Abstract
Current elevated concentrations of ozone in the atmosphere, as they are observed during summer seasons, can cause severe effects on plant vegetation. This study was initiated to analyze whether ozone-stressed plants also transfer signals below ground and thereby alter the bacterial community composition in their rhizospheres. Herbaceous plants, native to Germany, with tolerance (Anthoxanthum odoratum, Achillea millefolium, Poa pratensis, Rumex acetosa, and Veronica chamaedrys) and sensitivity (Matricaria chamomilla, Sonchus asper, and Tanacetum vulgare) to ozone, raised in the greenhouse, were exposed in open-top chambers to two different ozone regimes, i.e., “summer stress” and a normal ozone background. DNA of bacterial cells from the rhizospheres was directly extracted, and partial sequences of the 16S rRNA genes were PCR amplified with primers targeting the following phylogenetic groups: Bacteria, α-Proteobacteria, Actinobacteria, and Pseudomonas, respectively. The diversity of the amplified products was analyzed by genetic profiling based on single-strand conformation polymorphism (SSCP). Neither the tolerant nor the sensitive plants, the latter with visible above-ground damage, showed ozone-induced differences in any of the SSCP profiles, with the single exception of Actinobacteria-targeted profiles from S. asper. To increase the stress, S. asper was germinated and raised in the continuous presence of an elevated level of ozone. SSCP profiles with Bacteria-specific primers combined with gene probe hybridizations indicated an ozone-related increase in a Xanthomonas-related 16S rRNA gene and a decrease in the respective gene from the plant plastids. The fact that only this latter unrealistic scenario caused a detectable effect demonstrated that ozone stress has a surprisingly small effect on the structural diversity of the bacterial community in rhizospheres.
Ozone (O3) is an air pollutant generated in the troposphere in a photochemical reaction by the action of sunlight on volatile organic compounds or oxides of nitrogen emitted by vehicles and industry. In fact, the concentration of tropospheric ozone has increased fivefold in the past hundred years (28). As ozone formation depends on solar energy, annual and diurnal patterns of various concentrations result in high concentrations during the growing season from May to September in Europe, especially in the afternoon hours. Ozone concentrations in the northern hemisphere typically range between 20 and 60 ppb (44). In addition, on calm, sunny summer days, the concentration may rise and reach levels of up to 250 ppb (44). Current ambient ozone in industrial countries may cause severe effects on plant vegetation like visible foliar injury or growth reduction or effects on resource allocation and reproduction (11). This can decrease yields of sensitive arable crop species up to the point of economical importance (14). Less is known about ozone effects on natural vegetation, but there is evidence that wild plant species can be as sensitive as crops (11).
Most ecological research on the impact of ozone on terrestrial ecosystems is limited to effects observed above ground. However, it can be assumed that below-ground processes may also be affected, as plants live in close association and interaction with soil microorganisms, particularly in the rhizospheres and during decay of the plant material (21, 25). Knowledge on the potential effects of ozone-stressed plants on soil is fairly limited to the fact that root growth seems to be more affected than shoot growth and that carbon allocation may change to the disadvantage of the roots (10). Ozone-stressed plants may have a different root morphology (19) and a decreased starch content in their roots (1, 10), probably leading to an altered rhizodeposition (30). At the ecosystem level, it was observed that total soil carbon formation in aspen and mixed aspen-birch forests was reduced by elevated ozone against a background of elevated carbon dioxide after an exposure of 4 years (23). A possible role of ozone stress altering soil processes was also reported in a study of ozone-stressed blue wild rye: in response to the ozone stress, soil bacterial biomass decreased, but fungal biomass increased (53).
The objective of this study was to elucidate the effect of ozone on the structural diversity of bacteria inhabiting the rhizosphere. As this habitat is mainly influenced by the carbon and energy sources released by the plants, it was suspected that an altered root metabolism of ozone-stressed plants would become evident in the selection of different bacterial communities. The structural composition of the bacterial communities was evaluated by a cultivation-independent approach, based on PCR-amplified partial 16S rRNA genes from DNA directly extracted from the rhizospheres (37). Single-strand conformation polymorphism (SSCP) was chosen as a genetic profiling technique to directly compare the community composition from different samples with each other and to allow the identification of differences by DNA sequencing and phylogenetic analyses (12, 40, 46). In previous studies, this approach allowed bacterial communities from rhizospheres of two crops grown in the same soil to be distinguished (38) and for the effects of genetically engineered plants on their associated bacterial communities to be evaluated (4, 39).
To study whether below-ground processes in natural ecosystems would be affected, a total of eight herbaceous plants native to Germany were grown in the same soil and exposed in open-top chambers (OTCs) to ozone concentrations that may occur on sunny summer days. In addition, controls were incubated with a normal background of ozone, as a complete lack of ozone in the atmosphere would have been unrealistic. According to their response to ozone, five of these species were considered ozone resistant (Anthoxanthum odoratum, Achillea millefolium, Poa pratensis, Rumex acetosa, and Veronica chamaedrys), with no visible injuries after the ozone incubations, while three species were ozone sensitive (Matricaria chamomilla, Sonchus asper, and Tanacetum vulgare), as detected by foliar injuries. In addition to these experiments, which represented real-world episodic ozone stress, one ozone-sensitive plant species, i.e., S. asper (5), was also grown and incubated under continuous levels of increased atmospheric ozone. This latter treatment was included to test the response of the bacterial community structure under more vigorous conditions.
MATERIALS AND METHODS
Plant species and experimental setup for ozone fumigation.
Three separate experiments were conducted during the three growing seasons from the years 2000 to 2002 in OTCs (diameter, 315 cm; height, 336 cm). In the first and second year, the plants were raised from seeds in the greenhouse in a potting soil; seedlings of uniform development were transplanted to mesocosms (pot containers; 21 cm in diameter) filled with a native soil (loamy sand, pH 7.4; 150 mg kg−1 phosphorus, 120 mg kg−1 potassium, 50 mg kg−1 magnesium) and amended with slow-release fertilizer at 1.5 g per mesocosm (Plantacote Depot 8 M, N:P:K 14-9-15; Spiess-Urania Chemicals, Hamburg, Germany). Each mesocosm received four individual plants. After 7 weeks (in 2000) or 4 weeks (in 2001), the mesocosms were transferred to OTCs and exposed to different ozone regimes for 8 h day−1, except for days when the photosynthetically active radiation was <500 μE m−2 s−1, e.g., days with permanent rain. The OTCs received either (i) nonfiltered ambient air plus 50 ppb ozone (NF + 50) to cause ozone stress on top of the natural diurnal fluctuations or (ii) charcoal-filtered air plus 25 ppb ozone (CF + 25) as a control treatment, representing a natural ozone background concentration. The ozone-dispensing and -monitoring system for ozone concentrations in the OTCs has been described by Weigel and Jäger (50). Two OTCs with replicate treatments were established; in each OTC, two replicate mesocosms were placed. Thus, for each treatment the study was based on 16 replicate plants.
Plant seeds of Anthoxanthum odoratum, Achillea millefolium, Rumex acetosa, and Veronica chamaedrys were obtained from Rieger-Hofmann (Blaufelden-Raboldshausen, Germany); Poa pratensis var. LATO was obtained from Saatzucht Steinach (Steinach, Germany); Sonchus asper and Matricharia chamomilla were obtained from Blauetikett-Bornträger (Offstein, Germany); and Tanacetum vulgare was from Suffolk Herbs, Monks Farm (Kelvedon, Colchester, Essex, United Kingdom).
A total of eight plant species were analyzed in this study and tested in an accompanying study with the same OTC system. These species were classified as ozone tolerant or ozone sensitive depending on the absence or occurrence of visible leaf injuries after ozone exposure in the OTCs (E. Bergmann and J. Bender, unpublished results). In the first year of the study (2000), the ozone-resistant plant species A. odoratum, A. millefolium, P. pratensis, R. acetosa, and V. chamaedrys were analyzed. These plants were exposed to ozone over a period of 5 weeks. In the following growing season, the ozone-sensitive species M. chamomilla, S. asper, and T. vulgare were exposed to ozone for 6 weeks.
In the third year of this study, only S. asper was selected; this species had already been raised from seeds under continuous fumigation (8 h day−1, except for days with sustained rainfall) with charcoal-filtered air (CF) as a control or CF plus approximately 50-ppb ozone (CF + 50). After 28 days in the control chamber, three pots were transferred to OTCs with CF + 50 to include the exposure that was applied in the previous years as a control stress treatment. Five plants were raised in each pot. For each treatment, three replicate mesocosms were analyzed, thus including a total of 15 individual plants. Samples of the rhizosphere microbial community were collected 63 days after germination. All plants were watered regularly.
Sampling of rhizosphere bacteria and DNA extraction.
At the end of the ozone exposure, bacterial cells were collected from the rhizosphere samples. In the course of a destructive harvest, the roots of an entire mesocosm were carefully separated from the surrounding soil. The core regions of the established root system of all plants within a pot were collected and combined for each species and mesocosm (four individual plants). Rough soil particles were removed by dipping the roots for 30 to 60 s into tap water. The bacterial cells were then detached from the roots and adhering rhizosphere soil by washing 15 to 20 g root material in 300 ml sterile saline solution (0.85% NaCl [wt/vol]) for 0.5 h at 4°C in an orbital shaker (GFL, Burgwedel, Germany) at 20 rpm. After removal of the root material, the cell suspensions were centrifuged at 4,100 × g for 30 min. The supernatants were discarded, and the pellets were stored at −70°C until DNA extraction.
The bacterial cell pellet was lysed using five subsequent cycles of freeze-thawing, followed by proteinase K treatment and DNA phenol-chloroform extraction, as described in more detail previously (40). The DNA was then purified with a DNA purification kit (Wizard-DNA purification; Promega, Mannheim, Germany) and stored at −20°C. Alternatively, in the third year of the study (S.asper experiment), total genomic DNA was extracted with the BIO101 FastDNA SPIN kit for soil (Qbiogene, Carlsbad, Calif.) according to the manufacturer's instructions, comprising a bead beater treatment. With both methods, a pellet of 0.1 to 0.2 g (wet weight) yielded 100 μl of DNA solution in the range of 10 to 100 ng DNA μl−1.
PCR amplification of partial 16S rRNA genes.
For genetic profiling, a partial sequence of the 16S rRNA gene was PCR amplified. This sequence was located between positions 519 to 926, according to the numbering of the respective gene of Escherichia coli (7). The PCR product, which included the variable regions V4 and V5, was amplified by PCR with primers, universal for Bacteria (Com1 and Com2-Ph) (36, 40) (Table 1). The Com2-Ph primer was phosphorylated at the 5′ end for the following single-strand digestion (40). A reaction mixture of 50 μl contained 1× PCR buffer, 1.5 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate, 0.5 μM each primer, 2.5 U Platinum Taq-Polymerase (Invitrogen, Karlsruhe, Germany), and 1 μl of template (1:10 dilutions of the extracted DNA in 10 mM Tris-HCl, pH 8.0) (35). The PCR conditions were as follows: initial denaturation for 3 min at 94°C; 30 cycles, each consisting of denaturation for 1 min at 94°C, primer annealing for 1 min at 50°C, and a 70-s extension at 72°C; and a final elongation step of 5 min at 72°C. The thermocycling was conducted with 200-μl tubes in a MWG Primus 96 cycler (MWG Biotech, Ebersbach, Germany). All primers used in this study were synthesized by MWG Biotech. To decrease minor variations due to PCR variability, three independent PCRs were performed; subsequently, the PCR products were combined.
TABLE 1.
Primer specificities and their sequences used for PCR amplification of partial 16S rRNA genes in this study
| Targeted phylogenetic group | Primer name (forward and reverse) | Corresponding position in the E. coli rrn genea | Base sequence (5′-3′) | Reference |
|---|---|---|---|---|
| Bacteria | Com-1 | 519-536 | CAGCAGCCGCGGTAATAC | 40 |
| Com2-Ph | 907-926 | CCGTCAATTCCTTTGAGTTT | 40 | |
| Actinobacteria | F243HGC | 226-243 | GGATGAGCCCGCGGCCTA | 18 |
| R1387 | 1378-1401 | CGGTGTGTACAAGGCCCGGGAACG | 18 | |
| α-Proteobacteria | F203α | 175-218 | CCGCATACGCCCTACGGGGGAAAGATTTAT | 16 |
| R1492 | 1492-1513 | TACGG(C/T)TACCTTGTTACGACTT | 51 | |
| Pseudomonas | F311Ps | 290-311 | CTGGTCTGAGAGGATGATCAGT | 32 |
| R1459Ps | 1459-1478 | AATCACTCCGTGGTAACCGT | 32 |
According to Brosius et al. (7).
Partial rRNA genes from specific phylogenetic groups of bacteria were amplified by a nested PCR approach, described in detail previously (12). All specific primers are listed in Table 1. Briefly, in a first PCR, specific primers were used to target the DNA of a selected group. These PCR products then served as templates for a second amplification using the universal primers (Com1 and Com2-Ph). Thus, independent of the targeted phylogenetic group, PCR products from homologous regions of the 16S rRNA genes were generated, allowing a direct comparison of SSCP profiles with each other.
For the group-specific PCR amplifications, a reaction mixture of 25 μl was prepared with 1× PCR buffer, 1.5 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate, 0.5 μM each primer, 1.25 U Platinum Taq-Polymerase, and 0.5 μl template DNA (1:10 dilution of the original DNA solution). Thermal cycling started with an initial denaturation for 3 min at 94°C, followed by 25 cycles each consisting of denaturation at 94°C for 1 min; primer annealing, depending on the chosen primer pair (Table 1); a 2-min extension at 72°C; and a final elongation for 10 min at 72°C. To amplify the 16S rRNA genes of α-Proteobacteria, an annealing temperature of 56°C and an incubation time of 1 min were chosen. For the group of Actinobacteria, the annealing temperature was 63°C for 1 min. To obtain amplicons of the genus Pseudomonas, each cycle included an annealing of 2 min at 63°C. All PCR products were analyzed for their expected size and purity by electrophoresis on 1% agarose gels stained with ethidium bromide (35). The second PCR amplification was done with primers Com1 and Com2-Ph as described in the paragraph above, but here only with 25 cycles.
SSCP.
All PCR products subjected to SSCP analysis were purified with QIAquick spin columns (QIAGEN, Hilden, Germany) and quantified with PicoGreen reagent for double-stranded DNA (Molecular Probes, Eugene, Oreg.) according to the manufacturers' instructions. For single-strand digestion, up to 1,000 ng of DNA was digested for 45 min at 37°C in a total volume of 50 μl, with 2.5 U lambda exonuclease in 1× exonuclease buffer (New England Biolabs, Beverly, MA). The digestion was stopped with the first step of the purification with spin columns of the MinElute DNA cleanup system (QIAGEN).The columns were eluted with 10 μl of the supplied buffer; afterwards, 9 μl of SSCP loading buffer (10 mM NaOH, 0.25% xylene cyanol [wt/vol], 0.25% bromophenol blue [wt/vol], and 95% formamide [vol/vol]) was added to the eluate. The samples were denatured (2 min at 95°C, following 3 min on ice) directly before 6 μl of sample was loaded into each well. Based on the DNA concentrations of the PCR products, each lane of an SSCP gel received the same amount of total DNA to increase comparability of the SSCP profiles on a gel with each other.
The SSCP analysis was carried out following the protocol described in more detail previously (12). Briefly, the samples were separated on 0.4-mm-thick, 21-cm-long nondenaturing polyacrylamide gels of 0.625× MDE (Cambrex, East Rutherford, NJ) in 1× Tris-borate-EDTA (35). Electrophoresis was carried out for 17 h at 400 V (maximum, 8 mA) and at 20°C in a Macrophor (Amersham Bioscience, Freiburg, Germany). The SSCP profiles were visualized by silver staining (3).
Cloning and sequencing of partial 16S rRNA genes.
The sequences of 16S rRNA genes were determined either (i) directly from PCR products amplified from total bacterial DNA extracted from the rhizospheres or (ii) from bands selected from SSCP profiles. The PCR products from community DNA were cloned into competent cells of E. coli JM109 using the pGEM-T Vector system from Promega (Mannheim, Germany). The cloned PCR products were amplified with vector-specific primers as previously described (36). The amplified products were analyzed by amplified ribosomal DNA restriction enzyme analysis (ARDRA) (48). ARDRA groups were defined on the basis of DNA patterns obtained from two restriction endonucleases, i.e., AluI (Amersham Bioscience) and TacI (New England Biolabs, Beverly, MA).
Bands of SSCP profiles were cut out from dried gels, and the DNA was recovered by the crush and soak procedure (35). The DNA was then amplified by PCR with the primers Com1 and Com2-Ph, as described above. The PCR products were purified with QIAquick spin columns (QIAGEN; samples taken in 2000) or NucleoSpin Extract spin columns (Machery & Nagel; samples taken in 2002) and cloned as described above. The cloned DNA sequences were amplified with vector-specific primers (36), and the products were further amplified with Com1 and Com2-Ph. These products were subjected to another SSCP procedure to compare their electrophoretic mobility with the bands from the original community profiles. Only cloned PCR products in the expected positions were sequenced by cycle sequencing (36). The corresponding forward and reverse sequences were aligned with Consed software (http://www.phrap.org) to generate consensus sequences, which were then imported into the ARB software environment (24) for alignment and deletion of the primers. The sequences were compared to public databases using FASTA (http://www.ebi.ac.uk/fasta33/) or blastn (http://www.ncbi.nlm.nih.gov/blast/) and analyzed for chimera with the respective tool provided by the Ribosomal Database Project (8).
Gene probes and Southern blotting of SSCP gels.
The Southern blotting procedure for SSCP gels as used in this study has been described in detail previously (12, 37). Briefly, both plates enclosing the SSCP gel were treated with Repel-Silane (Amersham Bioscience). Sample preparation and electrophoresis were performed as described above, except that the gels were not stained after the electrophoretic run. Instead, the DNA separated by SSCP was transferred from the gels to positively charged nylon membranes (Hybond N+; Amersham Bioscience) by electroblotting using a Multiphor II (Amersham Bioscience) with a NovaBlot transfer unit, according to the manufacturer's instructions. The transfer conditions were set to 0.8 mA cm−2 for 5 h with 1× Tris-borate-EDTA. Membranes were dried at room temperature and DNA was cross-linked to the membrane with UV light (266 nm; FluoLink) at 1.5 J cm−2.
Specific probes to target the 16S rRNA gene between E. coli position 537 and 906 (7) were designed with the ARB software environment (24) and evaluated with OLIGO 6 software (MBI, Cascade, Colo.) and the Probe Match tool of the Ribosomal Database Project (8). Oligonucleotide probes were purchased from MWG Biotech and directly labeled with alkaline phosphatase using the AlkPhos Direct Labeling and Detection system (Amersham Bioscience). Membranes were hybridized in tubes, 4 cm in diameter and 30 cm long, with 20 ml hybridization buffer supplemented with 0.5 M NaCl and 100 ng labeled probe. The tubes were incubated in a hybridization oven (Oncor Appligene) for approximately 16 h at the temperature specified for each probe (Table 2).
TABLE 2.
Description of gene probes developed and applied in this study
| Probe name (target position)a | Targeted phylogenetic group | Nucleotide sequence (5′-3′) | Hybridization temp (°C) |
|---|---|---|---|
| STEN838 (838-857) | Xanthomonadaceae | GATACTGGGCACCAAGTTGTG | 40 |
| OXAL635 (635-656) | Oxalobacteriaceae | TTCTAGCCTTGCAGTCTCCATC | 45 |
| ORGA620 (620-637) | Plastids | ACCGCCTGTCCAGAGTTG | 40 |
According to E. coli position (7).
Stringency washes were performed with the buffers described in the AlkPhos protocol. For each washing step, 50 ml buffer was preheated to the desired temperature. The membranes were washed twice with the primary wash buffer at the hybridization temperature for 10 min. Two washes followed, each for 5 min at the hybridization temperature reduced by 10°C, in double-concentrated secondary wash buffer. Finally, the membranes were washed twice in secondary wash buffer at the hybridization temperature reduced by 15°C. Detection solution (CDP-Star) was spread on the membranes at a concentration of 40 μl cm−2 and incubated for 5 min. The membranes were placed into a plastic bag, which was subsequently sealed. For detection of the chemiluminescent signal, Hyperfilm ECL (Amersham Bioscience) was placed on top of the membrane in a film cassette and exposed for up to 17 h. The films were developed with LX 24 and then fixed with AL 4 (both from Kodak, Stuttgart, Germany).
Digital image and statistical analyses of SSCP profiles.
To calculate similarities between different SSCP profiles, GelCompar II software (Applied Bio-maths, Kortrijk, Belgium) was used. For each gel, individual background subtraction and median filtering with least-square filtering were applied as suggested by the software. The profiles were normalized with help of the SSCP migration markers (Fig. 1), and the Pearson correlation was used to calculate a similarity matrix based on pairwise comparison of densitometric curves. Mean similarity values and the respective standard deviations were calculated for replicate samples or for groups that were compared to each other (e.g., treatment compared to control). Statistical significance of variance was evaluated by one-way analysis of variance (ANOVA) using SigmaStat 2.03 software for Windows (SPSS Inc., San Rafael, CA) by comparing mean similarity values of different treatments to the values generated from all treatments to be compared.
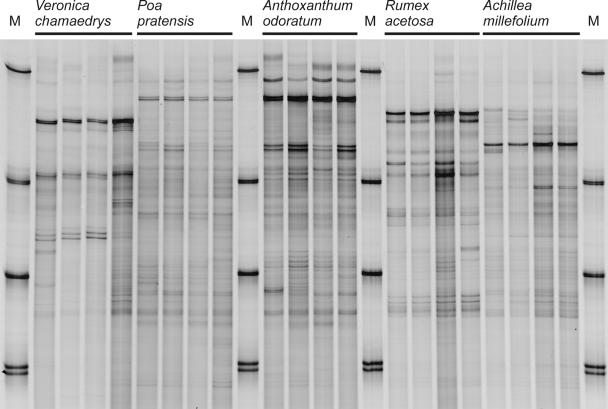
FIG. 1.
SSCP profiles of rhizosphere bacterial communities from the ozone control treatment (CF + 25) of Veronica chamaedrys, Poa pratensis, Anthoxanthum odoratum, Rumex acetosa, and Achillea millefolium. Four independent replicates are shown for each species. The 16S rRNA genes were PCR amplified with the universal primers Com1 and Com2-Ph. M, lanes with SSCP migration markers, which consisted of PCR-amplified rRNA genes from pure cultures of (from top to bottom) Bacillus licheniformis, Rhizobium trifolii, Flavobacterium johnsoniae, and Rhizobium radiobacter (double band).
For comparison of the intensities of single bands in silver-stained SSCP gels and on the Southern blotted membranes, band intensities were quantified by the tool included in GelCompar. Intensities of the ozone treatments were compared to the values of the controls by pairwise comparison with a t test (SigmaStat for Windows).
Calculation of phylogenetic trees.
Phylogenetic trees were calculated with ARB software (24). The ARB database (ssjun02.arb) was updated with the closest related 16S rRNA gene sequences of the respective target sequences found by database searches. Maximum-likelihood trees (FastDNA ML) were calculated from selected sequences of at least 1,000 nucleotides by applying the respective column filters. Conservation filters of 50% minimal similarity were generated for each tree on the respective sequences with >1,000 nucleotides, with all options at the “don't use when maximum” setting. Shorter sequences were added to the trees with the parsimony interactive option, including local optimization and the respective conservation filter.
Nucleotide sequence accession number.
The sequences determined in this study have been deposited in the GenBank database under the accession numbers AJ874129 to AJ874261 and AJ937631 to AJ937668.
RESULTS
Evaluation of the PCR-SSCP method.
The yield of PCR-amplifiable DNA obtained from rhizosphere samples ranged from 0.2 to 1.5 μg DNA g−1 of fresh roots, depending on the plant species, their respective total root morphology, and biomass. Repeated PCR amplifications of template DNA from the same DNA extract, as well as repeated SSCP analyses of the same PCR products, produced highly similar fingerprint profiles, indicating that the approach was well reproducible (data not shown). In addition, the differences between the replicate samples (pots) from the same treatments within an OTC and between OTCs were small, confirming a high level of reproducibility of the entire procedure (Fig. 1 and 2). Mean similarity values of SSCP profiles from independent replicates analyzed by pairwise comparisons ranged from 76.3% ± 7.0% (Poa pratensis) to 94.1% ± 1.8% (Tanacetum vulgare), depending on the plant species. Only the replicate profiles of Matricaria chamomilla incubated under ozone stress (NF + 50) showed lower mean similarities to each other (54.5% ± 19.2%).
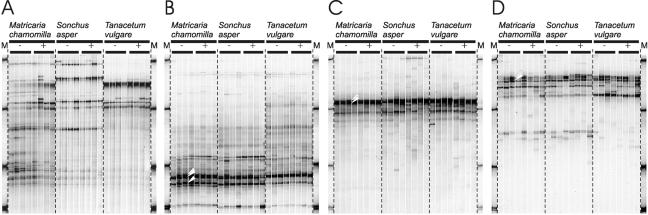
FIG. 2.
SSCP profiles of rhizosphere bacterial communities from Matricaria chamomilla, Sonchus asper, and Tanacetum vulgare from the ozone control treatments (−, CF + 25) and treatments with ozone-enriched air (+, NF + 50), four replicates each treatment. These gels were normalized for the position of the SSCP migration markers (see Fig. 1 for markers). The profiles were generated by PCR of 16S rRNA genes from the following groups according to different primer specificities (Table 1), i.e., Bacteria (A), α-Proteobacteria (B), Actinobacteria (C), and Pseudomonas (D). The arrowheads indicate sequences similar to the following species, from top to bottom: Caulobacter henricii, Rhizobium giardinii, and Devosia riboflavina (B); Streptomyces lincolnensis and Streptomyces turgidiscabies (C); and Pseudomonas sp. (D).
To prove the specificity of the nested PCR approach for targeting different phylogenetic groups, amplicons from the second, nonspecific PCR were cloned and differentiated by ARDRA, and different ARDRA types were sequenced. All clones predicted to be among the α-Proteobacteria (39 clones) affiliated within this group (AJ874253 to AJ874255 and AJ937631 to AJ937641). The accuracy of the approach targeting Pseudomonas (30 clones) was 87% (AJ 874258 and AJ937642 to AJ937653), with all non-Pseudomonas sequences affiliating with the Xanthomonadaceae (AJ937654 to AJ937655).The PCR targeting the Actinobacteria (34 clones) had an accuracy of 77% for this phylum (AJ874256, AJ874253, and AJ937656 to AJ937662); all sequences outside of the Actinobacteria belonged to the phylum Verrucomicrobia and within this phylum only to the family Verrucomicrobiaceae (AJ937663 to AJ937668). An in silico analysis using the ARB program confirmed the potential detection of all nontarget sequences reported here (data not shown). Despite the detection of nonspecific sequences, the chosen primer pairs of this study were regarded as sufficiently specific to distinguish the effect of ozone stress on different phylogenetic groups within the Bacteria.
Effects of ozone on rhizosphere bacterial communities of tolerant plant species.
The ozone-tolerant plant species (Anthoxanthum odoratum, Achillea millefolium, Poa pratensis, Rumexacetosa, and Veronica chamaedrys) exhibited no symptoms in response to an incubation with daily ozone episodes (NF + 50) over a period of 5 weeks (data not shown). The SSCP analysis of the rhizosphere bacterial community, based on the universal primers (Com1 and Com2-Ph) targeting all Bacteria, generated profiles which consisted of 15 to 30 detectable bands, depending on the plant species. The profiles from bacterial communities of ozone-stressed plants (NF + 50) were indistinguishable from those of the unstressed controls (CF + 25), as judged on the basis of visual inspection of the profiles and on ANOVA of similarity values between densitometric curves (Fig. 1; data of the stress treatment not shown). The SSCP profiles were highly specific for each plant species, indicating the sensitivity of this approach to detect structural differences in the bacterial rhizosphere communities.
To characterize the diversity of bacteria that was detected with the SSCP profiles, dominant bands from those profiles were chosen for DNA sequencing. From a total of 124 clones selected, most sequences (53.2%) fell into the phylum Proteobacteria; among these, most were attributed to members of the α-Proteobacteria (26.6% of all clones analyzed), while 9.7% were related to the β-Proteobacteria and 6.9% were related to the γ-Proteobacteria (see the supplemental material). Bacteroidetes (19.4%) and Actinobacteria (8.9%) were other prominent phyla that contributed to the profiles. About 60% of the sequences affiliated with sequences from noncultivated organisms. Interestingly, 50% of all sequences were most closely related to 16S rRNA genes that had been detected in bacteria from soil or sediment, while of the other 50%, almost half affiliated with sequences obtained from rhizosphere bacteria (see the supplemental material).
Rhizosphere bacterial communities of ozone-sensitive plants.
Before the ozone exposure, all plant species included were healthy with no leaf injuries. After an exposure to daily elevated ozone episodes over a period of 6 weeks, however, all plants from the elevated ozone treatments showed visible injuries on the leaves, as well as an increased ratio of chlorotic and senescent leaves compared to plants taken from OTCs with the control treatment (CF + 25) (data not shown). The SSCP profiles generated from the Bacteria-specific amplicons did not differ between the controls and the ozone-stressed plants (ANOVA of mean similarity values; P < 0.05), despite ozone-specific damage to the individual plants (Fig. 2A). As found with the ozone-tolerant plants, each of the ozone-sensitive plants harbored a structurally different bacterial community. The complexity of the profiles with 15 to 30 bands was similar to that of the ozone-tolerant plants.
Nested-PCR approaches were applied to increase the sensitivity of detection for specific members of phylogenetic groups suspected to be quantitatively and possibly qualitatively important, i.e., α-Proteobacteria, Actinobacteria, and Pseudomonas. The use of this approach reduced the complexity of the profiles to <15 distinguishable bands. With each targeted phylogenetic group, specific profiles were obtained (Fig. 2B to D). Pairwise comparisons of the similarity matrices with ANOVA (P < 0.05) indicated a response to ozone exposure only for S.asper-derived profiles targeting Actinobacteria. All other profiles were unaffected.
Interestingly, the SSCP profiles that were generated by nested PCR were less specific for each plant species than the profiles for the Bacteria, as indicated by higher similarity values for the respective specific profiles from all plants than those calculated for the Bacteria-specific profiles (data not shown). Thus, bacteria from other than the selected phylogenetic groups were more sensitive to different host plants. The profiles targeting the α-Proteobacteria (Fig. 2B) exhibited some specificity for T. vulgare compared to the other two species, the latter being more similar to each other. On the other hand, several dominant bands from the group-specific profiles were seen in the Bacteria-specific profiles, indicating their quantitative contribution to total bacterial diversity.
Sequencing of the dominant bands found in the α-Proteobacteria indicated the prevalence of bacteria that could be attributed to the genera Caulobacter (AJ874255), Rhizobium (AJ874254), and Devosia (AJ8742453) (bands marked in Fig. 2B). These dominant bands were also detected in the profiles of the universal analysis (Fig. 2A) where they were strong in the profiles of M. chamomilla, but weaker in those of S. asper and T. vulgare. The dominant bands from the SSCP profiles targeting the Actinobacteria (Fig. 2C) were closely related or identical to 16S rRNA genes of Streptomyces lincolnensis (AJ874256) and Streptomyces turgidiscabies (AJ874257). Comparable bands were detected in the profiles of the universal analysis of all plant species, indicating that they contributed to the dominant members of the total bacterial rhizosphere community. In contrast, the dominant bands of the analysis targeting Pseudomonas (Fig. 2D) were hardly detectable in profiles obtained with universal primers, suggesting a quantitatively minor role of this group in the total rhizosphere bacterial community. Sequencing of the dominant bands, indicated in Fig. 2D, revealed a single sequence (AJ874258) which was identical to the respective gene of Pseudomonas fluorescens, Pseudomonas tolaasii, and Pseudomonas corrugata, respectively.
Effect of continuous ozone exposure on rhizosphere bacteria.
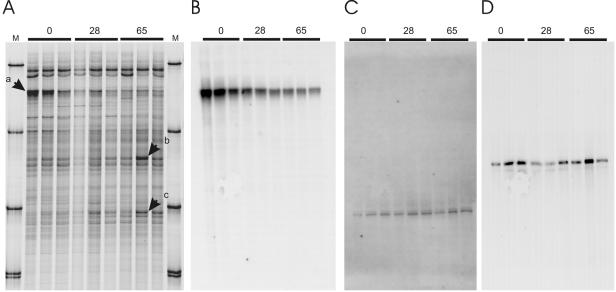
The ozone-sensitive species S. asper was successfully raised from seeds in the OTCs under continuous ozone fumigation (CF + 50). As expected, all plants exposed to ozone showed visible injuries in contrast to the healthy plants of the control treatment. The different SSCP profiles generated with the Bacteria-specific primers from the communities of the rhizospheres from ozone-stressed and control plants were very similar and not distinguishable by statistical methods (ANOVA; P < 0.05). However, some minor variations occurred in the intensities of three single bands (Fig. 3A). Band a in the upper part of the profiles was stronger in the samples of the control experiment (t test; P < 0.05), whereas the intensities of band b in the middle of the profile and band c at the bottom seemed to increase with prolonged ozone exposure (Fig. 3A). However, these latter differences could statistically not be confirmed.
FIG. 3.
SSCP profiles of PCR-amplified rRNA genes of bacterial communities from the rhizosphere of Sonchus asper germinated and raised from seeds under ozone exposure (65 days of exposure) or in the control chamber (0 days of exposure) or germinated from seeds in the control chamber and moved after 37 days to the chamber with elevated tropospheric ozone levels (28 days of exposure); three independent replicates of each. (A) Silver-stained SSCP gels; (B to D) Southern blot analysis with the ORGA620 probe (B), STEN838 probe (C), and the OXAL635 probe (D). For description of lanes M, see the legend to Fig. 1.
All three bands were chosen for further identification by being cloned and sequenced. The sequence of band a (clone 306_a_1; AJ874259) affiliated with the 16S rRNA genes of chloroplasts from Atropa belladonna (AJ316582) and Panax ginseng (AY582139), both with 99.5% similarity. It should be noted that sequences from the plastids of S. asper or other plants of its family (Asteraceae) or order (Asterales) were missing in the public database. The sequence from band b (clone 306_e_5; AJ874261) was identical to several different sequences within the β-Proteobacteria, among them the three species Janthinobacterium agaricidamnosum, Massilia timonae, and Oxalobacter sp. p8E, as illustrated in the supplemental material. The sequence from band c (clone 306_f_1; AJ874260) was 98.7% similar to a sequence of an uncultured clone of a Xanthomonas-related organism (AF467297) (see the supplemental material). The similarity to the closest cultivated isolate, i.e., Xanthomonas campestris (AF290420), was only 95.7%.
As band intensities are only an indicator of abundance but not proof (37), the relevant SSCP gel was blotted onto a membrane and successively hybridized with gene probes, specifically developed from the three respective sequences (see Materials and Methods). Labeled probes discovered the corresponding bands in all lanes and were specific only for these targeted bands. The probe ORGA620 which targeted the 16S rRNA gene of plastids (band a) revealed that the intensity of the bandin fact decreased with prolonged ozone exposure (t test) (Fig. 3B). In contrast, the intensity of band b, which seemed to increase under ozone exposure in the silver-stained SSCP profiles, did not hybridize accordingly with probe OXAL635. Thus, it was likely that the band variation was caused by an additional sequence that was not detected with the selected gene probe (Fig. 3D). However, the hybridization with probe STEN838 targeting the Xanthomonas-like sequence retrieved from band c in fact detected a slight increase in the intensity of this band with exposure to ozone (t test; P < 0.05) (Fig. 3C). Thus, the quantitative stimulation of this as-yet-uncultured Xanthomonas-related organism by ozone stress was confirmed.
DISCUSSION
Ozone has been considered the most important phytotoxic air pollutant in industrialized countries like the United States and Europe (28, 44); thus, it is important to understand whether ozone also affects below-ground processes, i.e., those mediated by the soil microbial community. Ozone itself is a bactericidal compound widely used for decontamination (17). However, a direct effect of ozone on soil microorganisms can only be of minor importance, as soil and vegetation remove ozone from the atmosphere, producing a vertical gradient of decreasing ozone concentrations towards the ground (47) and preventing the penetration of ozone into the soil to any appreciable extent (6).
The rhizosphere represents a microbial habitat in soil that is most strongly influenced by plants. It has been shown in several studies that the composition of root exudates, which mainly serve as carbon and energy sources of bacteria and fungi, has a strong influence on the structural diversity of bacteria inhabiting this ecological niche (22, 31, 38, 40, 42). Following the conclusion of Hofstra and coworkers (19) that below-ground processes may be changed by elevated levels of trophospheric ozone before symptoms on plants become detectable, this study first examined the structural diversity of bacteria from the rhizospheres of ozone-tolerant plants. The ozone stress chosen in this study corresponded with the normal elevated ozone concentrations that can be found in the summer in Germany (nonfiltered air plus 50-ppb ozone). As a control, these plants were compared to charcoal-filtered air plus 25 ppb of ozone to avoid the detection of ozone stress caused by a lack of ozone. It turned out that the elevated levels of ozone did not select for a different bacterial community composition, as seen by a diversity of approximately 30 different SSCP bands representing different ribosomal rRNA genes. Despite known biases generated by the use of PCR amplifications of partial rRNA genes from mixed templates (13, 36, 45, 49), it can be assumed that most of the amplified rRNA genes were derived from bacteria with a considerable, if not dominant, abundance in the rhizospheres. In contrast to the lack of detection of an effect caused by ozone exposure, the profiling technique was sensitive enough to detect differences between the four plant species investigated in this context.
Plant species-specific effects were also seen in profiles from ozone-sensitive plants incubated under the same conditions. As expected, these plants exhibited typical ozone-specific leaf injuries and chlorosis, as they have been described in more detail elsewhere (5). With the focus on the bacterial diversity in the rhizospheres, however, these differences between ozone-stressed and unstressed controls were generally not seen below ground. Neither Bacteria-specific profiles nor those with a more narrow phylogenetic range picked up an ozone-triggered difference, with the exception of the Actinobacteria in the rhizosphere of S. asper. These differences were picked up by statistical analysis of digitalized SSCP profiles, but they could not be attributed to the presence or absence of specific bands. All selected phylogenetic groups chosen in this study, i.e., Actinobacteria, α-Proteobacteria, and Pseudomonas, can typically be found as members of the bacterial communities in rhizospheres (15, 18, 20, 37). However, other equally important groups, i.e., the β-Proteobacteria and γ-Proteobacteria other than Pseudomonas-Xanthomonas or Bacteroidetes were left out of this study; sequencing of several SSCP bands from the Bacteria-specific profiles indicated their abundance in the rhizospheres of the selected plants of this study. Thus, it may be possible that ozone-specific effects on those groups were missed. However, dramatic quantitative changes are unlikely, as the profiles encompassing all Bacteria were also unaffected.
The result that the ozone-damaged plants harbored the same structural diversity as the controls was quite surprising, as it is well documented that ozone stress affects the carbon allocation between shoot and root (1, 10), as well as rhizodeposition (29, 30). Obviously, for the herbaceous plants tested in this study, these signals were not sufficient to modify the structural composition of the bacterial community. In some recent studies based on genetic profiling of PCR-amplified 16S rRNA genes, it could be shown that this technique is sensitive enough to distinguish even between bacterial communities of the same plant but of different ages (4, 16, 38, 42). Thus, it can be assumed that the applied technique was sufficiently sensitive to detect a substantially different root exudation. This assumable minor effect of ozone is corroborated by a study of microbial communities found beneath trees after incubation at elevated levels of tropospheric ozone over a period of 3 years (34). In that study, phospholipid fatty acid (PFLA) analysis of the microbial communities revealed that the relative proportions of gram-positive and gram-negative indicator PFLAs were not affected. In contrast, a reduction of fungal PFLAs was observed in that study, indicating that fungi are possibly more responsive in the rhizospheres of ozone-stressed plants than bacteria.
Provoked by the inertia of the bacterial community structure in response to ozone stress under a bad but realistic scenario, another experiment was included to increase the vigorousness of ozone stress. The ozone-sensitive plant S. asper was raised under continuous presence of increased levels of ozone in a background of charcoal-filtered air (CF + 50), whereas the control received ozone-free air. It should be noted that for S.asper, in contrast to other plants, a lack of ozone does not result in any symptoms of stress (5). An intermediate treatment was included with plants germinated and grown for 28 days in ozone-free air and then transferred to continuous ozone exposure. Despite visible injuries under ozone exposure, only 3 bands of a total of >35 bands of the SSCP profiles presumably changed, with 2 bands increasing in intensity with ozone and 1 band decreasing. Gene probe hybridization revealed that the band decreasing in intensity was related to plastids and a band increasing in intensity was related to a member of the Xanthomonas group. The gene probe targeting the third band did not respond to the treatment, indicating that the evaluation of band intensities may not always be sufficient for quantitative interpretations of genetic profiles. The changing band intensity may have been caused by additional comigrating sequences, as has been described in other studies based on SSCP or denaturing gradient gel electrophoresis (37, 41).
Plastids in root cells typically store starch as an energy reservoir. The decrease in the plastids of root cells from ozone-stressed S. asper plants is in accordance with the observation that was made with two different tree species, in which a decrease in starch content of the roots was detected in response to ozone stress (2, 9). The only bacterium that was detected to be more abundant in the rhizospheres of ozone-stressed plants was a member of the Xanthomonas group. Although phylogenetic affiliation can at best be suggestive of the biology of an organism, the increase in abundance of a Xanthomonas, a genus that harbors plant pathogenic bacteria (33, 43), raises the question of an elevated sensitivity to pathogens of the plant in response to ozone. On the other hand, it has been reported that ozone may reduce the infection severity of bacterial plant diseases (27), which is in agreement with the observation that ozone triggered the pathogen defense pathway (52). Thus, an increase in a single organism from the Xanthomonas group may simply be the effect of accelerated senescence triggered by ozone stress, as it has been described at the symptomatic level for another herbaceous plant (26).
Supplementary Material
Acknowledgments
We thank Elke Bergmann and Jürgen Bender for their kind help and assistance concerning all botanical aspects of this project; we thank both of them, as well as Hans-Joachim Weigel, for discussions. Carina Trenkler and Evelin Schummer provided valuable technical assistance, which we gratefully acknowledge.
This work was supported by the European Commission (BIOSTRESSproject; contract no. EVK2-CT-1999-00040).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Andersen, C. P. 2000. Ozone stress and changes below-ground: linking root and soil processes. Phyton 40:7-12. [Google Scholar]
- 2.Andersen, C. P., R. Wilson, M. Plocher, and W. E. Hogsett. 1997. Carry-over effects of ozone on root growth and carbohydrate concentrations of ponderosa pine seedlings. Tree Physiol. 17:805-811. [DOI] [PubMed] [Google Scholar]
- 3.Bassam, B. J., G. Caetano-Anolles, and P. M. Gresshoff. 1991. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal. Biochem. 196:80-83. [DOI] [PubMed] [Google Scholar]
- 4.Baumgarte, S., and C. C. Tebbe. 2005. Field studies on the environmental fate of the Cry1Ab Bt toxin produced by transgenic maize (MON810) and its effect on bacterial communities in the maize rhizosphere. Mol. Ecol. 14:2539-2551. [DOI] [PubMed] [Google Scholar]
- 5.Bergmann, E., J. Bender, and H. J. Weigel. 1999. Ozone threshold doses and exposure-response relationships for the development of ozone injury symptoms in wild plant species. New Phytol. 144:423-435. [DOI] [PubMed] [Google Scholar]
- 6.Blum, U., and D. T. Tingey. 1977. Study of potential ways in which ozone could reduce root-growth and nodulation of soybean. Atmos. Environ. 11:737-739. [Google Scholar]
- 7.Brosius, J., T. J. Dull, D. D. Sleeter, and H. F. Noller. 1981. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. [DOI] [PubMed] [Google Scholar]
- 8.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coleman, M. D., R. E. Dickson, J. G. Isebrands, and D. F. Karnosky. 1995. Carbon allocation and partitioning in aspen clones varying in sensitivity to tropospheric ozone. Tree Physiol. 15:593-604. [DOI] [PubMed] [Google Scholar]
- 10.Cooley, D. R., and W. J. Manning. 1987. The impact of ozone on assimilate partitioning in plants—a review. Environ. Pollut. 47:95-113. [DOI] [PubMed] [Google Scholar]
- 11.Davison, A. W., and J. D. Barnes. 1998. Effects of ozone on wild plants. New Phytol. 139:135-151. [Google Scholar]
- 12.Dohrmann, A. B., and C. C. Tebbe. 2004. Microbial community analysis by PCR-single-strand conformation polymorphism (PCR-SSCP), p. 809-838. In G. A. Kowalchuk, F. J. de Bruijn, I. M. Head, A. D. Akkermans, and J. D. van Elsas (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 13.Farrelly, V., F. A. Rainey, and E. Stackebrandt. 1995. Effect of genome size and rrn gene copy number on PCR amplification of 16S ribosomal RNA genes from a mixture of bacterial species. Appl. Environ. Microbiol. 61:2798-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuhrer, J., L. Skarby, and M. R. Ashmore. 1997. Critical levels for ozone effects on vegetation in Europe. Environ. Pollut. 97:91-106. [DOI] [PubMed] [Google Scholar]
- 15.Germida, J. J., and S. D. Siciliano. 2001. Taxonomic diversity of bacteria associated with the roots of modern, recent and ancient wheat cultivars. Biol. Fertil. Soils 33:410-415. [Google Scholar]
- 16.Gomes, N. C. M., H. Heuer, J. Schonfeld, R. Costa, L. MendoncaHagler, and K. Smalla. 2001. Bacterial diversity of the rhizosphere of maize (Zea mays) grown in tropical soil studied by temperature gradient gel electrophoresis. Plant Soil 232:167-180. [Google Scholar]
- 17.Guzel-Seydim, Z. B., A. K. Greene, and A. C. Seydim. 2004. Use of ozone in the food industry. Lebensm. Wiss. Technol. 37:453-460. [Google Scholar]
- 18.Heuer, H., M. Krsek, P. Baker, K. Smalla, and E. M. H. Wellington. 1997. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 63:3233-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofstra, G., A. Ali, R. T. Wukasch, and R. A. Fletcher. 1981. The rapid inhibition of root respiration after exposure of bean (Phaseolus vulgaris L.) plants to ozone. Atmos. Environ. 15:483-487. [Google Scholar]
- 20.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kogel-Knabner, I. 2002. The macromolecular organic composition of plant and microbial residues as inputs to soil organic matter. Soil Biol. Biochem. 34:139-162. [Google Scholar]
- 22.Kowalchuk, G. A., D. S. Buma, W. de Boer, P. G. L. Klinkhamer, and J. A. van Veen. 2002. Effects of above-ground plant species composition and diversity on the diversity of soil-borne microorganisms. Antonie Leeuwenhoek 81:509-520. [DOI] [PubMed] [Google Scholar]
- 23.Loya, W. M., K. S. Pregitzer, N. J. Karberg, J. S. King, and C. P. Giardina. 2003. Reduction of soil carbon formation by tropospheric ozone under increased carbon dioxide levels. Nature 425:705-707. [DOI] [PubMed] [Google Scholar]
- 24.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynch, J. M. 1990. The rhizosphere. Wiley, New York, N.Y.
- 26.Manning, W. J. 1978. Chronic foliar ozone injury: effects on plant root development and possible consequences. Calif. Air Environ. 7:3-4. [Google Scholar]
- 27.Manning, W. J., and A. von Tiedemann. 1995. Climate-change—potential effects of increased atmospheric carbon-dioxide (CO2), ozone (O3), and ultraviolet-B (Uv-B) radiation on plant-diseases. Environ. Pollut. 88:219-245. [DOI] [PubMed] [Google Scholar]
- 28.Marenco, A., H. Gouget, P. Nedelec, J. P. Pages, and F. Karcher. 1994. Evidence of a long-term increase in tropospheric ozone from Pic du Midi data series—consequences: positive radiative forcing. J. Geophys. Res. Atmos. 99:16617-16632. [Google Scholar]
- 29.McCool, P. M., and J. A. Menge. 1983. Influence of ozone on carbon partitioning in tomato—potential role of carbon flow in regulation of the mycorrhizal symbiosis under conditions of stress. New Phytol. 94:241-247. [Google Scholar]
- 30.McCrady, J. K., and C. P. Andersen. 2000. The effect of ozone on below-ground carbon allocation in wheat. Environ. Pollut. 107:465-472. [DOI] [PubMed] [Google Scholar]
- 31.Miethling, R., G. Wieland, H. Backhaus, and C. C. Tebbe. 2000. Variation of microbial rhizosphere communities in response to crop species, soil origin, and inoculation with Sinorhizobium meliloti L33. Microb. Ecol. 40:43-56. [DOI] [PubMed] [Google Scholar]
- 32.Milling, A., K. Smalla, F. X. Maidl, M. Schloter, and J. C. Munch. 2004. Effects of transgenic potatoes with an altered starch composition on the diversity of soil and rhizosphere bacteria and fungi. Plant Soil 266:23-39. [Google Scholar]
- 33.Palleroni, N. J. 1981. Introduction to the family Pseudomonadaceae, p. 655-665. In M. P. Starr, H. Stolp, H. G. Trüper, A. Balows, and H. G. Schlegel (ed.), The prokaryotes. Springer-Verlag, Berlin, Germany.
- 34.Phillips, R. L., D. R. Zak, W. E. Holmes, and D. C. White. 2002. Microbial community composition and function beneath temperate trees exposed to elevated atmospheric carbon dioxide and ozone. Oecologia 131:236-244. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Schmalenberger, A., F. Schwieger, and C. C. Tebbe. 2001. Effect of primers hybridizing to different evolutionarily conserved regions of the small-subunit rRNA gene in PCR-based microbial community analyses and genetic profiling. Appl. Environ. Microbiol. 67:3553-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmalenberger, A., and C. C. Tebbe. 2003. Bacterial diversity in maize rhizospheres: conclusions on the use of genetic profiles based on PCR-amplified partial small subunit rRNA genes in ecological studies. Mol. Ecol. 12:251-261. [DOI] [PubMed] [Google Scholar]
- 38.Schmalenberger, A., and C. C. Tebbe. 2003. Genetic profiling of noncultivated bacteria from the rhizospheres of sugar beet (Beta vulgaris) reveal field and annual variability but no effect of a transgenic herbicide resistance. Can. J. Microbiol. 49:1-8. [DOI] [PubMed] [Google Scholar]
- 39.Schmalenberger, A., and C. C. Tebbe. 2002. Bacterial community composition in the rhizosphere of a transgenic, herbicide resistant maize (Zea mays) and comparison to its non-transgenic cultivar Bosphore. FEMS Microbiol. Ecol. 40:29-37. [DOI] [PubMed] [Google Scholar]
- 40.Schwieger, F., and C. C. Tebbe. 1998. A new approach to utilize PCR-single-strand-conformation polymorphism for 16s rRNA gene-based microbial community analysis. Appl. Environ. Microbiol. 64:4870-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sekiguchi, H., N. Tomioka, T. Nakahara, and H. Uchiyama. 2001. A single band does not always represent single bacterial strains in denaturing gradient gel electrophoresis analysis. Biotechnol. Lett. 23:1205-1208. [Google Scholar]
- 42.Smalla, K., G. Wieland, A. Buchner, A. Zock, J. Parzy, S. Kaiser, N. Roskot, H. Heuer, and G. Berg. 2001. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl. Environ. Microbiol. 67:4742-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Starr, M. P. 1981. The genus Xanthomonas, p. 742-763. In M. P. Starr, H. Stolp, H. G. Trüper, A. Balows, and H. G. Schlegel (ed.), The prokaryotes. Springer-Verlag, Berlin, Germany.
- 44.Stockwell, W. R., G. Kramm, H. E. Scheel, V. A. Mohnen, and W. Seilev. 1997. Ozone formation, destruction and exposure in Europe and the United States, p. 1-38. In H. Sandermann, A. R. Wellburn, and L. R. Heath (ed.), Ozone and forest decline: a comparison of controlled chamber and field experiments. Springer-Verlag, Berlin, Germany.
- 45.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tebbe, C. C., A. Schmalenberger, S. Peters, and F. Schwieger. 2001. Single-strand conformation polymorphism (SSCP) for microbial community analysis, p. 161-175. In P. A. Rochelle (ed.), Environmental molecular microbiology: protocols and applications. Horizon Scientific Press, Wymondham, United Kingdom.
- 47.Turner, N. C., P. E. Waggoner, and S. Rich. 1974. Removal of ozone from atmosphere by soil and vegetation. Nature 250:486-489. [Google Scholar]
- 48.Vaneechoutte, M., R. Roussau, P. De Vos, M. Gillis, D. Janssens, N. Paepe, A. De Rouck, T. Fiers, G. Claeys, and K. Kesters. 1992. Rapid identification of bacteria of the Comamonadaceae with amplified ribosomal DNA-restriction analysis (ARDRA). FEMS Microbiol. Lett. 93:227-234. [DOI] [PubMed] [Google Scholar]
- 49.von Wintzingerode, F., U. B. Göbel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 50.Weigel, H. J., and H. J. Jäger. 1988. Ecotoxicological aspects of air-pollutants. 2. Design of an open-top field chamber system to investigate the effects of air-pollutants on plants. Landbauforsch. Volk. 38:182-195. [Google Scholar]
- 51.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wohlgemuth, H., K. Mittelstrass, S. Kschieschan, J. Bender, H. J. Weigel, K. Overmyer, J. Kangasjarvi, H. Sandermann, and C. Langebartels. 2002. Activation of an oxidative burst is a general feature of sensitive plants exposed to the air pollutant ozone. Plant Cell Environ. 25:717-726. [Google Scholar]
- 53.Yoshida, L. C., J. A. Gamon, and C. P. Andersen. 2001. Differences in above-and below-ground responses to ozone between two populations of a perennial grass. Plant Soil 233:203-211. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.