Abstract
Poplar, a plant species frequently used for phytoremediation of groundwater contaminated with organic solvents, was inoculated with the endophyte Burkholderia cepacia VM1468. This strain, whose natural host is yellow lupine, contains the pTOM-Bu61 plasmid coding for constitutively expressed toluene degradation. Noninoculated plants or plants inoculated with the soil bacterium B. cepacia Bu61(pTOM-Bu61) were used as controls. Inoculation of poplar had a positive effect on plant growth in the presence of toluene and reduced the amount of toluene released via evapotranspiration. These effects were more dramatic for VM1468, the endophytic strain, than for Bu61. Remarkably, none of the strains became established at detectable levels in the endophytic community, but there was horizontal gene transfer of pTOM-Bu61 to different members of the endogenous endophytic community, both in the presence and in the absence of toluene. This work is the first report of in planta horizontal gene transfer among plant-associated endophytic bacteria and demonstrates that such transfer could be used to change natural endophytic microbial communities in order to improve the remediation of environmental insults.
Phytoremediation of highly water-soluble and volatile organic xenobiotic compounds, such as benzene, toluene, ethylbenzene, and xylene, is often limited due to insufficient degradation of the pollutants by plants and their rhizospheres. This can result in phytotoxicity of these compounds and their metabolites or in volatilization of the compounds through the leaves, potentially resulting in new environmental problems (21). We recently demonstrated that phytoremediation of volatile and water-soluble organic xenobiotic compounds can be improved by using recombinant endophytic bacteria modified to contain the appropriate degradation pathway (3). Endophytic bacteria equipped with a toluene degradation pathway were able to reduce toluene phytotoxicity and evapotranspiration from their yellow lupine host plants.
Endophytic bacteria colonize internal plant tissues of healthy plants without causing symptoms of disease. These bacteria have been found in numerous plant species, and most of them are members of common soil bacterial genera, such as Pseudomonas, Burkholderia, Bacillus, and Azospirillum (10). It seems reasonable to hypothesize that endophytic bacteria possessing the genetic information required for efficient degradation of a pollutant can promote degradation as the pollutant moves through the plant vascular system. Especially in trees, such as poplar, which is used to develop phytoremediation strategies for organic contaminants, the time between uptake of molecules by the roots and arrival of the molecules in the leaves is several hours to days (11) as the compounds travel through the vascular system. The fast growth of poplar trees and their large transpiration potential make them “trees of choice” for phytoremediation purposes (16).
Although application of engineered endophytic bacteria to improve phytoremediation of volatile organic contaminants has several obvious advantages over application of engineered rhizosphere bacteria or genetic engineering of the plant's metabolism, several obstacles have to be overcome before this technology can move toward application (14). The major concerns include the persistence and stability of the engineered organisms and their degradation capabilities in field-grown plants, as phytoremediation projects can conceivably last decades. As long as a selection pressure is present, there is an advantage for the endophytic community members that possess the appropriate degradation characteristics. However, this is no guarantee that strains of an inoculum will become an integrated part of the endogenous endophytic community. Horizontal gene transfer has been shown to play an important role in allowing a microbial community to rapidly adapt to a new environmental stress (4), and we have speculated that it could play an important role in adapting the endogenous endophytic community (20); rather than integrating a new bacterium into a stable community, the degradation pathway is transferred among the members of the community.
To test this hypothesis, we inoculated cuttings of Populus trichocarpa × deltoides cv. Hoogvorst with the endophytic strain Burkholderia cepacia VM1468, whose natural host is yellow lupine, and with B. cepacia BU61, a soil isolate. Both strains possess the pTOM-Bu61 plasmid that constitutively expresses toluene degradation. The constitutive expression of the tom operon avoids the need for an in planta toluene concentration above the induction threshold and, if required, also opens the possibility for trichloroethylene degradation without toluene cometabolization (18). Analysis of the endophytic community showed that neither VM1468 nor BU61 became successfully established at detectable levels in the community. However, endogenous endophytic bacteria successfully acquired the tom operon, which is required for toluene metabolism, via horizontal gene transfer.
MATERIALS AND METHODS
Strain construction.
The constitutively expressed toluene degradation pathway of B. cepacia BU61 (17), a derivative of B. cepacia G4, was introduced by conjugation into B. cepacia BU0072 (19), a nickel- and kanamycin-resistant derivative of B. cepacia L.S.2.4. Conjugation and selection of the transconjugants were performed as described previously (3). Transconjugants were selected on the basis of the acquired Nir Kmr Tol+ phenotype on 284 minimal medium (12) supplemented with 1 mM NiCl2 and 100 μg/ml kanamycin while the plates were incubated under a toluene atmosphere as a carbon source. The presence of the nre Ni resistance operon and the pTOM plasmid in the transconjugants was confirmed by PCR using nre- and pTOM-specific primers, respectively. The primers specific for tomA4 (accession number AF319657), forward primer 5′-GTTGCCCTCAAACCCTACAA-3′ (position 3323) and reverse primer 5′-AGGGGCTGAATGTTGAGTTG-3′ (position 3780), amplified a 458-bp fragment; the primers specific for nreB (accession number L31491), forward primer 5′-GGATTACCGAGCCAGTTTCA-3′ (position 2421) and reverse primer 5′-GGTGTCTGCGTCATCGAATA-3′ (position 3445), amplified a 1,025-bp fragment. In addition to the presence of nre and pTOM, BOX-PCR (22) was used to confirm the genetic background of the transconjugants. A representative transconjugant, B. cepacia strain VM1468, which grew under the appropriate selective conditions (1 mM NiCl2, 100 μg/ml kanamycin) with toluene as the sole carbon source, was selected for further study.
Microbial inoculation of poplar cuttings.
Cuttings were taken from P. trichocarpa × deltoides cv. Hoogvorst. The cuttings were 40 cm long and had a diameter of approximately 1 cm. The cuttings were surface disinfected with 75% ethanol and placed with their bottom ends in tap water for 4 weeks to allow establishment of roots, after which root inoculation was performed.
B. cepacia strains VM1468 and BU61, which were used as inocula, were grown for 24 h in 284 gluconate medium at 30°C on a rotary shaker. After the cultures reached an optical density at 660 nm of ∼1, which corresponded to 109 CFU/ml, cells were harvested by centrifugation and subsequently washed twice with 10 mM MgSO4. The inoculum was prepared by suspending the cells to a final concentration of 108 CFU/ml in a solution comprised of 1 liter of half-strength sterile Hoagland's nutrient solution (3) to which 200 ml of 284 gluconate medium was added. Cuttings were placed with their roots in the inoculum for 72 h, while control plants were placed in the same solution without bacteria for 3 days. Subsequently, the cuttings were planted in 4-liter pots which were filled with nonsterile sandy soil and saturated with half-strength Hoagland's solution. Before planting the weights, numbers of roots and leaves, and root lengths of the cuttings were determined. The plants were allowed to stabilize for 2 weeks under greenhouse conditions, during which they were watered every other day with half-strength Hoagland's solution or distilled water; after this they were challenged with toluene.
Effect of toluene on the growth of poplar cuttings.
Starting 2 weeks after they were transferred into sandy soil, the poplar cuttings inoculated with B. cepacia VM1468 or BU61 or the noninoculated control plants were watered for 10 weeks with half-strength Hoagland's solution containing 500 mg liter−1 toluene, which resulted in gradual exposure of the plants to toluene. At the beginning of the experiment, the weight of a cutting together with the mass of its pot plus the sandy soil irrigated with Hoagland's solution to field capacity was adjusted to 4,800 g. Every other day, pots with cuttings were weighed and irrigated until they were their original weight with the half-strength Hoagland's solution to which toluene was added at a concentration of 500 mg liter−1. Control experiments without toluene were also set up.
In order to prevent growth of algae, the pots were covered with dark gray plastic foil. For each treatment 10 replicas were established. After 10 weeks the cuttings were harvested, and the following growth parameters were determined: total biomass, number of roots, root weight and length, number of young twigs, number of leaves, leaf surface area, and total leaf weight.
Recovery of inoculated bacteria.
At the end of the toluene exposure experiment plants were harvested, and the microbial colonization of the plants was examined. In order to examine the rate of successful inoculation, rhizosphere, root, stem, twig, and leaf samples were taken from inoculated and control plants that had been watered with Hoagland's solution containing either 0 or 500 mg liter−1 toluene. Roots and leaves were vigorously washed in distilled water for 5 min and surface sterilized for 10 min in a solution containing 2% (wt/vol) active chloride added as an NaOCl solution (Fluka) and supplemented with 1 droplet of Tween 80 (Merck) per 100 ml of solution; stems and twigs were vigorously washed in distilled water for 5 min and sterilized for 5 min in a solution containing 1% active chloride supplemented with 1 droplet of Tween 80 (Merck) per 100 ml of solution. After surface sterilization leaves, roots, stems, and twigs were rinsed three times for 1 min in sterile water and dried on sterile filter paper. The water from the third rinse was plated on 869 medium (12) as a control for sterility. After surface sterilization plant parts were macerated in 10 mM MgSO4 using a Polytron PT1200 mixer (Kinematica A6). Serial dilutions were made, and 100-μl samples were plated on 284 medium to which either gluconate or toluene was added as a carbon source in order to test for the presence of the inoculated strains, as well as other cultivable bacteria. The rhizosphere samples were serially diluted in 10 mM MgSO4 and plated on the same media. After 7 days of incubation at 30°C the number of CFU was determined and expressed per gram of plant tissue or rhizosphere soil.
Morphologically different bacteria that grew on toluene as a sole carbon source were purified three times on 284 gluconate medium or on 10-fold-diluted 869 medium, after which growth on toluene was confirmed, as was the irability to grow autotrophically with CO2 as the carbon source. Tol+ strains were genetically fingerprinted using BOX-PCR (22). The bacteria that exhibited different BOX-PCR patterns and that were able to grow on toluene as a sole carbon source were tested for the presence of the pTOM plasmid by PCR with tomA4-specific primers and were further identified by sequencing their 16S rRNA genes using the standard 26F-1392R primer set (1).
Toluene evapotranspiration.
Poplar cuttings (P. trichocarpa × deltoides cv. Hoogvorst), inoculated as described above, were used to evaluate the phytotoxicity and in planta degradation of toluene. Noninoculated plants were examined as controls. After 7 weeks of growth under greenhouse conditions, the cuttings were carefully taken out of the jars, and their roots were vigorously rinsed in sterile water to remove bacteria from the surface. Subsequently, plants were grown hydroponically in a two-compartment glass cuvette system (height, 60 cm; diameter, 25 cm) (3). Each cuvette compartment was inserted into a flowthrough system using a synthetic air source (Air Liquide) with an inflow of 3 liters per h on one side and with two linked Chromosorb 106 traps (capped sample tubes [Perkin-Elmer] and Chromosorb 60/80 [Alltech]) on the outflow. A column with CaCl2 was placed between the plant cuvettes and the Chromosorb traps in order to prevent water condensation in the traps. The lower compartment was filled with 2.5 liters of half-strength sterile Hoagland's solution to which 100 mg liter−1 toluene was added at the beginning of the experiment. The Chromosorb traps were changed at regular intervals, and they were analyzed by gas chromatography-mass spectrometry (GC-MS) with an ATD400 automatic thermal desorption system, an Auto System XLL gas chromatograph, and a Turbo mass spectrometer (Perkin-Elmer). To avoid gas exchange between the upper and lower compartments, the compartments were separated by a glass plate with an insert through which the stem of the plant was introduced. Each cuvette contained one plant, and the space around the stem was made gas tight with a Polyfilla exterior mixture (Polyfilla), so that the shoots in the upper compartment and the roots in the lower compartment were completely separated; this allowed no gas exchange between the shoots and the roots except through the stem. The upper compartment, the glass plate, and the lower compartment were sealed with Apiezon (Apiezon Products M&I Materials Ltd.).
Cuvettes with plants were placed in a growth chamber with a constant temperature of 22°C and a cycle consisting of 14 h of light (photosynthetically active radiation; 165 μmol m−2 s−1) and 10 h of darkness. The whole experiment was performed for 96 h, and the toluene concentrations in the traps were determined by GC-MS. The amount of evapotranspired toluene was calculated per unit of leaf surface area. All experiments were performed in triplicate to allow statistical analysis of the data using analysis of variance.
RESULTS AND DISCUSSION
Toluene phytotoxicity: effect on the growth of poplar cuttings.
Poplar cuttings inoculated with B. cepacia strain VM1468 or BU61 were grown in conditions under which they were watered with half-strength Hoagland's solution to which 500 mg liter−1 toluene was added. This resulted in step-wise wash-in of toluene. This high toluene concentration was chosen so that there would be clear effects of toluene toxicity in a relatively short time. Experiments with noninoculated plants and plants grown in the absence of toluene were performed as controls.
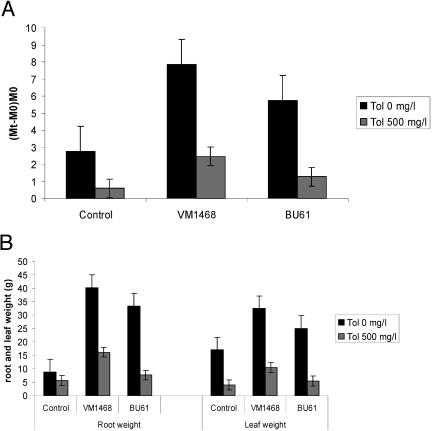
For all plants growth indexes were calculated by determining the difference between a plant's fresh weight at the onset of the experiment and the weight after 10 weeks of growth in the presence or absence of toluene. The results (Fig. 1A) show that plants inoculated with either bacterial strain produced more biomass than the noninoculated plants, even in the absence of toluene. All plants exposed to toluene exhibited a reduction in growth. However, when plants inoculated with B. cepacia VM1468 were challenged with toluene during the 10 weeks of growth, they produced nearly four times more biomass than the noninoculated plants produced. Inoculation of plants with B. cepacia BU61 also had a positive effect on plant growth in the presence of toluene. The biomass produced by these plants was two times the biomass produced by the control plants but only one-half the biomass produced by plants inoculated with VM1468.
FIG. 1.
Plant growth parameters for poplar cuttings calculated after 10 weeks of growth in the presence or absence of toluene (Tol) (0 or 500 mg liter−1). Poplar cuttings inoculated with B. cepacia strain VM1468 of BU61 or noninoculated control plants were grown in conditions under which they were watered with half-strength Hoagland's solution to which 500 mg liter−1 toluene was added, resulting in a step-wise wash-in of toluene. (A) Growth indexes were calculated as (Mt − M0)/M0 after 10 weeks of growth in the presence or absence of toluene (0 or 500 mg liter−1), where M0 is the plant weight (in grams) before addition of toluene and Mt is the plant weight (in grams) 10 weeks after toluene addition. (B) Root and leaf weight were determined after 10 weeks of growth in the presence or absence of toluene (0 or 500 mg liter−1). The data are the means of 10 replicates; standard deviations are indicated by error bars. The statistical significance of the results was confirmed at the 5% level using a two-way analysis of variance model, separately exploring treatment (bacterial inocula) and toluene doses.
All plants exposed to toluene produced significantly less root biomass than plants grown in the absence of toluene produced (Fig. 1B). After toluene exposure the root biomasses for the noninoculated plants and the plants inoculated with B. cepacia BU61 did not differ significantly. Plants inoculated with B. cepacia VM1468 produced about twice as much root biomass (Fig. 1B).
The effects of toluene exposure and microbial inoculation were also significant when leaf parameters were analyzed. After toluene treatment, plants inoculated with B. cepacia VM1468 formed 30 to 40% more leaves than plants inoculated with B. cepacia BU61 formed and about 60% more leaves than noninoculated control plants formed (results not shown). Without toluene treatment, the leaf weight was highest for plants inoculated with B. cepacia VM1468, followed by BU61 and noninoculated plants (Fig. 1B). A similar trend was observed after the plants were exposed to toluene.
Toluene evapotranspiration.
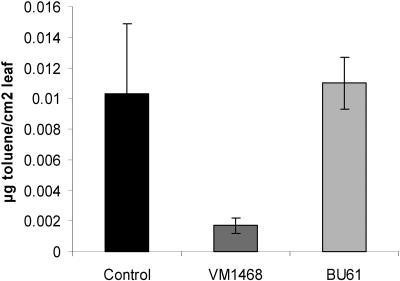
Seven weeks after inoculation with B. cepacia VM1468 or BU61, poplar plants were challenged by adding toluene at a subphytotoxic concentration (100 mg liter−1). After 96 h the total amount of toluene that evapotranspired through the aerial parts of the plants (upper compartment) was measured using GC-MS. The results for toluene evapotranspiration, expressed on the basis of leaf surface area, are shown in Fig. 2. Compared to noninoculated plants or plants inoculated with B. cepacia BU61, poplar cuttings inoculated with B. cepacia VM1468 released about five times less toluene through the leaves. It is important to notice that plants inoculated with B. cepacia VM1468 or BU61 had similar biomasses (63 g ± 5 g), making it very unlikely that the observed differences in toluene evaporation resulted from binding of toluene to plant biomass. This illustrates that inoculation with endophytic bacteria possessing the right degradation pathway not only protects plants against toluene phytotoxicity but also reduces the evapotranspiration of the pollutant into the air.
FIG. 2.
Total amount of toluene (in micrograms) released from poplar into the upper cuvette compartment. Toluene was adsorbed with Chromosorb traps, and the toluene concentration was determined by GC-MS. The amount of evapotranspired toluene was calculated per square centimeter of leaf area. The data are the means of three replicates; standard deviations are indicated by error bars.
Recovery of inoculated bacteria.
All previous experiments indicated that B. cepacia VM1468 is able to protect poplar against toluene phytotoxicity. Since the natural host plant of B. cepacia VM1468 is yellow lupine, we decided to examine the colonization of poplar by this bacterium. To do this, P. trichocarpa × deltoides cv. Hoogvorst cuttings were inoculated with B. cepacia VM1468 and grown for 12 weeks under greenhouse conditions in the presence or absence of 500 mg liter−1 toluene, after which the rhizosphere, roots, stems, twigs, and leaves were sampled to study the presence of the inoculum. Plants that did not receive an inoculum and plants inoculated with B. cepacia BU61 were used as controls. Seven days after samples were plated on different media, the total numbers of bacteria, their morphology, and their ability to grow on toluene as a sole carbon source were determined (Table 1). The number of CFU was calculated per gram (fresh weight) of plant material or rhizosphere soil.
TABLE 1.
Recovery of cultivable bacteria from P. trichocarpa × deltoides cv. Hoogvorst cuttings which were inoculated with B. cepacia VM1468 or BU61a
| Inoculum | Sample | Toluene concn (mg/liter) | No. of cells/g (fresh wt) on:
|
|
|---|---|---|---|---|
| 284 + gluconate | 284 + toluene | |||
| None | Rhizosphere | 0 | 2.5 × 107 | 2.2 × 108 |
| None | Root | 0 | 9.6 × 109 | 0 |
| None | Stem | 0 | 1.1 × 109 | 0 |
| None | Twig | 0 | 4.5 × 108 | 0 |
| None | Leaf | 0 | 5.9 × 107 | 0 |
| None | Rhizosphere | 500 | 2.1 × 107 | 8.9 × 107 |
| None | Root | 500 | 3.3 × 108 | 0 |
| None | Stem | 500 | 3.1 × 108 | 0 |
| None | Twig | 500 | 3.2 × 108 | 0 |
| None | Leaf | 500 | 2.9 × 106 | 0 |
| VM1468 | Rhizosphere | 0 | 2.1 × 107 | 1.2 × 107 |
| VM1468 | Root | 0 | 4.3 × 108 | 3.8 × 108 |
| VM1468 | Stem | 0 | 5.2 × 108 | 9.2 × 106 |
| VM1468 | Twig | 0 | 3.2 × 104 | 3.7 × 104 |
| VM1468 | Leaf | 0 | 6.8 × 102 | 0 |
| VM1468 | Rhizosphere | 500 | 1.7 × 107 | 1.2 × 107 |
| VM1468 | Root | 500 | 5.6 × 108 | 5.6 × 107 |
| VM1468 | Stem | 500 | 4.6 × 107 | 2.2 × 108 |
| VM1468 | Twig | 500 | 3.6 × 108 | 2.1 × 106 |
| VM1468 | Leaf | 500 | 1.2 × 108 | 0 |
| BU61 | Rhizosphere | 0 | 5.4 × 106 | 3.2 × 107 |
| BU61 | Root | 0 | 4.8 × 108 | 2.5 × 108 |
| BU61 | Stem | 0 | 6.6 × 108 | 2.1 × 108 |
| BU61 | Twig | 0 | 0 | 0 |
| BU61 | Leaf | 0 | 6.4 × 107 | 0 |
| BU61 | Rhizosphere | 500 | 7.0 × 106 | 7.5 × 106 |
| BU61 | Root | 500 | 2.7 × 108 | 1.6 × 108 |
| BU61 | Stem | 500 | 3.2 × 108 | 2.9 × 108 |
| BU61 | Twig | 500 | 3.7 × 108 | 4.1 × 108 |
| BU61 | Leaf | 500 | 2.7 × 107 | 0 |
The microbial populations from noninoculated plants were analyzed as controls. Bacteria were isolated on 284 medium plates to which toluene or gluconate was added as a sole carbon source. The numbers of bacteria isolated on the different growth media are expressed per gram (fresh weight) of rhizosphere soil or plant material.
Cultivable endophytic bacteria were found in all parts of inoculated and noninoculated poplar plants. For noninoculated poplar, only one type of rhizosphere bacteria was found to be able to grow on toluene as a sole carbon source, while no cultivable endophytic bacteria able to grow on toluene as a sole carbon source were observed. For plants inoculated with B. cepacia VM1468 and BU61, both rhizosphere and endophytic bacteria able to grow on toluene as a sole carbon source were isolated (Table 1). This result indicates that the presence of Tol+ endophytic bacteria depends on the presence of the inoculum, despite the fact that Tol+ bacteria could be isolated from the rhizosphere of noninoculated control plants. In addition, toluene selection pressure had no effect on the numbers of Tol+ bacteria in the endophytic community.
Interestingly, based on their morphology, different endophytic bacteria were found to grow on toluene as a carbon source. Since no toluene-degrading endophytes were found in the control plants, this strongly suggests that horizontal gene transfer of the tom operon occurred from B. cepacia VM1468 and BU61 to the natural endophytic communities of poplar. To test this hypothesis, morphologically different bacteria growing on media with toluene as a sole carbon source were characterized further. Their DNAs were extracted, the strains were fingerprinted using BOX-PCR, and PCR with tomA4-specific primers was used to confirm the presence of the tom operon. In addition, the 16S rRNA genes of strains that had different BOX-PCR fingerprints were PCR amplified, cloned, and sequenced. As a control to demonstrate horizontal gene transfer of the tom operon, we also tried to identify the corresponding recipients among the cultivable bacteria from the noninoculated control plants. The results for representative strains are shown in Table 2. The most common species found were Enterobacter sp., Pseudomonas putida, and Stenotrophomonas maltophilia. All bacteria tested showed BOX-PCR fingerprints that differed from those of the B. cepacia VM1468 and BU61 donor strains; this included B. cepacia T675, which was isolated from poplar inoculated with strain VM1468. This finding indicates that the inoculum strains were only a minor fraction of the endogenous endophytic community of poplar, if they were able to establish themselves at all. Strains with identical BOX-PCR patterns could be found among rhizosphere and endophytic isolates that had acquired the tom toluene degradation pathway by horizontal gene transfer. For the S. maltophilia and Enterobacter sp. transconjugants, we were able to isolate the corresponding recipient strains from the rhizospheres of noninoculated control plants.
TABLE 2.
Representative rhizosphere and endophytic strains found in association with poplar plants that were inoculated with B. cepacia BU61 or VM1468
| Straina | Origin | Species | Toluene phenotype | tomA4 PCR | BOX-PCR pattern |
|---|---|---|---|---|---|
| BU61 | Control | B. cepacia | Tol+ | + | Unique |
| VM1468 | Control | B. cepacia | Tol+ | + | Unique |
| W604 | Root, VM1468 inoculum | Enterobacter sp. | Tol+ | + | Similar to S662, S671, R558-1 |
| W607 | Root, VM1468 inoculum | Stenotrophomonas maltophilia | Tol+ | + | Similar to S649, W635, R551-3 |
| W619 | Root, VM1468 inoculum | Pseudomonas putida | Tol+ | + | Similar to W630, W645, S672, R599 |
| W630 | Root, BU61 inoculum | Pseudomonas putida | Tol+ | + | Similar to W619, W645, S672, R599 |
| W633 | Root, BU61 inoculum | Ochrobactrum sp. | Tol+ | + | Unique |
| W635 | Root, BU61 inoculum | Stenotrophomonas maltophilia | Tol+ | + | Similar to W607, S649, R551-3 |
| S645 | Stem, VM1468 inoculum | Pseudomonas putida | Tol+ | + | Similar to W619, W630, S672, R599 |
| S649 | Stem, VM1468 inoculum | Stenotrophomonas maltophilia | Tol+ | + | Similar to W607, R551-3 |
| S656 | Stem, VM1468 inoculum | Stenotrophomonas maltophilia | Tol+ | + | Similar to R572, R591 |
| S662 | Stem, VM1468 inoculum | Enterobacter sp. | Tol+ | + | Similar to W604, S671, R558-1 |
| S671 | Stem, BU61 inoculum | Enterobacter sp. | Tol+ | + | Similar to W604, S662, R558-1 |
| S672 | Stem, BU61 inoculum | Pseudomonas putida | Tol+ | + | Similar to W619, W630, S645, R599 |
| T675 | Twig, VM1468 inoculum | B. cepacia | Tol+ | + | Unique |
| R571 | Rhizosphere, VM1468 inoculum | Pseudomonas putida | Tol+ | + | Unique |
| R572 | Rhizosphere, VM1468 inoculum | Stenotrophomonas maltophilia | Tol+ | + | Similar to S656, R591 |
| R591 | Rhizosphere, BU61 inoculum | Stenotrophomonas maltophilia | Tol+ | + | Similar to R572, S656 |
| R599 | Rhizosphere, BU61 inoculum | Pseudomonas putida | Tol+ | + | Similar to W619, W630, S645, S672 |
| R551-2 | Rhizosphere, no inoculum | Pseudomonas putida | Tol+ | − | Unique |
| R551-3 | Rhizosphere, no inoculum | Stenotrophomonas maltophilia | Tol− | − | Similar to W607, W635, S649 |
| R557-2 | Rhizosphere, no inoculum | Pseudomonas sp. | Tol− | − | Unique |
| R558-1 | Rhizosphere, no inoculum | Enterobacter sp. | Tol− | − | Similar to W604, S662, S671 |
| R558-2 | Rhizosphere, no inoculum | Pseudomonas putida | Tol− | − | Unique |
Strains were selected from 284 medium plates with toluene as the sole carbon source. Distinct strains from various samples were compared based on their different BOX-PCR patterns. Strains were identified based on the sequences of their 16S rRNA genes. PCR was used to confirm the presence of the tomA4 gene. Cultivable bacteria from noninoculated control plants that were isolated on 284 gluconate medium plates were characterized as controls.
No Tol+ endophytes were found in the absence of the inocula, despite the presence in the rhizosphere of a Tol+ P. putida strain, R551-2. This strain is closely related to P. putida KT2440 (a sequenced plasmid-free derivative of MT-2 [13]), a strain known to efficiently colonize the rhizosphere of crop plants (5). R551-2 is able to grow on toluene as a carbon source but, as confirmed by PCR, lacks the tomA4 toluene ortho-monooxygenase gene found in B. cepacia G4.
Conclusions.
After inoculation of poplar with B. cepacia strains BU61 and VM1468 a positive effect on plant growth was observed compared to the growth of noninoculated controls, even in the absence of toluene. Endophytic bacteria have been shown to have plant growth-promoting activity that could be responsible for the observed plant growth-promoting effects on the inoculated plants, even in the absence of toluene. These plant growth-promoting effects often result from the production of phytohormones or enzymes involved in growth regulator metabolism, such as ethylene, 1-aminocyclopropane-1-carboxylic acid deaminase, auxins, indoleacetic acid, or cytokinins (2, 6-9).
In the presence of toluene inoculation with strain VM1468 resulted in reductions in toluene phytotoxicity and evapotranspiration compared to the results of control experiments. This led us to believe that the endophytic bacterium B. cepacia VM1468, whose natural host is yellow lupine, was able to successfully colonize poplar and protect the host plant against toluene toxicity. When analyzing the microbial communities of poplar inoculated with B. cepacia strains BU61 and VM1468, we were unable to identify these strains among the cultivable strains but found that both strains had successfully transferred their toluene degradation pathway, encoded by the tom operon, to members of the endogenous plant-associated bacterial communities. This horizontal gene transfer seems to be specific for plasmid pTOM-Bu61 and happens both in the presence and in the absence of toluene.
Despite the fact that both B. cepacia BU61 and VM1468 were able to transfer the pTOM-Bu61-encoded tom operon to the endogenous microbial populations associated with poplar, major differences were observed. Plants inoculated with the endophyte VM1468 suffered less from toluene toxicity and released less toluene into the environment, indicating that their microbial communities were better adapted to degrade toluene than plants inoculated with soil isolate BU61. This observation might be explained by the endophytic characteristics of strain VM1468; we hypothesize that strain VM1468 can enter poplar, in which it is able to directly transfer the pTOM-Bu61-encoded tom operon to the endogenous endophytic community, despite the fact that the strain is unable to eventually establish itself. On the other hand, strain BU61 is unable to enter poplar as an endophyte. Therefore, BU61 can act only as a donor to transfer the tom operon to bacteria present in the poplar rhizosphere. Some rhizosphere bacteria, such as S. maltophilia and Enterobacter sp., can also colonize poplar as endophytes, and therefore the transfer of the tom operon into the endophytic community with BU61 as the donor strain depends on the efficiency of endophytic colonization by the transconjugants. Thus, although inoculation of poplar with B. cepacia BU61 eventually results in transfer of the tom operon into the endogenous endophytic community of poplar, it takes more time than direct transfer from an endophytic donor strain.
Toluene and xylene degradation by P. putida MT-2 is encoded by the two xyl operons located on pWW0. Horizontal gene transfer and pWW0-mediated retrotransfer of chromosomal markers have been reported in the rhizosphere among members of the genus Pseudomonas (15). Due to the presence of P. putida strain R551-2 (Tol+) in the rhizosphere, one would thus expect to find Tol+ endophytes in the noninoculated control plants. It is possible that the heterologous expression range of the xyl operon is limited to Pseudomonas. This is in contrast to the expression range of the tom operon; tom-mediated growth on toluene was observed for bacteria belonging to different branches of the Proteobacteria. It is also striking that only P. putida R551-2 is able to use toluene as a sole carbon source, despite the presence of P. putida strains R557-2 and R558-2 in the poplar rhizosphere. This might indicate that horizontal gene transfer of the P. putida R551-2 toluene degradation pathway, even to other P. putida strains, is very inefficient.
The observation that there is horizontal gene transfer opens the possibility of direct adaptation of a plant's endogenous endophytic population without the need of first selecting the appropriate endophytic microorganisms from the plant species of interest. However, in order to be successful, the genetic information encoding the desired metabolic properties should be present on a broad-host-range plasmid that can be efficiently transferred within the endophytic community, and it should have a broad expression range. If this is the case, the combination of horizontal gene transfer and heterologous expression has several obvious advantages over our original approach (3), in which an endophytic strain is optimized in a laboratory setup before it is introduced into its host plant: there is no need to isolate plant-specific endophytic bacteria, there is no need for genetic manipulation of isolated plant-specific endophytes, and there is no need to establish the endophytic inoculum in the plant's endogenous endophytic community as the genetic information will be transferred to many members of the endogenous endophytic population.
Acknowledgments
The European Commission under the Fifth Framework Programme, Quality of Life, supported this work by providing grant QLK3-2000-00164 entitled “ENDEGRADE.” This work was also supported by Ford Motor Company (Genk Plant in Belgium and the Environmental Quality Office Europe in Köln, Germany), which also provided experimental sites. D.V.D.L. and S.T. were supported by Laboratory Directed Research and Development funds at the Brookhaven National Laboratory under contract with the U.S. Department of Energy.
We thank J. Czech and R. Carleer for help with toluene analysis.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arshad, M., and W. T. Frankenberger. 1991. Microbial production of plant hormones. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 3.Barac, T., S. Taghavi, B. Borremans, A. Provoost, L. Oeyen, J. V. Colpaert, J. Vangronsveld, and D. van der Lelie. 2004. Engineered endophytic bacteria improve phytoremediation of water-soluble, volatile, organic pollutants. Nat. Biotechnol. 22:583-588. [DOI] [PubMed] [Google Scholar]
- 4.Dong, Q. H., D. Springeal, D., Schoeters, G. Nuyts, M. Mergeay, and L. Diels. 1998. Horizontal transfer of bacterial heavy metal resistance genes and its applications in activated sludge systems. Water Sci. Technol. 37:465-468. [Google Scholar]
- 5.Espinosa-Urgel, M., R. Kolter, and J. L. Ramos. 2002. Root colonization by Pseudomonas putida: love at first sight. Microbiology 148:341-343. [DOI] [PubMed] [Google Scholar]
- 6.Glick, B. R. 2004. Bacterial ACC deaminase and the alleviation of plant stress. Adv. Appl. Microbiol. 56:291-312. [DOI] [PubMed] [Google Scholar]
- 7.Glick, B. R., C. B. Jacobson, M. K. Schwarze, and J. J. Pasternak. 1994. 1-Aminocyclopropane-1-carboxylic acid deaminase mutants of the plant growth promoting rhizobacterium Pseudomonas putida GR 12-2 do not stimulate canola root elongation. Can. J. Microbiol. 40:911-915. [Google Scholar]
- 8.Glick, B. R., D. M. Penrose, and J. Li. 1998. A model for the lowering of plant ethylene concentrations by plant growth promoting bacteria. J. Theor. Biol. 190:63-68. [DOI] [PubMed] [Google Scholar]
- 9.Kuklinsky-Sobral, J., W. L. Araujo, R. Mendes, I. O. Geraldi, A. A. Pizzirani-Kleiner, and J. L. Azevedo. 2004. Isolation and characterization of soybean-associated bacteria and their potential for plant growth promotion. Environ Microbiol. 6:1244-1251. [DOI] [PubMed] [Google Scholar]
- 10.Lodewyckx, C., J. Vangronsveld, F. Porteous, E. R. B. Moore, S. Taghavi, M. Mergeay, and D. van der Lelie. 2002. Endophytic bacteria and their potential applications. Crit. Rev. Plant Sci. 21:583-606. [Google Scholar]
- 11.McCrady, J., C. McFarlane, and F. Lindstrom. 1987. The transport and affinity of substituted benzenes in soybean stems. J. Exp. Bot. 38:1875-1890. [Google Scholar]
- 12.Mergeay, M., D. Nies, H. G. Schlegel, J. Gerits, P. Charles, and F. Van Gijsegem. 1985. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J. Bacteriol. 162:328-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. Martins dos Santos, D. E. Fouts, S. R. Gill, M. Pop, M. Holmes, L. Brinkac, M. Beanan, R. T. DeBoy, S. Daugherty, J. Kolonay, R. Madupu, W. Nelson, O. White, J. Peterson, H. Khouri, I. Hance, P. Chris Lee, E. Holtzapple, D. Scanlan, K. Tran, A. Moazzez, T. Utterback, M. Rizzo, K. Lee, D. Kosack, D. Moestl, H. Wedler, J. Lauber, D. Stjepandic, J. Hoheisel, M. Straetz, S. Heim, C. Kiewitz, J. A. Eisen, K. N. Timmis, A. Dusterhoft, B. Tummler, and C. M. Fraser. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799-808. [DOI] [PubMed] [Google Scholar]
- 14.Newman, L. A., and C. M. Reynolds. 2005. Bacteria and phytoremediation: new uses for endophytic bacteria in plants. Trends Biotechnol. 23:6-8;discussion 8-9. [DOI] [PubMed] [Google Scholar]
- 15.Ronchel, M. C., M. A. Ramos-Diaz, and J. L. Ramos. 2000. Retrotransfer of DNA in the rhizosphere. Environ. Microbiol. 2:319-323. [DOI] [PubMed] [Google Scholar]
- 16.Schnoor, L. J., A. L. Licht, C. S. McCutchon, N. L. Wolfe, and H. L. 1995. Carreira. Phytoremediation of organic and nutrient contaminants. Environ. Sci. Technol. 29:318A-323A. [DOI] [PubMed] [Google Scholar]
- 17.Shields, M. S., and M. J. Reagin. 1992. Selection of a Pseudomonas cepacia strain constitutive for the degradation of trichloroethylene. Appl. Environ. Microbiol. 58:3977-3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shields, M. S., M. J. Reagin, R. R. Gerger, R. Campbell, and C. Somerville. 1995. TOM, a new aromatic degradative plasmid from Burkholderia (Pseudomonas) cepacia G4. Appl. Environ. Microbiol. 61:1352-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taghavi, S., H. Delanghe, C. Lodewyckx, M. Mergeay, and D. van der Lelie. 2001. Nickel-resistance-based minitransposons: new tools for genetic manipulation of environmental bacteria. Appl. Environ. Microbiol. 67:1015-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Lelie, D., T. Barac, S. Taghavi, and J. Vangronsveld. 2005. Response to Newman: new uses of endophytic bacteria to improve phytoremediation. Trends Biotechnol. 23:8-9. [Google Scholar]
- 21.van der Lelie, D., J. P. Schwitzguebel, D. J. Glass, J. Vangronsveld, and A. Baker. 2001. Assessing phytoremediation's progress in the United States and Europe. Environ. Sci. Technol. 35:446A-452A. [DOI] [PubMed] [Google Scholar]
- 22.Vinuesa, P., J. L. Rademaker, F. J. de Bruijn, and D. Werner. 1998. Genotypic characterization of Bradyrhizobium strains nodulating endemic woody legumes of the Canary Islands by PCR-restriction fragment length polymorphism analysis of genes encoding 16S rRNA (16S rDNA) and 16S-23S rDNA intergenic spacers, repetitive extragenic palindromic PCR genomic fingerprinting, and partial 16S rDNA sequencing. Appl. Environ. Microbiol. 64:2096-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]