (79) |
| 1445 | Transposase, Synechocystis sp. strain PCC 6803 | 5e-60 | 40 (56) |
| 1444 | Putative hydrolase, Streptomyces coelicolor A3 | 1e-136 | 45 (60) |
| 1443 | Putative β-glucosidase, Bifidobacterium longum | 1e-160 | 83 (89) |
| 1442 | Transcriptional regulator, Kineococcus sp. | 3e-19 | 33 (55) |
| 1441 | Putative transferase, Bifidobacterium longum | 7e-10 | 66 (70) |
| 1440 | Glycosyl hydrolase, Caulobacter crescentus | 3e-75 | 36 (41) |
| 1439 | ABC transporter, Enterococcus faecalis | 1e-124 | 45 (50) |
| 1438 | ABC transporter, Enterococcus faecalis | 1e-75 | 52 (67) |
| 1437 | ABC transporter, Enterococcus faecalis | 2e-100 | 63 (75) |
| 1436 | Putative transposase, Bifidobacterium longum DJO10A | 1e-67 | 84 (88) |
| 1435 | Putative transposase, Corynebacterium efficiens | 2e-79 | 40 (52) |
| 1432 | Putative transposase and inactivated derivatives, IS30 Bifidobacterium longum DJO10A | 3e-30 | 75 (82) |
| 1431 | Site-specific recombinase (Tn5401), Bacillus thuringiensis | 1e-08 | 31 (47) |
| 1430 | Holin, Enterococcus faecalis | 4e-05 | 44 (60) |
| 1429 | Lysin, Lactococcus phage P335 | 2e-30 | 40 (51) |
ORFs are displayed in bold when they are part of the putative transposon element.
Analysis of the next 20,529-bp DNA segment shows intriguing links. Database matches indicated that this segment carries, in addition to mobile elements, sequences that appear to be of bacterial origin (Table 1), such as a complete ABC transporter system, a β-glucosidase, and glycosyl hydrolase encoding genes. ORF 1443 specifies a protein containing a glycosyl-hydrolase family 3N terminal domain (PFAM 00933), which is typically found in β-glucosidases involved in the hydrolysis of terminal β-glucose residues, such as in sucrose metabolism (11). The protein encoded by ORF 1443 is predicted to contain a signal peptide sequence typically found in extracellular enzymes. The 20.5-kb DNA segment is delineated by two direct repeats (CCCGAACCAAAC) and an integrase gene (ORF 1431) at one side, which may be responsible for the insertion of this presumptive composite mobile element (Fig. 1).
The presence of such a chimeric mobile element inserted into a prophage genomic sequence is not unusual. In Streptococcus a prophage genomic sequence has been described that carries a transposon element encompassing the mefA gene which confers a macrolide resistance phenotype to the bacterial host (3). Moreover, the staphylococcal prophage φPV83-pro possesses a transposon element which carries the exofoliative toxin-encoding gene (46). The G+C content of the transposon genes was nearly identical to that of the prophage genes and to the rest of the host-genome. Moreover the analysis of the dinucleotide frequencies of the transposon sequences did not reveal any differences with that found in the prophage sequences, thus indicating that the transposon sequences may have coevolved with the prophage sequences.
The transposon element in Bbr-1 prophage is followed by ORFs 1430 and 1429 whose predicted products exhibit significant similarity to a holin and a lysin, respectively, of Enterococcus faecalis V583, which suggests that they represent the lysis module of the Bbr-1 prophage.
Transcription analysis.
Total RNA was isolated from B. breve UCC 2003 cells and separated on a denaturing agarose gel, blotted and phage transcripts were revealed by Northern blot hybridization using specific DNA probes. The 3′ position of the mRNA was deduced from the estimated size of the mRNAs and from the presence of predicted rho independent terminator structures.
Previous transcription analysis of prophages from lactic acid bacteria revealed that only genes in the vicinity of both attachment sites were expressed (36, 38-40, 45). Therefore, we concentrated on the transcription analysis of the two ends of the Bbr-1 prophage genome. A small and prominent transcript of 370 bp corresponding to ORF 1452b, which resembled a transcriptional regulator, was detected (Fig. 3a). In contrast, no transcripts were detected with probes covering the prospective structural genes or the phage lysis module (Fig. 3a).
FIG. 3.
Transcription analysis of prophage-like element Bbr-1. Panel a: Northern blot hybridization of total RNA from B. breve UCC 2003 isolated at optical densities of 0.6, 0.8, and 1.1 (lanes 1 to 3, respectively). Blots were probed with radiolabeled PCR products indicated at the bottom of each blot and corresponding to those indicated as horizontal bars in panel b. The sizes calculated for the hybridization signals are provided in bp. Panels b to d: summary of the transcription data. b) Prophage genome map. c) Location of the PCR probes used in Northern experiments. d) Location of transcripts on the genome map. The arrows point the to 3′end of the mRNA. The length of the arrow is proportional to the length of the mRNA estimated from the Northern blots. The size of mRNAs is indicated above the arrows.
Furthermore, we analyzed the transcription of some genes belonging to the putative transposon by Northern blot hybridization (Fig. 3). A probe corresponding to ORF 1443 yielded a transcript of 2.1 kb. Its estimated length and the presence of a predicted rho independent terminator structure located immediately downstream of ORF1443 indicate that this gene is monocistronic. The expression of this gene, which encodes a putative β-glucosidase, may have a selective value for the host by providing specific sugar-degrading capabilities to the lysogenic cell. No transcripts were detected using probes encompassing ORFs 1444, 1441, or 1440 or ORF 1438 (Fig. 3).
Distribution of Bbr-1 prophage-like elements in bifidobacteria.
We selected six B. breve strains and three strains of a closely related species (B. longum) to determine the distribution of Bbr-1-like prophages in bifidobacterial strains. All these strains were assayed by molecular fingerprinting using PFGE. The nine strains represented nine different SpeI restriction patterns in PFGE, which were different from the pattern of the reference strain UCC 2003 (Fig. 4). Two Bbr-1-specific probes located in the structural part of the phage genome (ORFs 1430 and 1449), hybridized with a 188-kb fragment from UCC 2003 DNA (data not shown). A weak 51-kb hybridization signal was observed in B. breve JCM 7019 strain when a probe specific for ORF 1430 was used, which suggests the presence of a prophage-like sequence that shares at least some DNA homology with Bbr-1 (data not shown). This finding is in agreement with the failure to generate a PCR product using B. breve JCM 7019 chromosomal DNA as a template and PCR primers flanking the putative attB site of this strain, which may indicate that a prophage is integrated in this site (Fig. 2c). The other B. breve strains tested were shown to contain an intact attB site (Fig. 2c). Moreover, the PFGE profiles of these strains tested did not reveal any hybridization signal, suggesting that these strains do not contain Bbr-1 prophage sequences.
FIG. 4.
Distribution of Bbr-1 prophage-like element in B. breve and B. longum strains. Pulsed-field gel of SpeI digested DNA from the indicated B. breve and B. longum strains stained with ethidium bromide (negative of the photo). The size of the DNA markers is given. The position of the restriction fragment containing the prophage-like element Bbr-1 is indicated with a black arrow, whereas the cross-hybridizing band is indicated with a white arrow.
Bbr-1 prophage-like element induction.
In order to determine whether the Bbr-1 prophage-like element is a cryptic or an active phage element its inducibility was tested by exposing the B. breve UCC 2003 culture to mitomycin C. In the case of an excised and circularized phage genome, a PCR product of 600 bp should be obtained with primers placed in the left- and right-most parts of the prophage genome and running outwards from the prophage. Furthermore, due to the expected variability in PCR conditions when using complete bacterial cells, we wanted to ensure that the lack of any PCR product was attributable to the absence of circularized phage DNA target rather than a failure of the amplification reaction.
To distinguish between these two possibilities, the use of a second pair of primers (1451-1, and 1449-2) targeting a 2,000-bp region within the prophage genome was used as positive control. As assessed by PCR, no spontaneous induction of prophage-like element Bbr-1 was detected during growth of strain UCC 2003 (Fig. 5b, lanes 5 and 10). Moreover, DNA isolated from cells upon addition of different concentrations of mitomycin C yielded amplicons only with primers placed within prophage sequences (positive control), whereas no specific amplicons were achieved using primers running out of the prophage genome, indicating that no free circularized phage particles are present (Fig. 5b, lanes 6 to 9). Furthermore, no PCR products suggestive of prophage excision were obtained with primers running out of the prophage genome in the culture supernatants after filtration (Fig. 5b, lanes 1 to 4).
FIG. 5.
Alignment of region surrounding the attB sites from different bifidobacterial strains and Thermosynechococcus elogantus (panel a). Bb, B. breve; Bl, B. longum; Bal, B. animalis subsp. lactis; Baa, B. animalis subsp. animalis; Bs, B. suis; Bi, B. infantis; Ba, B. adolescentis and Ts, Thermosynechococcus elogantus. The putative attB sites for Bbr-1 (attBBbr-1) and Bl-1 (attBBl-1) prophage-like elements are underlined. Panel b: mitomycin C induction experiments. Amplification of prophage sequences from UCC 2003 culture filtrate supernatants (lanes 1 to 5) and from UCC 2003 cells (lanes 6 to 9) after mitomycin C treatment. Amplification of prophage sequences from DJO10A culture filtrate supernatants (lanes 11 to 13) and from DJO10A cells (lanes 14 to 15) after mitomycin C or hydrogen peroxide treatment. In lanes 1 to 10, primers targeting the ORFs 1451 to 1449 (positive control) and the phage DNA with primers running out of the int gene and the rightmost part of Bbr-1 prophage-like element (1 kb expected, nothing observed). In lanes 11 to 17, primers targeting ORF 26 (positive control) and the phage DNA with primers running out of the int gene and the rightmost part of Blj-1 prophage (Blj reverse int and Blj forward cI). Lane MK, DNA molecular markers (Gibco, Brl). Lanes 1 and 6, after addition of 1 μg/ml of mitomycin C; lanes 2 and 7, after addition of 2 μg/ml of mitomycin C; lanes 3, 8, and 11, after addition of 3 μg/ml of mitomycin C; lanes 4, 9, 12, and 14 after addition of 5 μg/ml of mitomycin C; lanes 13 and 15 after addition of 2 mM hydrogen peroxide; lanes 5, 10, and 16, without any addition of mitomycin C; lane 17, negative control. Panel c: aligned nucleotide sequences of the regions containing attL, attR, attP, and attB of Blj-1 phage. The bacterial sequences outside of the Blj-1 attB site are boxed. The conserved region representing the core sequences of att sites are highlighted. The putative attB site for Bbr-1 (attBBbr-1) and Bl-1 (attBBl-1) prophage-like elements is underlined.
Mitomycin C induction experiments did not result in the lysis of the lysogenic cell nor was any effect on cell growth observed. Attempts to detect viable bacteriophages using plaque and spot assays did not reveal the presence of phage-like particles in the culture supernatants upon mitomycin C induction. The absence of either spontaneous or mitomycin C-mediated prophage induction in UCC 2003 strain is not an unusual finding. Previous studies using Lactobacillus johnsonii NCC 533 (39) and Lactobacillus plantarum WCFS1 (40) have shown that none of their prophages could be induced. Furthermore, only one of the many lambda-like prophages in Escherichia coli strain O157:H7 (28) and only one out of the four prophages reported in Streptococcus pyogenes (13) could be induced. However, we cannot rule out that in their natural environment Bbr-1 prophage may be activated under physiological conditions other than mitomycin induction.
Integration site and bioinformatic analysis of prophage-like element Bl-1.
Database matches allowed a tentative subdivision of the Bl-1 prophage genome into modules. The modular organization was atypical for temperate phages of gram-positive bacteria (9): the integrase gene and the repressor gene were located at the boundaries of the Bl-1 genome (Fig. 1a). A similar genomic architecture was also found in the B. longum Blj-1 prophage-like element which may suggest a new lineage of temperate phage.
Prophage-like element Bl-1 is located between the metA gene on one side and a gene encoding a hypothetical protein on the other side, while being flanked by a 35-bp repeat (Fig. 2b). Using primers targeting the sequences of the bacterial genes flanking the prophage, we amplified a 750-bp DNA segment from many B. longum strains (Fig. 2d). This amplicon contained the 35-bp sequence that overlaps the 3′ end of a tRNAMet. The sequences to the right and left of this deduced attB were very similar to those abutting the likely attL and attR sites. Interestingly, the predicted integration site of Bl-1 prophage-like element is highly similar to that of Bbr-1 prophage, reflecting a common target and similar mechanism of integration and excision. This notion is confirmed by the fact that the integrases of Bl-1, Blj-1, and Bbr-1 prophage-like elements share 53% amino acid identity (75% at the C terminus). The insertion of Bl-1 and Bbr-1 prophage-like elements in the 3′end of tRNAMet gene should be of little impact on bacterial survival because the acceptor stem of tRNA is reconstituted after phage integration.
The Bl-1 integrase gene is preceded by a predicted holin and a lysin gene with sequence similarities to the Bacillus halodurans phage C-125 (Table 2) and to the lactococcal bacteriophage BK5-T. Of note, the deduced lysin sequence contains a typical amidase motif (PFAM01510).
TABLE 2.
Database matches for Bl-1
| ORF | Representative similarity to protein in database | e value | % Identity (% similarity) |
|---|---|---|---|
| 386 | Tyrosine recombinase xerD, Sinorhizobium meliloti | 2e-17 | 30 (48) |
| 385 | Holin protein, Bacillus halodurans C-125 | 1e-09 | 45 (68) |
| 384 | Putative amidase, Streptomyces coelicolor A3 | 5e-07 | 32 (46) |
| 381 | Putative minor tail protein, Streptococcus pyogenes | 1e-13 | 25 (42) |
| 380 | gp40 bacteriophage φBT1 | 2e-04 | 32 (53) |
| 376 | Prohead protease, prophage lambda Ba04 | 1e-06 | 27 (45) |
| 375 | Portal protein, Salmonella enterica serovar Typhimurium phage ST6 | 4e-19 | 27 (45) |
| 374 | Putative phage large terminase subunit, Corynebacterium sp. | 6e-19 | 31 (46) |
| 372 | Small terminase subunit, Lactobacillus casei bacteriophage | 1e-3 | 27 (44) |
| 376 | Putative repressor bacteriophage A118, Listeria innocua | 3e-04 | 41 (64) |
Further upstream, putative tail morphogenesis genes (encoding a minor tail component, and a prohead protease) and genes putatively involved in DNA packaging (encoding small and large terminase, as well as a portal protein) were identified (Fig. 1a). Their closest relatives are from phages infecting low-G+C bacteria like Streptococcus pyogenes, Lactobacillus bacteriophage LL-H, Lactobacillus phage A2, prophage lambda Ba4, and phages infecting high-G+C bacteria such as Mycobacterium and Corynebacterium (Table 2).
Genome analysis of the Blj-1 prophage-like element.
The predicted prophage-like element Blj-1 has a genome size of 36,690 bp, extending from ORF 1 (integrase) to ORF 52 (cI repressor). This diagnosis was based on the observation that ORF 1 and ORF 52 are flanked downstream by a 46-bp repeat which is highly similar to the integration sites of Bl-1 prophage-like elements. Using primers targeting the sequences of the bacterial genes flanking the prophage, we obtained a 750-bp PCR product apparently from a nonlysogenic B. longum DJ1O10A cell (data not shown), which contained the 46-bp attB sequence that, similar to the 35-bp repeat in B. longum NCC 2705 strain, overlaps the 3′ end of a tRNAMet. The extent of nucleotide sequence similarity shared among all three bifidoprophage varies considerably (Fig. 1). Interestingly, regions of homology between both B. longum prophage-like elements are very short. In contrast, Blj-1 exhibits greater homology to the B. breve Bbr-1 prophage-like element.
Database matches allowed a functional annotation of some of the identified ORFs of the Blj-1 prophage genome (Table 3). The Blj-1 integrase gene is preceded by a predicted holin and a lysin gene with sequence similarities to the Enterococcus faecalis V583 prophage and to the lactococcal bacteriophage TP901-1. The putative lysis module is followed by three ORFs encoding hypothetical proteins and by ORF 7, which shares 25% amino acid identity with a reverse transcriptase of Bordetella bacteriophage BPP-1 (22). Of note, Liu et al. (22) recently described a group of Bordetella species-infecting phages that undergo a unique template-dependent, reverse transcriptase-mediated tropism-switching phenomenon. Central to this process is a reverse transcriptase-mediated exchange between two repeats, one serving as a donor template (TR) located in the intergenic region preceding the reverse transcriptase-encoding gene and the other as a recipient of variable sequence information (VR) located in the putative tail fiber gene (mtd). Tropism switching results from the introduction of adenine substitutions at defined positions in the VR segment (22).
TABLE 3.
Database matches for Blj-1
| ORF | Similarity to representative phage protein in database | e value | % Identity (% similarity) |
|---|---|---|---|
| 1 | Tyrosine recombinase xerD, Sinorhizobium meliloti | 1e-13 | 29 (42) |
| 3 | Holin, Enterococcus faecalis V583 | 5e-08 | 50 (63) |
| 4 | Lysin, Lactococcus phage P335 | 2e-34 | 43 (59) |
| 7 | Reverse transcriptase, Bordetella phage BPP-1 | 4e-10 | 25 (43) |
| 9 | Putative neck passage, Lactococcus phage LC3 | 1e-3 | 39 (63) |
| 12 | gp47, Lactococcus bacteriophage TP901-1 | 5e-11 | 25 (42) |
| 14 | Tail protein, Enterococcus faecalis V583 | 6e-52 | 30 (52) |
| 17 | Major tail protein, prophage Streptococcus lambda Sa1 | 1e-2 | 39 (63) |
| 18 | Putative structural protein phage associated, Streptococcus pyogenes | 1e-1 | 31 (40) |
| 21 | Hypothetical protein rtp34, Lactococcus phage rlt | 2e-34 | 28 (48) |
| 22 | Hypothetical protein, Lactococcus phage φLC3 | 6e-09 | 33 (46) |
| 24 | gp7, mycobacteriophage Che9d | 4e-35 | 34 (48) |
| 26a | gp5, mycobacteriophage Che9d | 0.83 | 28 (40) |
| 26 | gp4, mycobacteriophage Che9d | 1e-34 | 30 (47) |
| 27 | Putative terminase, mycobacteriophage TM4 | 5e-69 | 37 (53) |
| 29 | gp3, Lactococcus bacteriophage φ31 | 2e-3 | 49 (56) |
| 33 | WhiB1, putative transcriptional regulator, Mycobacterium tuberculosis | 6e-07 | 47 (56) |
| 38 | gp52 protein, Yersinia pestis phage φA 1122 | 1e-08 | 47 (58) |
| 39 | Putative oligoribonuclease, Diachasmimorpha entomopoxvirus | 4e-06 | 28 (46) |
| 40 | gp70, mycobacteriophage Che8 | 4e-07 | 33 (47) |
| 44 | Single-stranded DNA binding protein, Mycobacterium sp. | 7e-36 | 61 (83) |
| 49 | Membrane protease subunits, Nostoc punctiforme | 6e-08 | 26 (47) |
| 52 | Putative repressor protein phage associated, Streptococcus pyogenes M1 GAS | 1e-02 | 33 (54) |
Blj-1 sequence analysis revealed two direct repeats resembling the Bordetella TR and VR segments at the 3′ end of ORF 8 and in the intergenic region between ORF 7 and ORF 8, respectively (Fig. 1b). The VR cassette carries 13 adenine substitutions and the canonical TTCT motif at its 3′ end (22) for replacement of VR by mutated TR segment (Fig. 1b). Consequently, in accordance with what was previously described for Bordetella bacteriophage BBP-1 (22), it is plausible that ORF 7 and ORF 8 of the Blj-1 prophage-like element may represent a diversity-generating retroelement in B. longum DJO10A.
Further database matches allowed the identification of a likely tail morphogenesis and tail fiber genes on the basis of observed homology to such genes in low-G+C lactococcal and streptococcal phages. In contrast, the putative DNA packaging and head morphogenesis genes (from ORF 24 to ORF 27) are closely related to the homologous genes of mycobacteriophage (Table 3 and Fig. 1). A tRNA gene carrying an anticodon (GCT) specific for a Ser is located downstream of the putative DNA packaging modules, in a gene constellation reflective of many low-G+C prophages (40, 41). Interestingly, this tRNA specifies a codon that is prominent in the Blj-1 genome, and the second most prominent in the DJO10A genome. Furthermore, as described by Tomita et al. (35), the tRNASer GCT possesses a 7-methylguanosine which should be able to translate all five codons of serine and thus the tRNASer carried by Blj-1 prophage would be able to increase the translation capacity of DJO10A.
The remainder of the Blj-1 genome, with a few exceptions, contains ORFs encoding hypothetical proteins with no significant homology to known protein sequences. One exception is represented by ORF 33 which encodes a putative regulator protein similar to a protein in Mycobacterium spp. as well as in the mycobacteriophage Che9d. Other genes with database matches are ORF 39, whose deduced product resembles an oligoribonuclease involved in mRNA decay (29), and ORF 44, the protein product of which shares 61% amino acid identity with a single-stranded DNA binding protein from Mycobacterium as well as one from B. longum NCC 2705. The rightmost part of the Blj-1 genome is occupied by two adjacent but divergently oriented genes (ORFs 51 and 52) that may represent the likely genetic switch structure, with the protein encoded by ORF 52 showing database matches to a repressor protein from Enterococcus. Moreover, the ORF 52 product contains a helix turn helix motif which is commonly found in repressor proteins.
Blj-1 prophage induction.
The inducibility of Blj-1 prophage was assessed by exposing the B. longum DJO10A to 1 to 5 μg of mitomycin C or to 2 mM of hydrogen peroxide (Fig. 5b and data not shown). In order to identify an excised and circularized Blj-1 phage genome, an identical PCR strategy used for testing the Bbr-1 inducibility, was employed. A PCR primer pair (Blj reverse int and Blj forward cI) were designed at each border of the Blj-1 prophage forwarding the prophage sequences. Moreover, another PCR primer pair (26-1 and 26-2) which target ORF 26 was used as a positive PCR control. Strikingly, amplicons of 777 bp were achieved with primers placed at the border of the Blj-1 prophage, which indicates that a circularized Blj-1 bacteriophage was obtained after either mitomycin C or hydrogen peroxide treatment (Fig. 5b, lanes 13 and 15). Whereas no specific 777-bp PCR product was achieved when DNA extracted from an uninduced DJO10A culture was used (Fig. 5b, lane 16). A 610-bp amplicon generated by using the 26-1/26-2 primer pair, which corresponds to the positive PCR control, was obtained for all samples (Fig. 5b, lanes 11 to 16).
When the 777-bp amplicon, which should correspond to the Blj-1 attP site, was sequenced and aligned with the Blj-1 attL, attR, and attB sites, a common 46-bp core region was found (Fig. 5c). In this common core, DNA strand exchange is expected to occur during phage genome integration into the bacterial chromosome.
Attempts to detect viable Blj-1 bacteriophages using plaque assays did not reveal the presence of clear plaques, which may be due to the difficulty to find a suitable indicator B. longum strain or that despite induction no intact phage particles are produced. Ten different B. longum strains, which include the JCM 7055, JCM 7052, JCM 7053, JCM 7050, JCM 7056, CCUG 15137, CCUG 30698, CCUG 15137, CIP 64.63, and LMG 11589, were used in these experiments.
Analysis of the conservation of Bbr-1 and Bl-1 attB sites among bifidobacteria.
Three primer pairs (B1-B2, L1-L2, and Bal1-Bal2) allowed the PCR amplification of fragments containing attB from Bifidobacterium infantis, Bifidobacterium suis, and Bifidobacterium adolescentis and from two phylogenetically distant taxa, B. animalis subsp. lactis and B. animalis subsp. animalis. An alignment of these sequences indicated positions in the minimal site that can tolerate base changes (Fig. 5a). With the exception of B. animalis subsp. lactis and B. animalis subsp. animalis, the attB sequences are located at the 3′ end of a tRNAMet. Interestingly, we have found that in B. suis and B. animalis subsp. lactis a gene encoding an integrase gene flanks the attachment sites (attL) (Fig. 5a and data not shown).
We speculate that the integrase gene is a remnant of an ancient prophage element which had been targeted by a deletion process. A similar situation has been reported in a previous study (36), in which a deletion process starting from the lysis/structural gene region was shown to eliminate large parts of streptococcal prophage sequences, whereas the integrase gene was conserved. Of note, the use of a tRNAMet as attB site for bifidoprophage like elements represents the first case of such tRNA anticodon as a target for phage integration in gram-positive bacteria (6-8).
Database searches for attachment site sequences revealed that these are highly conserved in a phylogenetic unrelated bacterium, Thermosynechococcus elogantus (26), but surprisingly not in other sequenced members of the Actinobacteridae group. Similar to bifidobacteria, in T. elogantus an integrase gene belonging to a likely transposon element is located in the vicinity of the potential integration site sequences.
The highly conserved attB sequences in bifidobacteria taken together with the presence of mobile elements (prophages and transposon elements) in the flanking regions may indicate that these sequences represent a common target site for the acquisition of mobile foreign DNA. Thus, we propose that a selection pressure may have conserved specific integration sites, thereby promoting genome variability and genome shaping in bifidobacterial taxa.
Phylogenetic analysis of the Bbr, Bl-1, and Blj-1 prophage-like elements.
Recently, a sequence-based taxonomic system has been established for inferring phylogeny among phages and prophages (31) through the generation of a proteomic tree. This system is based on the overall relatedness of both complete phage genomes and prophages identified within complete bacterial genomes (10) without considering morphology or genome size (31). The database consists of 16,260 proteins from 375 genomes.
We performed such a proteomic tree analysis (Fig. 6) using this database which was updated with the Bbr-1, Bl-1, and Blj-1 sequences. A striking finding was that the Bbr-1, Bl-1, and Blj-1 sequences were not grouped together in this system, and that they did not appear to be related to other phages of high-G+C bacteria (e.g., mycobacteriophages). However, they did exhibit a close phylogenetic relationship with phage infecting low-G+C bacteria (e.g., lactococcal and staphylococcal phages) (Fig. 6). It is tempting to speculate that these bacteria have shared the same ecological niche (e.g., the animal gastrointestinal tract) during their evolution and consequently exchanged sequences within these groups. This may therefore represent evidence pointing to an ancient exchange of DNA sequences between low- and high-G+C bacteria.
FIG. 6.
Phage proteomic tree illustrating the relationship between Bbr-1, Bl-1, and Blj-1 prophage-like elements and other sequenced phages and prophages. The tree is based on the more closely related phage and prophage sequences of Bbr-1, Bl-1, and Blj-1 deduced on the basis of a previous proteomic tree based on 375 sequenced phage genomes and prophages (data not shown). The high- and low-G+C phages are indicated for correlation to reference 31.
Possible exchange of phage sequences between two gastrointestinal tract commensals (Lactobacillus plantarum and Streptococcus pyogenes) has already been described (40). The clustering of these bifidoprophagelike elements with low-G+C phages may also be explained by the latter's originally infecting the ancestor of high-G+C gram-positive bacteria, which is in agreement with the concept based on the evolutionary origin of high-G+C gram-positive bacteria from low-G+C ancestors (15). The reduced size of the Bbr-1 and Bl-1 prophage-like elements may be another sign of the ancient origin of these phage sequences, since on an evolutionary time scale, a series of events leading from the accumulation of mutations to massive loss of prophage DNA and ultimate disappearance of the prophage is well documented (9, 10).
Conclusions.
This paper describes for the first time the occurrence and comparative analysis of prophage-like sequences in a number of bifidobacteria. The genomes of B. breve UCC 2003, B. longum NCC 2705, and B. longum DJO10A all contain a prophage-like element. Their similarities at both the DNA and protein levels imply homology among genes of double-stranded DNA phages spanning a broad phylogenetic range of host bacteria. The sequence matches reported here establish genetic connections not only with the lambdoid phages of high-G+C bacteria such as Mycobacterium, Streptomyces, and Corynebacterium species, but also with phages of low-G+C bacteria (e.g., Lactococcus). The homologies found here support previous results (16, 30) indicating the possibility of common ancestry among double-stranded DNA phages.
We believe that this may argue for horizontal exchange of sequences among the ancestors of the contemporary phages as was previously proposed (16, 30). This exchange presumably happens most often when two phage genomes find themselves in the same host, either as two coinfecting phages or, perhaps of more importance, as a single phage infecting a cell that carries one or more prophages. While horizontal exchange is recently recognized to play a crucial role in the evolution of bacterial genomes (27), the extensive mosaicism of phage genomes illustrates the powerful creativity of this process.
The bifidobacterial prophage-like elements studied here showed a significant diversity, but the extent of bifidophage diversity remains unclear and many more genomes will have to be sequenced before such questions can be addressed.
Although the Bbr-1 and Bl-1 prophage-like elements appear to represent two deficient bacteriophages, they could constitute functional satellite phages which can become mobile in a manner similar to that described for the cryptic mycophages Rv1 and Rv2 (16). In contrast, the Blj-1 prophage was shown to be inducible following mitomycin or hydrogen peroxide treatment, and future work will be carried out to isolate Blj-1 phage particles.
It is of interest that each of the three prophage-like elements was integrated into a tRNAMet, which has not previously been identified as an attB site in gram-positive bacteria (6-8).
Analysis of the distribution of attB in many bifidobacterial species highlighted positions where sequence variation may be tolerated in sites that are active for recombination. The use of these conserved sequences and the int gene sequences of bifidophages might allow the construction of a very efficient recombination module, analogous to the Streptomyces integrating plasmid pSE211 (5). Such a recombination module may represent the ideal source for the construction of vector integration systems that enable the food-grade introduction of foreign DNA sequences in single copy at a specific site within the host chromosome without disturbing any host function, similar to systems developed for lactic acid bacteria such as Lactobacillus (24) and for high-G+C bacteria such as Streptomyces and Mycobacterium (12, 19).
With the prospect of identifying other bifidobacterial prophages, it will be possible to address questions concerning horizontal versus vertical DNA transfer within different species of Bifidobacterium.
Acknowledgments
This work was financially supported by the Department of Agriculture and Food FIRM Program under National Development Plan 2000-2006, by the Higher Education Authority Programme for Research in Third Level Institutions (PRTLI3), by the Science Foundation Ireland Alimentary Pharmabiotic Centre located at University College Cork, by a Marie Curie Development Host Fellowship (HPMD-2000-00027) to M.V., and by Dairy Management Inc., USA.
We thank Stephen McGrath for helpful and constructive discussions.
REFERENCES
- 1.Ackermann, H. W. 2001. Frequency of morphological phage descriptions in the year 2000. Brief review. Arch. Virol. 146:843-857. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. Schaffer, A. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banks, D. J., S. F. Porcella, K. D. Barbian, J. M. Martin, and J. M. Musser. 2003. Structure and distribution of an unusual chimeric genetic element encoding macrolide resistance in phylogenetically diverse clones of group A Streptococcus. J. Infect. Dis. 188:1898-1908. [DOI] [PubMed] [Google Scholar]
- 4.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:138-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, D. P., K. B. Idler, and L. Katz. 1990. Characterization of the genetic elements required for site-specific integration of plasmid pSE211 in Saccharopolyspora erythraea. J. Bacteriol. 172:1877-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brussow, H. 2001. Phages of dairy bacteria. Annu. Rev. Microbiol. 55:283-303. [DOI] [PubMed] [Google Scholar]
- 7.Campbell, A. M. 1992. Chromosomal insertion sites for phages and plasmids. J. Bacteriol. 174:7495-7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, A. M. 2002. Preferential orientation of natural lambdoid prophages and bacterial chromosome organization. Theor. Popul. Biol. 61:503-507. [DOI] [PubMed] [Google Scholar]
- 9.Canchaya, C., C. Proux, G. Fournous, A. Bruttin, and H. Brüssow. 2003. Prophage genomics. Microbiol. Mol. Biol. Rev. 67:238-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casjens, S. 2003. Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol. 49:277-300. [DOI] [PubMed] [Google Scholar]
- 11.Chinchetru, M. A., J. A. Cabezas, and P. Calvo. 1989. Purification and characterization of a broad specificity β-glucosidase from sheep liver. Int. J. Biochem. 21:469-476. [DOI] [PubMed] [Google Scholar]
- 12.Combes, P., R. Till, S. Bee, and M. C. M. Smith. 2002. The Streptomyces genome contains multiple pseudo-attB sites for the phage C31-encoded site-specific recombination system. J. Bacteriol. 184:5746-5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foley, S., S. Lucchini, M. C. Zwahlen, and H. Brussow. 1998. A short noncoding viral DNA element showing characteristics of a replication origin confers bacteriophage resistance to Streptococcus thermophilus. Virology 250:377-387. [DOI] [PubMed] [Google Scholar]
- 15.Gupta, R. S. 2001. The branching order and phylogenetic placement of species from completed bacterial genomes, based on conserved indels found in various proteins. Int. Microbiol. 4:187-202. [DOI] [PubMed] [Google Scholar]
- 16.Hendrix, R. W., M. C. M. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huber, K. E., and M. K. Waldor. 2002. Filamentous phage integration requires the host recombinases XerC and XerD. Nature 417:656-659. [DOI] [PubMed] [Google Scholar]
- 18.Lawrence, J. G. 2002. Gene transfer in bacteria: speciation without species? Theor. Popul. Biol. 61:449-460. [DOI] [PubMed] [Google Scholar]
- 19.Lee, M. H., L. Pascopella, W. R. Jacobs, and G. F. Hatfull. 1991. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis and bacilli Calmette-Guerin. Proc. Natl. Acad. Sci. USA 88:3111-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindqvist, B. H., G. Deho, and R. Calendar. 1993. Mechanisms of genome propagation and helper exploitation by satellite phage P4. Microbiol. Rev. 57:683-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lipman, D. J., and W. R. Pearson. 1985. Rapid and sensitive protein similarity searches. Science 227:1435-1441. [DOI] [PubMed] [Google Scholar]
- 22.Liu, M., R. Deora, S. R. Doulatov, M. Gingery, F. A. Eiserling, A. Preston, D. J. Maskell, R. W. Simons, P. A. Cotter, J. Parkhill, and J. F. Miller. 2002. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science 295:2091-2094. [DOI] [PubMed] [Google Scholar]
- 23.Lowe, T. M., S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin, M. C., J. C. Alonso, J. E. Suarez, and M. A. Alvarez. 2000. Generation of food-grade recombinant lactic acid bacterium strains by site-specific recombination. Appl. Environ. Microbiol. 66:2599-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moreau, S., V. Leret, C. Le Marrec, H. Varangot, M. Ayache, S. Bonnassie, C. Blanco, and A. Trautwetter. Prophages distribution in coryneform bacteria. Res. Microbiol. 146:493-505. [DOI] [PubMed]
- 26.Nakamura, Y., T. Kaneko, S. Sato, M. Ikeuchi, H. Katoh, S. Sasamoto, A. Watanabe, M. Iriguchi, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. Complete genome structure of the thermophilic cyanobacterium Thermosynechoccus elongatus BP-1. DNA Res. 31:123-130.
- 27.Ochman, H., J. G. Lawerence, and E. A. Groismann. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 28.Ohnishi, M., K. Kurokowa, and T. Hayashi. 2002. Diversification of Escherichia coli genomes: are bacteriophages the major contributors? Trends Microbiol. 9:481-485. [DOI] [PubMed] [Google Scholar]
- 29.Ohnishi, Y., Y. Nishiyama, R. Sato, S. Kameyama, and S. Horinouchi. An oligoribonuclease gene in Streptomyces griseus. J. Bacteriol. 182:4647-4653. [DOI] [PMC free article] [PubMed]
- 30.Pedulla, M. L., M. E. Ford, J. M. Houtz, T. Karthikeyan, C. Wadsworth, J. A. Lewis, D. Jacobs-Sera, J. Falbo, J. Gross, N. R. Pannunzio, W. Brucker, V. Kumar, J. Kandasamy, L. Keenan, S. Bardarov, J. Kriakov, J. G. Lawrence, W. R. Jacobs, Jr., R. W. Hendrix, and G. Hatfull. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171-182. [DOI] [PubMed] [Google Scholar]
- 31.Rohwer, F., and R. Edwards. 2002. The phage proteomic tree: a genome based taxonomy for phage. J. Bacteriol. 184:4529-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 33.Schell, M. A., M. Karmirantzou, B. Snel, B. Berger, D. Vilanova, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, D. R. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sgorbati, B., M. B. Smiley, and T. Sozzi. 1983. Plasmids and phages in Bifidobacterium longum. Microbiologica. 6:169-173. [PubMed] [Google Scholar]
- 35.Tomita, K., T. Ueda, and K. Watanabe. 1998. 7-methylguanosine at the anticodon wobble position of squid mitochondrial tRNASer GCU: molecular basis for assignment of AGA/AGG codons as serine in invertebrate mitochondria. Biochim. Biophys. Acta 1399:78-82. [DOI] [PubMed] [Google Scholar]
- 36.Ventura, M., A. Bruttin, C. Canchaya, and H. Brüssow. 2002. Transcription analysis of Streptococcus thermophilus phages in the lysogenic state. Virology 302:21-32. [DOI] [PubMed] [Google Scholar]
- 37.Ventura, M., and R. Zink. 2002. Specific identification and molecular typing analysis of Lactobacillus johnsonii by using PCR-based methods and pulsed-field gel electrophoresis. FEMS Microbiol. Lett. 217:141-154. [DOI] [PubMed] [Google Scholar]
- 38.Ventura, M., C. Canchaya, D. Pridmore, and H. Brussow. 2003. Integration and distribution of Lactobacillus johnsonii prophages. J. Bacteriol. 185:4603-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ventura, M., C. Canchaya, D. Pridmore, and H. Brussow. 2004. The prophages of Lactobacillus johnsonii NCC 533: comparative genomics and transcription analysis. Virology. 320:229-242. [DOI] [PubMed] [Google Scholar]
- 40.Ventura, M., C. Canchaya, M. Kleerebezem, W. M. de Vos, R. J. Siezen, and H. Brussow. 2004. The prophage sequences of Lactobacillus plantarum strain WCFS1. Virology. 316:245-255. [DOI] [PubMed] [Google Scholar]
- 41.Ventura, M., C. Canchaya, R. Zink, G. F. Fitzgerald, and D. van Sinderen. 2004. Characterization of the groEL and groES loci in Bifidobacterium breve UCC 2003: genetic, transcriptional and phylogenetic analysis. Appl. Environ. Microbiol. 70:6197-6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ventura, M., D. van Sinderen, G. F. Fitzgerald, R. Zink. 2004. (review) Insights into the taxonomy, genetics and physiology of bifidobacteria. Antonie van Leeuwenhoek 86:205-223. [DOI] [PubMed] [Google Scholar]
- 43.Ventura, M., M. Elli, R. Reniero, R. Zink. 2001. Molecular microbial analysis of Bifidobacterium isolates from different environments by the species-specific amplified ribosomal DNA restriction analysis (ARDRA). FEMS Microb. Ecol. 36:113-121. [DOI] [PubMed] [Google Scholar]
- 44.Ventura, M., R. Reniero, and R. Zink. 2001. Specific identification and targeted characterization of Bifidobacterium lactis from different environmental isolates by a combined multiplex-PCR approach. Appl. Environ. Microbiol. 67:2760-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ventura, M., S. Foley, A. Bruttin, S. Chibani Chennoufi, C. Canchaya, and H. Brüssow. 2002. Transcription mapping as a tool in phage genomics: the case of the temperate Streptococcus thermophilus phage Sfi21. Virology 296:62-76. [DOI] [PubMed] [Google Scholar]
- 46.Zou, D., J. Kanedo, S. Narita, and Y. Kamio. 2000. Prophage ΦPV83-pro carrying Panteon-Valentine leukocidin genes, on the Staphylococcus aureus P83 chromosome: comparative analysis of the genome structures of ΦPV83-pro, ΦPVL, Φ11, and other phages. Biosci. Biotechnol. Biochem. 64:2631-2643. [DOI] [PubMed] [Google Scholar]