Abstract
Several asco-, basidio-, and zygomycetes isolated from an agricultural field were shown to be able to hydroxylate the phenylurea herbicide isoproturon [N-(4-isopropylphenyl)-N′,N′-dimethylurea] to N-(4-(2-hydroxy-1-methylethyl)phenyl)-N′,N′-dimethylurea and N-(4-(1-hydroxy-1-methylethyl)phenyl)-N′,N′-dimethylurea. Bacterial metabolism of isoproturon has previously been shown to proceed by an initial demethylation to N-(4-isopropylphenyl)-N′-methylurea. In soils, however, hydroxylated metabolites have also been detected. In this study we identified fungi as organisms that potentially play a major role in the formation of these hydroxylated metabolites in soils treated with isoproturon. Isolates of Mortierella sp. strain Gr4, Phoma cf. eupyrena Gr61, and Alternaria sp. strain Gr174 hydroxylated isoproturon at the first position of the isopropyl side chain, yielding N-(4-(2-hydroxy-1-methylethyl)phenyl)-N′,N′-dimethylurea, while Mucor sp. strain Gr22 hydroxylated the molecule at the second position, yielding N-(4-(1-hydroxy-1-methylethyl)phenyl)-N′,N′-dimethylurea. Hydroxylation was the dominant mode of isoproturon transformation in these fungi, although some cultures also produced traces of the N-demethylated metabolite N-(4-isopropylphenyl)-N′-methylurea. A basidiomycete isolate produced a mixture of the two hydroxylated and N-demethylated metabolites at low concentrations. Clonostachys sp. strain Gr141 and putative Tetracladium sp. strain Gr57 did not hydroxylate isoproturon but N demethylated the compound to a minor extent. Mortierella sp. strain Gr4 also produced N-(4-(2-hydroxy-1-methylethyl)phenyl)-N′-methylurea, which is the product resulting from combined N demethylation and hydroxylation.
Isoproturon [N-(4-isopropylphenyl)-N′,N′-dimethylurea] (Fig. 1) is a phenylurea herbicide that is used in Europe mainly for the control of annual grasses and broad-leaf weeds in winter cereals. Environmental concerns have arisen from the frequent finding of isoproturon in surface water and groundwater at concentrations exceeding the European Union limit for drinking water (0.1 μg liter−1) (12, 31, 32). Due to the widespread use and high water solubility of isoproturon, it is on the list of 33 priority substances that seriously threaten surface and groundwater set up by the European Union in the Water Framework Directive.
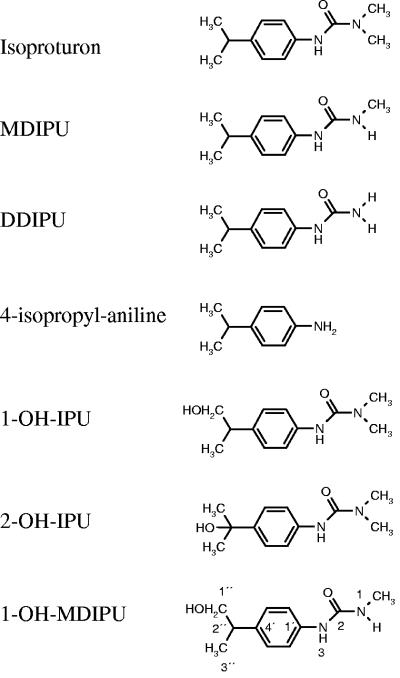
FIG. 1.
Chemical structure of isoproturon (IPU), MDIPU, DDIPU, 4-isopropylaniline, 1-OH-IPU, 2-OH-IPU, and 1-OH-MDIPU. The numbering system for 1H-NMR spectroscopy is shown for 1-OH-MDIPU.
Degradation of isoproturon in soil takes place primarily through microbial processes, although photodegradation of isoproturon has been demonstrated (17). Microbial degradation can lead to complete mineralization of isoproturon, which can mitigate the release of the herbicide to the environment, but it may also result in metabolites with possible detrimental properties. The degradation pathways have recently been reviewed by Sørensen and coworkers (29) and proceed either by consecutive N demethylation to N-(4-isopropylphenyl)-N′-methylurea (trivial name, monodesmethyl-isoproturon [MDIPU]) and N-(4-isopropylphenyl)urea (trivial name, didesmethyl-isoproturon [DDIPU]), followed by hydrolysis to 4-isopropylaniline, or directly from isoproturon to 4-isopropylaniline (Fig. 1). The microbial pathways have been studied mainly in bacterial cultures, and four strains able to mineralize isoproturon have been isolated by enrichment from soils previously treated with isoproturon (1, 5, 27, 30, 36). A Sphingomonas sp. isolated from a British soil seems to degrade isoproturon by initial N demethylation via MDIPU to DDIPU, followed by hydrolysis to 4-isopropylaniline before mineralization of the ring structure (30), while an Arthrobacter sp. isolated from the same field mineralizes the urea side chain directly and accumulates 4-isopropylaniline (5, 36). In soils treated with isoproturon, however, several hydroxylated metabolites have been detected. Based on these findings, a parallel pathway has been proposed (21, 26). This pathway is initiated by hydroxylation of the isopropyl side chain of isoproturon to N-(4-(1-hydroxy-1-methylethyl)phenyl)-N′,N′-dimethylurea (2-OH-IPU), which is subsequently N demethylated to 2-OH-MDIPU and 2-OH-DDIPU, and hydroxylated 4-isopropylaniline is the final known end product. To our knowledge, no bacteria able to perform this pathway have been isolated from soils, although products hydroxylated at the first position of the isopropyl side chain were produced in modest amounts by bacterial enrichments from soil [N-(4-(2-hydroxy-1-methylethyl)phenyl)-N′,N′-dimethylurea (1-OH-IPU)] (16) and by two species of soil algae [N-(4-(2-hydroxy-1-methylethyl)phenyl)-N′-methylurea (1-OH-MDIPU)] (20).
Fungi seem to have a widespread ability to transform isoproturon since many fungi are able to remove isoproturon from the medium when they are grown in liquid culture (3, 9, 14, 37). The products have not been characterized or quantified in detail, however, and most of the fungi studied thus far have not originated from agricultural soils where the herbicide was used. The objectives of this study were to isolate representatives of the fungal community from an agricultural soil, to study their potential for isoproturon transformation, and to identify the transformation products. Among the fungi that we isolated, asco-, basidio-, and zygomycetes were found to transform isoproturon mainly to 1-OH-IPU and 2-OH-IPU, thus indicating that soil fungi could be the source of the hydroxylated metabolites of isoproturon detected in environmental samples.
MATERIALS AND METHODS
Media and chemicals.
Mineral medium contained (per liter) 1.0 g (NH4)2SO4, 1.3 g K2HPO4, 0.5 g NaCl, 0.066 g MgSO4 · 7H2O, 0.04 mg CuSO4 · 5H2O, 0.021 mg ZnCl2, 0.041 mg CoCl2 · 6H2O, and 0.025 mg Na2MoO4 · 2 H2O; the pH was adjusted to 6.5 with 1 M HCl. Glucose medium was mineral medium with 5.0 g/liter glucose. Lignin-guaiacol-benomyl medium (35) contained (per liter) 0.5g KH2PO4, 0.2 g MgSO4 · 7H2O, 0.1 g NH4NO3, 0.1 g KCl, 0.02 g FeSO4 · 7H2O, 0.05 g Ca(NO3)2 · 4H2O, 2.0 g malt extract broth (Lab M, International Diagnostic Group, Lancashire, United Kingdom), and 15 g agar (Difco). After autoclaving and cooling to 55°C, the medium was supplemented with 4 mg of Benlate 50 (50% benomyl; Dupont) suspended in 2 ml of 1:1 acetone-70% ethanol, 1.0 g lignin (catalog no. 370959; Aldrich) dissolved in 5 ml of 1 M KOH, 0.4 ml guaiacol (catalog no. G5502; Sigma), 40 mg of tetracycline (catalog no. T3383; Sigma), 20 mg penicillin G (catalog no. 194537; ICN Biomedicals), and 20 mg of streptomycin (catalog no. 194541; ICN Biomedicals) dissolved in sterile distilled water.
Malt extract agar (MEA) contained (per liter) 10 g malt extract broth and 20 g agar; the pH was adjusted to 6 with 1 M NaOH. To prepare malt extract agar with antibiotics (MEA+a), MEA was autoclaved and cooled to 50°C, and then tetracycline, penicillin G, and streptomycin were added as described above for lignin-guaiacol-benomyl medium.
Malt extract broth contained (per liter) 10 g malt extract broth, pH 6. Spezieller Nährstoffarmer agar contained (per liter) 0.2 g sucrose, 0.2 g glucose, 1.0 g KNO3, 1.0 g KH2PO4, 0.5 g MgSO4 · 7H2O, 0.5 g NaCl, and 12 g agar.
The following analytical standards were purchased from Dr. Ehrenstorfer GmbH (Augsberg, Germany): isoproturon (CAS no. 34123-59-6), MDIPU (CAS no. 34123-57-4), DDIPU (CAS no. 56046-17-4), and 4-isopropylaniline (CAS no. 99-88-7).
Soil sampling and soil particle washing.
Soil samples were taken from the upper 10 cm of an agricultural field near Græse, Denmark (28) and were stored at 5°C for 3 days before use. Organic soil particles for the isolation of soilborne fungi were obtained as described by Thorn et al. (35). Thus, soil samples were sieved through a 2-mm sieve, and 5.0 g soil was added to a 1-liter sterile screw-cap bottle (Schott Duran) with 500 ml of 0.1% (wt/vol) Na4O7P2 · 10H2O to disperse soil clumps and colloids and shaken horizontally for 1 h at 4°C. The suspension was poured through a stack of two sieves with grids of 0.5 mm and 63 μm. After a brief rinse with cold tap water, the 0.5-mm sieve was removed, and the contents of the 63-μm sieve were washed for about 2 min under cold tap water. When the mineral fraction had settled and most of the water had run out, 1 ml was collected from the dense suspension of organic particles and used for isolation of fungi.
Isolation of fungi.
Three strategies were used to isolate fungi. Ascomycetes and zygomycetes were isolated by plating 200 μl of a 1 × 10−2 dilution of the soil organic particles on 20 MEA+a plates. The MEA+a plates were incubated in the dark at 10°C and screened for 3 weeks. Hyphae growing from particles were transferred to MEA plates. Fast-growing isolates were cut out from the MEA+a plates after isolation to prevent them from growing over isolates that emerged later.
Basidiomycetes were isolated by plating 300 μl of a 5 × 10−2 dilution of soil particles on lignin-guaiacol-benomyl medium and incubating the preparation in the dark at 10°C. The plates were screened regularly for 6 weeks for nonsporulating isolates that turned the agar red, thus indicating that guaiacol had been oxidized by laccase or peroxidase activity.
Yeasts were isolated by adding 0.5 g of soil to 1.5 ml sterile distilled water. A 100-μl aliquot of this suspension was inoculated into 250-ml Erlenmeyer flasks containing 100 ml of malt extract broth and incubated in the dark at 10°C and 150 rpm for 2 days. The medium was filtered through sterile filter papers to remove filamentous fungi, and 200 μl of the filtrate was plated on MEA+a. Yeast colonies were transferred to MEA plates after 4 days of incubation. All isolates were restreaked twice.
Characterization of fungi.
Fungal isolates were characterized on the basis of colony morphology, microscopy of spore structures. and nucleotide sequences of the internal transcribed spacers (ITS) of rRNA genes. Mycelium from cultures growing on Spezieller Nährstoffarmer agar was extracted in 40 μl Tris-EDTA buffer (pH 8.0) with 10 μl 20% Chelex. A 2-μl aliquot of the extract was used as the template for a PCR using primers ITS1F and ITS4 (10, 38). Amplification was performed with a thermocycler (PCT-200; MJ Research Inc., Massachusetts) using the PCR conditions described by Gardes and Bruns (10). PCR products were purified (StrataPrep PCR purification kit; catalog no. 211189-1; Stratagene) and sequenced by MWG-Biotech AG.
Degradation studies.
Ten fungi were examined for degradation of isoproturon. For the degradation studies, isoproturon was added to sterile 100-ml screw-cap bottles (Schott Duran) with Teflon-lined lids as described by Sørensen et al. (30). After this, 25 ml of a sterile glucose/mineral medium was added to obtain a final isoproturon concentration of 48 μmol/liter. The isoproturon was allowed to dissolve overnight before use.
The isolates identified as Mortierella sp. strain Gr4, Fusarium sp. strains Gr6 and Gr57, Phoma cf. eupyrena Gr61, Acremonium sp. strain Gr161, and basidiomycete strain Gr177 were inoculated as agar plugs from MEA plates, while Mucor sp. strain Gr22, yeast strain Gr86, Clonostachys sp. strain Gr141, and Alternaria sp. strain Gr174 were inoculated as 1-ml portions of spore suspensions prepared by vortexing sporulating mycelium or yeast colonies in 3 ml of distilled water containing 1 g/liter Tween 80 and 8.5 g/liter NaCl. Each fungus was inoculated into three of the isoproturon-containing 100-ml bottles, as well as into a bottle without isoproturon to enable differentiation between fungal exudates and isoproturon and its metabolites in the chromatographic analysis. The cultures were incubated in the dark at 20°C and 150 rpm for 25 days. A 1-ml sample was collected every fifth day with a syringe and passed through a polytetraflouroethylene filter (diameter, 17 mm; Titan 2 HPLC filter; 0.20-μm polytetrafluoroethylene membrane; catalog no. 42213-PC) into glass vials for high-performance liquid chromatography (HPLC).
The biomass of the fungi was determined at the end of the experiment (day 25) by filtering the mycelia onto filter paper disks. Media containing yeasts were centrifuged at 1,700 × g, and the supernatants were discarded. The samples were air dried to a constant weight in a fume hood.
Biosynthesis of hydroxylated metabolites.
A “resting cell” approach was used for the production of unknown metabolites for identification by nuclear magnetic resonance (NMR) spectroscopy. Pellets of the fungi were obtained by inoculating 100-ml screw-cap bottles (Schott Duran) with Teflon-lined lids containing 25 ml of malt extract broth with agar plugs from MEA plates and incubating the bottles in the dark at 20°C and 150 rpm for 1 week. The malt extract broth was removed from the culture bottles with a pipette, leaving the pellets. Isoproturon or MDIPU was added from a stock solution in ethanol to fresh sterile mineral medium at final concentrations of 50 mg/liter isoproturon or MDIPU and 2% (vol/vol) ethanol. The pellets were rinsed twice with 25 ml of sterile mineral medium, and then 15 ml of medium with isoproturon or MDIPU was added. All cultures were incubated at 20°C and 150 rpm in the dark and sampled for HPLC.
HPLC.
An HPLC system (1050 HP; Hewlett-Packard) with a UV/VIS detector was used with a Hypersil 5 μm C18 column (250 by 2 mm; Phenomenex). Fast routine HPLC analysis of isoproturon, MDIPU, DDIPU, and 4-isopropylaniline was performed using the isocratic method of Juhler et al. (13). For detection of hydroxylated metabolites, a gradient method was used with acetonitrile and water as eluents at a flow rate of 0.3 ml/min. The acetonitrile concentration was 15% (by volume) for the first 8 min, and after this it was raised linearly to 45% between min 8 and 12. Between min 23 and 24 the acetonitrile concentration was decreased to 15%. Equilibration time before the next injection was 7 min. The injection volume was 4 μl, and the column temperature was 45°C. Phenylureas were detected and quantified at 245 nm, and 4-isopropylaniline was detected and quantified at 200 nm. The retention times were as follows: 1-OH-MDIPU, 7.0 min; 2-OH-IPU, 9.8 min; 1-OH-IPU, 11.1 min; DDIPU, 17.8 min; MDIPU, 18.7 min; isoproturon, 20.0 min; and 4-isopropylaniline, 22.0 min.
Preparative HPLC was used to purify the unknown metabolites, and the identities were further verified by NMR spectroscopy. The preparative HPLC was performed using a gradient program with the same flow rate and eluents that were used for the analytical method. The acetonitrile concentration was 15% (by volume) for the first 8 min, and after this it was raised linearly to 45% between min 8 and 12 and to 70% between min 12 and 14. After 20 min, the acetonitrile concentration was decreased to 15%. Equilibration time before the next injection was 7 min. The injection volume was 50 μl, and samples from 10 injections were pooled for each compound.
NMR spectroscopy.
Samples of unknown compounds were dried under a stream of N2 and dissolved in CD3CN (99.7% 2H; Euriso-top) to concentrations of approximately 50 μmol liter−1 in a total volume of 600 μl. One-dimensional 1H spectra were recorded at 303 K with a Varian INOVA 750 with a 5-mm triple-resonance probe equipped with a Z-field gradient. The sweep widths were 10,000 Hz using 32,000 data points, and a total of 128 or 156 transients were accumulated. Chemical shifts were referenced to solvent [CD3CN: δ = 1.93 ppm], and spectra were transformed and analyzed using MestReC (Mestrec Research, Spain).
Nucleotide sequence accession numbers.
The sequences determined in this study have been deposited in the EMBL database under accession numbers AJ890432 to AJ890441.
RESULTS
More than 160 isolates were obtained from the Græse soil using the three isolation strategies. From these isolates, 10 fungi were selected to represent the zygo-, asco-, and basidiomycetes. The ITS rRNA genes of the 10 selected fungi were sequenced, and the fungi were identified to the genus level when possible (Table 1). A BLAST search of the ITS sequences confirmed the morphological identification except for the fungus morphologically identified as Acremonium sp. strain Gr161, for which no close matches were detected. Strain Gr177 could be identified as a basidiomycete by its clamp connections, and its sequence most closely resembled sequences of isolates of Coprinacea. Gr57 did not sporulate, but the closest matching sequence was the sequence from Tetracladium maxilliforme, an aquatic hyphomycete which is also often isolated from soils (7).
TABLE 1.
Fungal isolates and most closely related fungi from a BLAST search of the GenBank database
| Isolatea | Results of BLAST search (% identity) | N demethylation | Hydroxylation |
|---|---|---|---|
| Mortierella sp. strain Gr4 (zygo) | Mortierella cf. hyalina (91) | +b | + |
| Fusarium sp. strain Gr6 (asco) | Fusarium spp. (100) | − | − |
| Mucor sp. strain Gr22 (zygo) | Mucor hiemalis (100) | + | + |
| Gr57 (asco) | Tetracladium maxilliforme (99) | + | − |
| Phoma cf. eupyrena Gr61 (asco) | Phoma spp., Podaspora spp., and Didymella applanata (98)c | − | + |
| Yeast Gr86 (bas) | Cryptococcus sp. (100) | − | − |
| Clonostachys sp. strain Gr141 (asco) | Clonostachys spp., Nectria gliocladioides, and Bionectria ochroleuca (100)c | + | − |
| Acremonium sp. strain Gr161 (asco) | No sequences with >82% similarity | − | − |
| Alternaria sp. strain Gr174 (asco) | Alternaria spp. (100) | + | + |
| Basidiomycete strain Gr177 | Coprinus lagopus (93) | + | + |
Asco, ascomycete; zygo, zygomycete; bas, basidiomycete.
+, present; −, absent.
Didymella is a teleomorphic genus of Phoma, and Nectria and Bionectria are teleomorphic genera of Clonostachys.
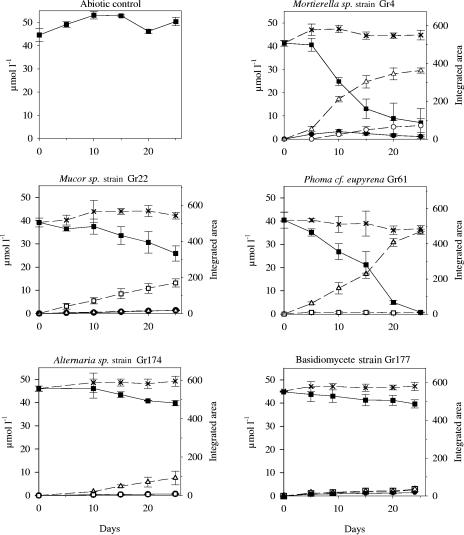
The most rapid degradation of isoproturon was seen with cultures of Mortierella sp. strain Gr4 and Phoma cf. eupyrena Gr61. In these cultures, about 80% of the isoproturon disappeared within 20 days (Fig. 2). This could not be explained by production of MDIPU, DDIPU, or 4-isopropylaniline since only a transient accumulation of MDIPU was observed with Mortierella sp. strain Gr4. However, gradient HPLC analysis revealed a peak at 11.1 min that accumulated with both fungi and a peak at 7.0 min with Mortierella sp. strain Gr4. There was less rapid disappearance of isoproturon with three other fungi. Mucor sp. strain Gr22 produced a peak at 9.8 min, while Alternaria sp. strain Gr174 produced a peak at 11.1 min and basidiomycete strain Gr177 produced a mixture of MDIPU and low concentrations of the compounds with peaks at 9.8 and 11.1 min. The integrated area for the unknown compounds at 7.0, 9.8, and 11.1 min mirrored the area of the disappearing isoproturon peak since the total integrated area was almost constant in the five cultures (Fig. 2). These compounds were not fungal exudates, since they were never observed in cultures to which isoproturon was not added. Isoproturon did not degrade in the abiotic control (Fig. 2). None of the remaining cultures exhibited degradation of isoproturon, and the average concentrations of isoproturon at day 25 ranged from 45.6 to 55.2 μmol/liter (coefficient of variation, <5.2%; n = 3 [n = 2 for Acremonium sp. strain Gr161]). Gr57 and Clonostachys sp. strain Gr141 produced traces of MDIPU, however. With Gr57 the average concentration was 1.0 μmol/liter at day 25, while with Gr141 the average concentration ranged from 1.5 to 2.3 μmol/liter from day 10 to day 25 (coefficient of variation, <74%; n = 3).
FIG. 2.
HPLC analysis of isoproturon and metabolites in liquid cultures. The data for solid lines with solid symbols can be read on both ordinates. The data for dashed lines with open symbols can be read only on the right ordinate (integrated area). ▪, isoproturon; •, MDIPU; ▵, 1-OH-IPU (retention time, 11.1 min); □, 2-OH-IPU (retention time, 9.8 min); ○, 1-OH-MDIPU (retention time, 7.0 min); ×, total integrated area. The error bars indicate ±1 standard deviation (n = 3).
Preparative HPLC was performed with media containing 50 mg/liter isoproturon incubated for 14 days with mycelia of Mucor sp. strain Gr22 or Phoma cf. eupyrena Gr61 and media with 50 mg/liter MDIPU incubated for 4 days with Mortierella sp. strain Gr4. By comparison with known NMR assignments we easily identified the HPLC peak at 9.8 min as 2-OH-IPU and the peak at 11.1 min as 1-OH-IPU (11). The peak at 7.0 min did not match any reported NMR assignments for isoproturon metabolites. Together with the typical shifts from the remaining protons of the molecule, the presence of methylene shifts at 3.50 ppm and 3.56 ppm showing vicinal coupling and two separate NH shifts at 7.03 ppm (N3H) and 5.00 (N1H), respectively, confirmed the identification of the HPLC peak at 7.0 min as 1-OH-MDIPU (Table 2).
TABLE 2.
1H-NMR data for 1-OH-MDIPUa
| 1H-NMR data |
|---|
| 7.30 (2H, AA′BB′, H2′/6′, J = 8.5) |
| 7.11 (2H, AA′BB′, H3′/5′, J = 8.5) |
| 7.03 (1H, s(br), N3H) |
| 5.00 (1H, s(br), N1H) |
| 3.56 (1H, ddd, H1", J = 10.4, 6.7, 4.9) |
| 3.50 (1H, ddd, H1", J = 10.4, 6.7, 4.9) |
| 2.78 (1H, hept, H2", J = 6.7) |
| 2.70 (3H, d, NCH3, J = 4.3) |
| 2.54 (1H, t, OH, J = 4.9) |
| 1.19 (3H, d, CH3", J = 6.7) |
| (1.93 CD3CN) |
| (2.10 water) |
Growth was observed for all fungi, and the biomasses ranged from 0.14 ± 0.04 to 2.0 ± 0.1 mg/ml (mean ± standard deviation; n = 3), so the lack of isoproturon degradation with some of the fungi was not due to an inability to grow in the medium.
DISCUSSION
The 10 fungi tested in the present study represent asco-, basidio-, and zygomycetes and are related to genera considered to be common soil saprophytes that are often isolated from agricultural soils (7, 19). Basidiomycetes, such as isolate Gr177, are rarely isolated from agricultural soils, but this may be because they are overlooked due to their low growth rates on agar (35).
Several of the fungal cultures rapidly transformed isoproturon with hydroxylated metabolites as the dominant products. To our knowledge, this is the first detailed demonstration of the hydroxylation of isoproturon by soil fungi. N demethylation has previously been reported to be the dominant pathway for fungal degradation of isoproturon, although it has been mentioned that unidentified products might be the result of hydroxylation (2, 3) and hydroxylated isoproturon has previously been suggested to be a fungal degradation product (4). The lignin and manganese peroxidases of the white rot fungus Phanerochaete chrysosporium have also been shown to produce unidentified products that have been suggested to be hydroxylated compounds (6).
In the present study, three isoproturon transformation processes were identified for the soil fungi: N demethylation to MDIPU and two types of hydroxylation, at the first and second positions on the isopropyl side chain, yielding 1-OH and 2-OH metabolites, respectively. Mortierella sp. strain Gr4 hydroxylated only at the first position. This process was dominant for Phoma cf. eupyrena Gr61 and Alternaria sp. strain Gr174, but these organisms also produced traces of 2-OH-IPU. Mucor sp. strain Gr22 hydroxylated mainly at the second position, while basidiomycete strain Gr177 produced equal amounts of 1-OH- and 2-OH-IPU, although it produced both at low concentrations. The only hydroxylating isolate that did not also N demethylate was Phoma cf. eupyrena Gr61 (Fig. 2). 1-OH-MDIPU is the product of combined N demethylation and hydroxylation and was produced only by Mortierella sp. strain Gr4. Isoproturon did not serve as an energy or nutrient source for the fungi, and the purpose of these fungal transformations is unknown. They may be elicited by a detoxification system, however.
A mass balance for the transformation of isoproturon could not be obtained because no authentic standards of the hydroxylated compounds were available. In all cultures, however, the total integrated area of the isoproturon and metabolite peaks remained close to constant (Fig. 2). Isoproturon, MDIPU, and DDIPU have similar responses at 245 nm for similar concentrations, and their UV spectra in the range from 200 to 300 nm are comparable to those of the hydroxylated metabolites. It is therefore reasonable to assume that the peak area of hydroxylated phenylureas explains the removal of isoproturon observed; i.e., hydroxylation was the major degradation process in this study. Sorption and mineralization are other possible means of isoproturon removal from the medium. P. chrysosporium mineralized 14C-labeled isoproturon and 3,4-dichloroaniline, the aniline metabolite of the phenylurea herbicide diuron, but only at a high temperature (39°C) and in a pure oxygen atmosphere (18, 24). No mineralization of isoproturon has been observed with soil fungi incubated under atmospheric air with 14C-labeled isoproturon (3). Only bacteria enriched from soil with a history of isoproturon application have otherwise been able to mineralize isoproturon (27, 29). Berger (3) reported that the mycelium sorbed about one-third of the isoproturon in liquid cultures of Phoma eupyrena and Cladosporium herbarum, while six other fungi did not sorb isoproturon. Sorption must have been a minor mechanism in our study, since almost all the disappearance of isoproturon can be explained by hydroxylation.
Isoproturon can be hydroxylated at two different positions on the isopropyl side chain, yielding 1-OH-IPU and 2-OH-IPU (Fig. 1). Only 2-OH metabolites have previously been detected in soil samples, however (8, 16, 22, 23, 25, 26, 34). In the studies in which 1-OH metabolites were analyzed, they were not detected (26, 33). In our study, in contrast, the soil fungi were able to produce both 1-OH and 2-OH metabolites. It is possible that the two types of metabolites are both produced in soils and that they have markedly different half-lives.
The initial transformation of phenylurea herbicides has previously been shown to be crucial for the rate of mineralization. A mixed bacterial culture from the Græse field mineralized MDIPU readily but isoproturon poorly, thus indicating that the mixed culture depends on the N demethylation being carried out by other organisms (28). In a study with Acinetobacter calcoaceticus, mineralization of the phenylurea chlortoluron was dependent on a transformation catalyzed by a cytochrome P450 monooxygenase. The bacterium was genetically engineered to express cytochrome P450 CYP105D1 and gained the ability to N demethylate and hydroxylate chlortoluron. The transformed strain was then able to mineralize and grow on chlortoluron (15). This raises the possibility of interactions between different microorganisms since it seems possible that the hydroxylated and N-demethylated metabolites of a phenylurea herbicide could serve as substrates for mineralizing microorganisms. Future research should clarify the potential of soils and bacterial isolates to mineralize fungal metabolites of isoproturon. This could determine the persistence of the metabolites and may provide new insight into interactions between fungi and bacteria in the degradation of the herbicide.
A large proportion of the fungi examined in this study transformed isoproturon to hydroxylated metabolites that are also found in environmental samples (25, 26). This suggests that soil fungi could be responsible for production of the hydroxylated metabolites detected in environmental samples. Any conclusion should be drawn cautiously, however, as the activity of the isolated fungi in soil is unknown, and the involvement of other soil organisms cannot be ruled out. The hydroxylated metabolites produced by fungi may be important from an environmental point of view as field data suggest that 2-OH-IPU has a greater tendency to leach from the soil than MDIPU (26). The higher mobility of hydroxylated metabolites underlines the need to evaluate the production of these compounds, their ecotoxicity, and their fate in soils.
Acknowledgments
A. H. Johnsen and A. Kastrup provided valuable assistance in the preliminary identification of metabolites.
S. Rønhede was funded by a grant from the Danish Technical Research Council.
REFERENCES
- 1.Bending, G. D., S. D. Lincoln, S. R. Sørensen, J. A. W. Morgan, J. Aamand, and A. Walker. 2003. In-field spatial variability in the degradation of the phenyl-urea herbicide isoproturon is the result of interactions between degradative Sphingomonas spp. and soil pH. Appl. Environ. Microbiol. 69:827-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger, B. M. 1999. Factors influencing transformation rates and formation of products of phenylurea herbicides in soil. J. Agric. Food Chem. 47:3389-3396. [DOI] [PubMed] [Google Scholar]
- 3.Berger, B. M. 1998. Parameters influencing biotransformation rates of phenylurea herbicides by soil microorganisms. Pestic. Biochem. Phys. 60:71-82. [Google Scholar]
- 4.Bolte, M. 2004. Environmental fate of pollutants: example of phenylurea pesticides. Actual. Chim. 277-278:33-39. [Google Scholar]
- 5.Cullington, J. E., and A. Walker. 1999. Rapid biodegradation of diuron and other phenylurea herbicides by a soil bacterium. Soil Biol. Biochem. 31:677-686. [Google Scholar]
- 6.Del Pilar Castillo, M., S. von Wirén-Lehr, I. Scheunert, and L. Torstensson. 2001. Degradation of isoproturon by the white rot fungus Phanerochaete chrysosporium. Biol. Fertil. Soils 33:521-528. [Google Scholar]
- 7.Domsch, K. H., W. Gams, and T. Anderson. 1980. Compendium of soil fungi, vol. 1. Academic Press, London, United Kingdom.
- 8.Elkhattabi, K., A. Bouhaouss, C. Perrin-Ganier, and M. Sciavon. 2004. Fate of isoproturon in two Moroccan soils. Agronomie 24:177-183. [Google Scholar]
- 9.Fournier, J. C., and G. Catroux. 1980. Use of strains of collected microorganisms for studies on pesticide biodegradability. Chemosphere 9:33-38. [Google Scholar]
- 10.Gardes, M., and T. D. Bruns. 1993. ITS primers with enhanced specificity for basidiomycetes—application for the identification of mycorrhizae and rusts. Mol. Ecol. 2:113-118. [DOI] [PubMed] [Google Scholar]
- 11.Glässgen, W. E., D. Komossa, O. Bohnenkamper, M. Haas, N. Hertkorn, R. G. May, W. Szymczak, and H. Sandermann. 1999. Metabolism of the herbicide isoproturon in wheat and soybean cell suspension cultures. Pestic. Biochem. Physiol. 63:97-113. [Google Scholar]
- 12.Johnson, A. C., T. J. Besien, C. L. Bhardwaj, A. Dixon, D. C. Gooddy, A. H. Haria, and C. White. 2001. Penetration of herbicides to groundwater in an unconfined chalk aquifer following normal soil applications. J. Contam. Hydrol. 53:101-117. [DOI] [PubMed] [Google Scholar]
- 13.Juhler, R. K., S. R. Sørensen, and L. Larsen. 2001. Analysing transformation products of herbicide residues in environmental samples. Water Res. 35:1371-1378. [DOI] [PubMed] [Google Scholar]
- 14.Khadrani, A., M. F. Seigle, R. Steiman, and T. Vroumsia. 1999. Degradation of three phenylurea herbicides (chlortoluron, isoproturon and diuron) by micromycetes isolated from soil. Chemosphere 38:3041-3050. [DOI] [PubMed] [Google Scholar]
- 15.Lamb, D. C., D. E. Kelly, S. Masaphy, G. L. Jones, and S. L. Kelly. 2000. Engineering of heterologous cytochrome P450 in Acinetobacter sp.: Application for pollutant degradation. Biochem. Biophys. Res. Commun. 276:797-802. [DOI] [PubMed] [Google Scholar]
- 16.Lehr, S., W. E. Glassgen, H. Sandermann, F. Beese, and I. Scheunert. 1996. Metabolism of isoproturon in soils originating from different agricultural management systems and in cultures of isolated soil bacteria. Int. J. Environ. Anal. Chem. 65:231-243. [Google Scholar]
- 17.Mansour, M., E. A. Feicht, A. Behechti, K. W. Schramm, and A. Kettrup. 1999. Determination photostability of selected agrochemicals in water and soil. Chemosphere 39:575-585. [DOI] [PubMed] [Google Scholar]
- 18.May, R. G., I. Sparrer, E. Hoque, and H. Sandermann. 1997. Mineralization of native pesticidal plant cell-wall complexes by the white-rot fungus, Phanerochaete chrysosporium. J. Agric. Food Chem. 45:1911-1915. [Google Scholar]
- 19.Morganjones, G., and K. B. Burch. 1988. Studies in genus Phoma. 10. Concerning Phoma eupyrena, an ubiquitous, soil-borne species. Mycotaxon 31:427-434. [Google Scholar]
- 20.Mostafa, F. I. Y., and C. S. Helling. 2001. Isoproturon degradation as affected by the growth of two algal species at different concentrations and pH values. J. Environ. Sci. Health Part B 36:709-727. [DOI] [PubMed] [Google Scholar]
- 21.Mudd, P. J., R. J. Hance, and S. J. L. Wright. 1983. The persistence and metabolism of isoproturon in soil. Weed Res. 23:239-246. [Google Scholar]
- 22.Perrin-Ganier, C., F. Schiavon, J.-L. Morel, and M. Schiavon. 2001. Effect of sludge-amendment or nutrient addition on the biodegradation of the herbicide isoproturon in soil. Chemosphere 44:887-892. [DOI] [PubMed] [Google Scholar]
- 23.Pieuchot, M., C. Perrin-Ganier, J. M. Portal, and M. Schiavon. 1996. Study on the mineralization and degradation of isoproturon in three soils. Chemosphere 33:467-478. [Google Scholar]
- 24.Sandermann, H., W. Heller, N. Hertkorn, E. Hoque, D. Pieper, and R. Winkler. 1998. A new intermediate in the mineralization of 3,4-dichloroaniline by the white rot fungus Phanerochaete chrysosporium. Appl. Environ. Microbiol. 64:3305-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schroll, R., and S. Kuhn. 2004. Test system to establish mass balances for C-14-labeled substances in soil-plant-atmosphere systems under field conditions. Environ. Sci. Technol. 38:1537-1544. [DOI] [PubMed] [Google Scholar]
- 26.Schuelein, J., W. E. Glaessgen, N. Hertkorn, P. Schroeder, H. Sandermann, and A. Kettrup. 1996. Detection and identification of the herbicide isoproturon and its metabolites in field samples after a heavy rainfall event. Int. J. Environ. Anal. Chem. 65:193-202. [Google Scholar]
- 27.Sebai, T. E., B. Lagacherie, G. Soulas, and F. Martin-Laurent. 2004. Isolation and characterisation of an isoproturon-mineralising Methylopila sp. TES from French agricultural soil. FEMS Microbiol. Lett. 239:103-110. [DOI] [PubMed] [Google Scholar]
- 28.Sørensen, S. R., and J. Aamand. 2001. Biodegradation of the phenylurea herbicide isoproturon and its metabolites in agricultural soils. Biodegradation 12:69-77. [DOI] [PubMed] [Google Scholar]
- 29.Sørensen, S. R., G. D. Bending, C. S. Jacobsen, A. Walker, and J. Aamand. 2003. Microbial degradation of isoproturon and related phenylurea herbicides in and below agricultural fields. FEMS Microbiol. Ecol. 45:1-11. [DOI] [PubMed] [Google Scholar]
- 30.Sørensen, S. R., Z. Ronen, and J. Aamand. 2001. Isolation from agricultural soil and characterization of a Sphingomonas sp. able to mineralize the phenylurea herbicide isoproturon. Appl. Environ. Microbiol. 67:5403-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spliid, N. H., and B. Køppen. 1998. Occurrence of pesticides in Danish shallow ground water. Chemosphere 37:1307-1316. [DOI] [PubMed] [Google Scholar]
- 32.Stangroom, S. J., C. D. Collins, and J. N. Lester. 1998. Sources of organic micropollutants to lowland rivers. Environ. Technol. 19:643-666. [Google Scholar]
- 33.Suhadolc, M., M. Schloter, R. Schroll, C. Bergmuller, S. V. Bolta, F. Lobnik, and D. Lestan. 2000. Fate of isoproturon in two heavy metal contaminated soils in a microcosm experiment. Fresenius Environ. Bull. 9:691-700. [Google Scholar]
- 34.Suhadolc, M., R. Schroll, A. Gattinger, M. Schloter, J. C. Munch, and D. Lestan. 2004. Effects of modified Pb-, Zn-, and Cd-availability on the microbial communities and on the degradation of isoproturon in a heavy metal contaminated soil. Soil Biol. Biochem. 36:1943-1954. [Google Scholar]
- 35.Thorn, R. G., C. A. Reddy, D. Harris, and E. A. Paul. 1996. Isolation of saprophytic basidiomycetes from soil. Appl. Environ. Microbiol. 62:4288-4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turnbull, G. A., J. E. Cullington, A. Walker, and J. A. W. Morgan. 2001. Identification and characterisation of a diuron-degrading bacterium. Biol. Fertil. Soils 33:472-476. [Google Scholar]
- 37.Vroumsia, T., R. Steiman, M. F. Seigle, G. J. L. Benoit, and A. Khadrani. 1996. Biodegradation of three substituted phenylurea herbicides (chlortoluron, diuron, and isoproturon) by soil fungi: a comparative study. Chemosphere 33:2045-2056. [DOI] [PubMed] [Google Scholar]
- 38.White, T. J., T. Bruns, P. Leeflang, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, New York, N.Y.