Abstract
Glucooligosaccharide oxidase from Acremonium strictum was screened for potential applications in oligosaccharide acid production and carbohydrate detection. This protein is a unique covalent flavoenzyme which catalyzes the oxidation of a variety of carbohydrates with high selectivity for cello- and maltooligosaccharides. Kinetic measurements suggested that this enzyme possesses an open carbohydrate-binding groove, which is mainly composed of two glucosyl-binding subsites. The encoding gene was subsequently cloned, and one intron was detected in the genomic DNA. Large amounts of active enzymes were expressed in Pichia pastoris, with a yield of 300 mg per liter medium. The protein was predicted to share structural homology with plant cytokinin dehydrogenase and related flavoproteins that share a conserved flavin adenine dinucleotide (FAD)-binding domain. The closest sequence matches are those of plant berberine bridge enzyme-like proteins, particularly the characteristic flavinylation site. Unexpectedly, mutation of the putative FAD-attaching residue, H70, to alanine, serine, cysteine, and tyrosine did not abolish the covalent FAD linkage and had little effect on the Km. Instead, the variants displayed kcat values that were 50- to 600-fold lower, indicating that H70 is crucial for efficient redox catalysis, perhaps through modulation of the oxidative power of the flavin.
Sugar oxidases and dehydrogenases that catalyze the oxidation of glucose and other carbohydrates into the corresponding lactones are widely distributed in nature. These enzymes have received great attention as potential diagnostic reagents and industrial biocatalysts. For example, glucose oxidase (GOX) is widely used in analytical biochemistry and in the food industry (21, 22). A search of the BRENDA enzyme database (23) revealed that most of these enzymes are specific for a variety of mono- and disaccharides, and only a few enzymes exhibit high specificity with oligosaccharides. The latter enzymes include galactose oxidase, cellobiose dehydrogenase (CDH), and glucooligosaccharide oxidase (GOOX). GOOX from Acremonium strictum was identified from sugar oxidase-producing microorganisms with the aim of identifying enzymes with potential applications in oligosaccharide acid production and alternative carbohydrate assays (16).
GOOX is a monomeric glycoprotein with a covalently linked flavin adenine dinucleotide (FAD) (16). It catalyzes the oxidization of a variety of carbohydrates with concomitant reduction of molecular oxygen to hydrogen peroxide. Screening of 30 monosaccharides and derivatives of these compounds showed that d-glucose is the only good substrate. In terms of the disaccharides, maltose, cellobiose, and lactose with reducing-end glucosyl residues linked by an α bond or β-1,4 bonds are good substrates, whereas disaccharides containing other linkage types are not. Moreover, GOOX can react with maltooligosaccharides composed of α-1,4-linked d-glucopyranosyl residues (up to at least seven units); hence, the name of this novel oxidase. The broad substrate specificity of GOOX, particularly with oligosaccharides, suggests that it may have great potential applicability.
GOOX shares some substrate specificity with GOX, CDH, and pyranose oxidase (POX), which exhibit significant structural homology (10, 11, 25). Because of the unique catalytic properties and potential industrial use of A. strictum GOOX, in the present study we isolated the native protein for measurement of the kinetic parameters and identification of internal peptide sequences, cloned and expressed the encoding gene, and analyzed the putative flavinylation residue by site-directed mutagenesis.
MATERIALS AND METHODS
Preparation, activity assay, and cyanogen bromide digestion of the native protein.
The methods used for fungal culture, protein isolation, and the enzyme activity assay were the methods reported previously (16). Briefly, the A. strictum T1 strain was grown in a wheat bran culture at 28°C for 5 to 7 days. The native GOOX from the culture broth was first fractionated by ammonium sulfate precipitation and purified by Fractogel DEAE-650, Toyopearl Phenyl-650, Biogel P-100, and Ultragel HA hydroxyapatite chromatography. The kinetic parameters were measured by determining H2O2 production by coupling to a peroxidase enzyme assay (Table 1). The enzyme was incubated in 50 mM Tris-HCl buffer (pH 8.0) containing various substrates, 2 U peroxidase, 0.1 mM 4-aminoantipyrine, and 1 mM phenol at 37°C, and the absorbance at 500 nm was monitored. One unit of enzyme activity was defined as the amount of enzyme that produced 1 μmol H2O2 per min. For cyanogen bromide digestion, the purified enzyme was dissolved in 70% (vol/vol) formic acid containing 1% (wt/vol) CNBr and incubated at room temperature for 24 h. The resulting peptide fragments were separated by 12% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), electroblotted onto a polyvinylidene difluoride membrane, and subjected to N-terminal sequencing with a Perkin-Elmer 477A sequencer (Table 2).
TABLE 1.
Substrate specificity of native GOOX
| Substrate | Relative activitya
|
Kmb (mM) | kcat (min−1) | kcat/Km (mM−1 min−1) | |
|---|---|---|---|---|---|
| 0.2 mM maltose | 10 mM maltose | ||||
| Glucose | 0.2 | 3.5 | 8.12 ± 0.16 | 546 ± 16 | 67 |
| Lactose | 6.4 | 3.8 | 0.066 ± 0.008 | 819 ± 25 | 12,400 |
| Maltose | 1.0 | 6.0 | 2.47 ± 0.05 | 531 ± 16 | 215 |
| Maltotriose | 1.1 | 6.2 | 1.11 ± 0.02 | 385 ± 12 | 348 |
| Maltotetraose | 0.4 | 4.3 | 2.51 ± 0.05 | 388 ± 12 | 155 |
| Maltopentaose | 0.2 | 2.6 | 10.6 ± 0.12 | 618 ± 18 | 58 |
| Maltohexaose | 0.2 | 3.1 | 2.60 ± 0.05 | 276 ± 11 | 106 |
| Maltoheptaose | 0.2 | 2.3 | 6.95 ± 0.15 | 415 ± 13 | 60 |
| Cellobiose | 7.7 | 2.8 | 0.048 ± 0.08 | 322 ± 11 | 6,710 |
| Cellotriose | 7.4 | 2.1 | 0.026 ± 0.002 | 776 ± 16 | 29,800 |
| Cellotetraose | 7.5 | 2.0 | 0.012 ± 0.002 | 200 ± 10 | 16,700 |
| Cellopentaose | 7.6 | 1.3 | 0.026 ± 0.003 | 239 ± 11 | 9,190 |
| Cellohexaose | 7.2 | 1.1 | 0.069 ± 0.007 | 222 ± 10 | 3,220 |
The enzyme activity with 0.2 mM maltose was defined as 1.
The kinetic parameter data are means ± standard errors for three independent experiments.
TABLE 2.
N-terminal and internal peptide sequences of native GOOX
| Protein or peptide | Sequencea |
|---|---|
| N terminus (amino acids 1-30) | NSINAXLAAADVEFHEEDSEGWDMDXTAFN |
| CNBr-1 (amino acids 245-269) | RLEINANALNWEGNFFGNAKDLKKI |
| CNBr-2 (amino acids 275-296) | KKAGGKSTISKLVETDWXGQIN |
X indicates an ambiguous residue. The underlined sequences were selected for designing the PCR primers.
Cloning of the GOOX-encoding gene.
Two degenerate primers, GOOX-5′ and GOOX-3′ (Table 3), were then designed based on partial peptide sequences. To perform the PCR, the reaction mixture was initially heat denatured at 94°C for 4 min, and this was followed by 30 cycles of 1 min at 94°C, 1 min at 52°C for annealing, and 1 min at 72°C for extension. The PCR products were cloned into the vector pCRII (Invitrogen), transformed into Escherichia coli INVαF′ cells, and selected on an agar plate containing ampicillin (100 μg/ml) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal).
TABLE 3.
Oligonucleotide primers used in this study
| Primer | Sequencea | Purpose |
|---|---|---|
| GOOX-5′ | GARTTYCAYGARGARGA | Degenerate PCR |
| GOOX-3′ | CCRAARAARTTNCCYTCCCARTT | Degenerate PCR |
| GOOX-RT1 | GTCCAGGCTTCATGGATCCAAGAAGCGCAACTC | Reverse transcription-PCR |
| GOOX-RT2 | AAGCTTCAAGTCTAAATCAG | Reverse transcription-PCR |
| GOOX-EX1 | GCTTCATGGATCCAGGAATTCAACTCAATCAACGCCTG | Expression in P. pastoris |
| GOOX-EX2 | TTCAAGTCTAAATCATCTAGATAGGCAATGGGCTCAAC | Expression in P. pastoris |
| H70A-FOR | AAGGGTGGTGGTGCCAGTTACGGTTCTTATGG | H70A mutant |
| H70A-REV | ATAAGAACCGTAACTGGCACCACCACCCTTGG | H70A mutant |
| H70S-FOR | AAGGGTGGTGGTTCCAGTTACGGTTCTTATGG | H70S mutant |
| H70S-REV | ATAAGAACCGTAACTGGAACCACCACCCTTGG | H70S mutant |
| H70C-FOR | AAGGGTGGTGGTTGCAGTTACGGTTCTTATGG | H70C mutant |
| H70C-REV | ATAAGAACCGTAACTGCAACCACCACCCTTGG | H70C mutant |
| H70Y-FOR | AAGGGTGGTGGTTACAGTTACGGTTCTTATGG | H70Y mutant |
| H70Y-REV | ATAAGAACCGTAACTGTAACCACCACCCTTGG | H70Y mutant |
R = A/G; Y = T/C.
A 744-bp DNA fragment was initially amplified from the A. strictum T1 chromosome, and this fragment was then used as a probe to screen a DNA library, which was constructed by ligating HindIII-digested DNA fragments into the pUC19 vector. Due to the presence of one putative intron in the genomic sequence encoding the GOOX protein, reverse transcription-PCR was employed to obtain cDNA for a second cloning step. The mycelium was first ground in liquid nitrogen, and total RNA was extracted. Total RNA (5 μg) that was treated with DNase I was reverse transcribed with a 20-μl mixture containing 25 pmol of oligo(dT)15, 20 mM dithiothreitol, each deoxynucleoside triphosphate at a concentration of 500 μM, and 200 U of reverse transcriptase (Invitrogen) at 37°C for 1 h. Two other primers, which defined the open reading frame of GOOX (GOOX-RT1 and GOOX-RT2) (Table 3), were designed for cDNA amplification. Gel-purified cDNA fragments were cloned into the pGEM-T vector (Promega).
Expression and isolation of the recombinant protein.
Primers GOOX-EX1 and GOOX-EX2 (Table 3) were used to amplify the entire GOOX-encoding gene. The 1.5-kb PCR products were cloned into the pPICZαA vector with fusion of the α-factor signal peptide at the N terminus for efficient secretion into the medium and c-myc and a His6 tag at the C terminus for protein isolation. The plasmid was confirmed by sequencing and was transformed into Pichia pastoris strain KM71 using electroporation. P. pastoris cells were grown in BMGY medium (Invitrogen) at 30°C to an optical density at 600 nm of 2 to 6. The cells were harvested by centrifugation at 1,500 × g for 5 min, and the cell pellets were resuspended in BMMY medium (Invitrogen) using one-fifth the original culture volume. To induce GOOX expression, the cultures were incubated for 2 to 5 days at 30°C with addition of 0.5% methanol every 24 h. The time course of the level of expression was monitored every 24 h using Western blot analysis and an activity assay. Large-scale high-density fermentation was carried out in a 5-liter BioFlo 3000 fermentor (New Brunswick Scientific).
Pichia cells were removed from the culture broth by centrifugation, and the medium was clarified by passage through a Millipore Opticap prefilter. The filtrate was concentrated to about 200 ml with a Millipore Pellicon filter unit and dialyzed against 10 mM Tris-HCl (pH 8.0). Initial attempts to purify the recombinant protein using an Ni-nitrilotriacetic acid (QIAGEN) column failed because this His6-tagged fusion protein did not bind to the resin. The enzyme was then isolated by using a Toyopearyl Phenyl-650 column with a linear 2.5 to 0 M ammonium sulfate gradient in 10 mM Tris-HCl (pH 8.0). The yellow protein fractions were pooled and concentrated by ultrafiltration using Amicon YM30 membranes. The protein concentrations were determined with a Coomassie protein assay kit (Pierce) and SDS-PAGE analysis with Coomassie blue R-250 staining and bovine serum albumin as the standard.
Site-directed mutagenesis.
The primers used for generation of the H70 mutants are listed in Table 3. Site-directed mutagenesis was carried out using a Quick-Change mutagenesis kit (Stratagene). The mismatching primers with the desired mutations were annealed to the pPICZαA vector containing the GOOX-encoding gene, and Pfu polymerase was used to replicate the whole plasmid. After PCR, wild-type DNA fragments were selectively digested by DpnI (Roche Molecular Biochemicals). The resultant mixture was used for transformation and amplification.
Nucleotide sequence accession number.
The nucleotide sequence determined in this study has been deposited in the GenBank database under accession number AY573966.
RESULTS AND DISCUSSION
Substrate specificity.
GOOX substrate specificity was screened in our previous studies, which revealed that only glucose, lactose, maltose, cellobiose, and maltooligosaccharides are good substrates (16). In the present study, we determined the kinetic parameters and showed that GOOX can also react with cellooligosaccharides having up to at least six glucosyl residues (Table 1). These kinetic results have a number of implications. First, d-glucose is the only monosaccharide substrate, suggesting that as observed with GOX, other monosaccharides and derivatives of these compounds may form either fewer bonds or unfavorable contacts with the amino acid residues surrounding the substrate-binding site (25). For example, the lack of the OH2 group at the C-2 position (OH2) in 2-deoxy-d-glucose and the exocyclic CH2OH group in d-xylose might miss some hydrogen bonds, while the axial OH2 in mannose, the axial OH3 in allose, the axial OH4 in galactose, and the equatorial NH2 in glucosamine might cause unfavorable contacts. In addition, modification at the C-1, C-2, and C-6 positions, such as phosphorylation and N-acetylation, might cause steric hindrance because of a lack of appropriate accommodation space. Second, the Km for cellobiose is 2 orders of magnitude lower than that for glucose and is similar to the Km values for cellooligosaccharides, implying that the carbohydrate-binding groove might consist mainly of two glucosyl-binding subsites. Cellobiose and lactose bind to GOOX with higher affinities than maltose binds, indicating that these two disaccharides have more contacts in the second glycosyl-binding subsites than maltose due to the stereochemistry of the glycosidic bonds (βversus α-1,4 bond). However, severe substrate inhibition by cellobiose and lactose was observed. Third, GOOX may possess an open carbohydrate-binding groove similar to that of CDH, so the non-reducing-end glucose residues can stick out into the solvent and be exposed on the protein surface. The crystal structures of GOX and POX have revealed a “size-exclusion mechanism,” in which the shape of the active site cavity is such that the enzyme can accept only monosaccharides (11, 25). On the other hand, CDH has an open carbohydrate-binding groove that allows accommodation of longer-chain oligosaccharides (10).
Catalytic mechanism.
After incubation of the substrate maltose with GOOX, the reaction product was first eluted from a thin-layer plate, purified with a μ-Spherogel column (Beckman),and then identified as maltobionic acid using infrared analysis, 13C and 1H nuclear magnetic resonance, and mass spectrometry. The absorption spectrum of the purified native GOOX had two maxima, at 380 nm and 444 nm, which are spectral characteristics typical of the oxidized FAD cofactor (Fig. 1). Addition of the substrate maltose or sodium hydrosulfite to the enzyme solution eliminated the peak at 444 nm, revealing production of the reduced FAD. Thus, like most of the reactions catalyzed by the FAD-dependent oxidases and dehydrogenases (6), the GOOX-catalyzed reaction may consist of two steps: oxidation of the reducing-end glucosyl residue to glucono-1,5-lactone by FAD and regeneration of the oxidized FAD by dioxygen. The glucono-1,5-lactone is then spontaneously hydrolyzed to gluconic acid. The optimal pH for activity of GOOX (pH 10) implies that a tyrosine residue may serve as a general base (16), whereas CDH and GOX utilize a histidine as a proton shuttle and hence the optimal pH is near pH 6 (10, 25). The similar kcat values of various substrates ranging from glucose to maltoheptaose (Table 1) imply that there is similar stereogeometry between the reactive reducing-end C-1 atom and the flavin N-5 atom. In addition to molecular oxygen, it was found that 2,6-dichlorophenol-indophenol can act as an efficient electron acceptor; this is not true of cytochrome c, phenazine methosulfate, or Fe(CN)63−. The apparent Km and kcat for 2,6-dichlorophenol-indophenol using 2 mM cellobiose as the electron donor were 0.85 mM and 6.2 min−1, respectively.
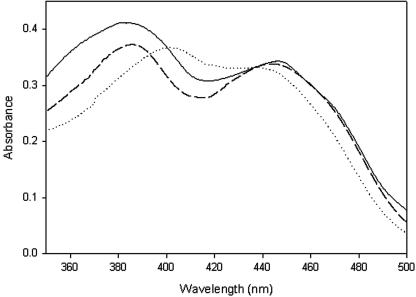
FIG. 1.
Flavin absorption spectra. The spectra of the native and wild-type recombinant enzymes and the H70Y mutant are indicated by solid, dashed, and dotted lines, respectively. The protein concentration was 2 mg/ml in 20 mM Tris-HCl (pH 8.0). The native and recombinant enzymes had similar spectra with two maxima, at 380 nm and 444 nm; the maximum wavelength was shifted from 380 nm to 400 nm in the H70 mutant.
Genomic and cDNA cloning.
To design degenerate PCR primers, the purified native protein was treated with cyanogen bromide to obtain two internal fragments, along with the known N-terminal 30 residues (Table 2). A 744-bp DNA fragment was first amplified from the chromosomal DNA of A. strictum T1 and used as a probe to isolate a 2,611-bp DNA fragment, which contained the complete GOOX-encoding gene, including one putative intron. Total RNA was then isolated and reverse transcribed for cDNA production. The GOOX-encoding gene consisted of a 1,500-bp open reading frame encoding 499 amino acids. Southern blot hybridization revealed that there was a single copy gene in the A. strictum genome.
Analysis of the deduced amino acid sequence predicted a processing site at the dibasic Lys-Arg sequence, and this resulted in a 474-residue mature protein, which was consistent with the N-terminal sequencing of the native protein. Several probable transcription elements were observed in the promoter region, including one CAAT element at positions −427 to −421, one TATA box at positions −108 to −102, and a Kozak consensus sequence flanking the start codon. One typical short fungal intron (53 bp) was detected in the genomic gene with consensus 5′ and 3′ splice sites and lariat sequences (GTPuNGPy, PyAG, and NNCTPuAPy, respectively) (12). The deduced molecular mass, 55,234 Da, is apparently less than the value obtained from size exclusion chromatography (∼61 kDa) (16). Based on this, a posttranslational modification, such as glycosylation, may be assumed, since three possible Asn glycosylation sites, N305, N341, and N394, were detected.
Recombinant protein.
Expression of the recombinant GOOX in E. coli was not successful, perhaps due to a need forposttranslational modifications. Instead, the gene was expressed in P. pastoris KM71 at 30°C, and the yield was 300 mg per liter medium in a bioreactor with automatic delivery of methanol. An unusually high absorbance at about 320 nm was observed for the freshly isolated protein, and the value decreased with time. This might have been due to the presence of some reduced FAD molecules and/or contaminants from the Pichia culture medium. The flavin absorption spectrum of the native GOOX, at wavelengths ranging from 350 nm to 500 nm, nearly overlaid the spectruma of the recombinant protein, with maxima at 380 nm and 444 nm (Fig. 1). When maltose was used as the substrate, the native GOOX had a Km of 2.47 mM, which was similar to that of the recombinant protein (2.29 mM) (Tables 1 and 4). However, the kcat for the native protein was 531 min−1, while that for the recombinant GOOX was 361 min−1. These different kcat values may have been due to different protein processing in distinct organisms (A. strictum versus P. pastoris) or to inaccurate measurements of the protein concentrations by the protein dye-binding assay.
TABLE 4.
Kinetic parameters of recombinant wild-type and mutant GOOXa
| Enzyme | Km (mM) | kcat (min−1) | kcat/Km (mM−1 min−1) |
|---|---|---|---|
| Wild type | 2.29 ± 0.11 | 360.8 ± 0.14 | 157.0 |
| H70A | 2.63 ± 0.10 | 6.8 ± 0.10 | 2.6 |
| H70S | 1.02 ± 0.05 | 0.57 ± 0.09 | 0.56 |
| H70C | 2.56 ± 0.08 | 5.2 ± 0.11 | 2.0 |
| H70Y | 2.52 ± 0.07 | 1.5 ± 0.10 | 0.6 |
The data are means ± standard errors for three independent experiments, in which the enzyme activity was measured using maltose as the substrate.
Sequence analysis.
To date, only three hypothetical fungal proteins exhibit high overall levels of sequence identity (40%) with GOOX. Interestingly, GOOX exhibits no similarity to GOX, CDH, and POX. In contrast, it exhibits significant sequence homology to several berberine bridge enzyme-like proteins, including Eschscholzia californica berberine bridge enzyme (EcBBE) (13), a hemp tetrahydrocannabinolic acid synthase (CsTHCAS) (24), pollen allergen group 4 from Bermuda grass (Cyn d 4) (15), a tobacco nectar protein (nectarin V, NlNEC5) with GOX activity (2), and a sunflower defense protein with carbohydrate oxidase activity (HaCHOX) (4) (Fig. 2). Lower levels of sequence similarity were also found with other flavoproteins, such as those encoded by some Streptomyces genes involved in the biosynthesis of antitumor antibiotics, a hexose oxidase from the seaweed Chondrus crispus (CcHEOX), and 6-hydroxy-d-nicotine oxidase from Arthrobacter oxidans (AoHDNO).
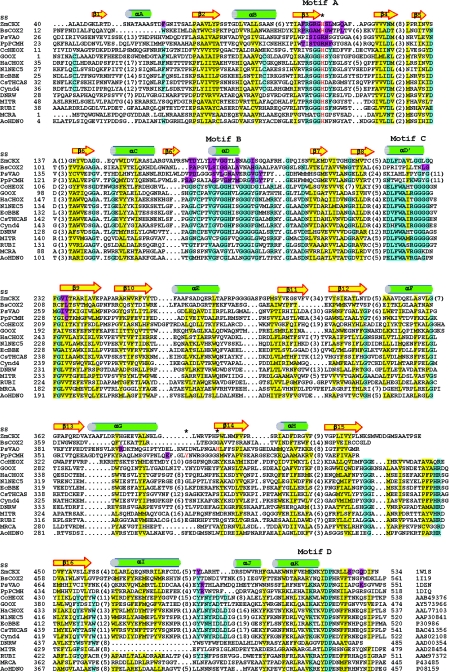
FIG.2.
Structure-based sequence alignment. The secondary structure elements for ZmCKX are labeled (SS). The protein accession numbers are indicated at the bottom on the right. The numbers of residues present in gaps are indicated in parentheses. The FAD-interacting residues in the known structures are indicted by magenta shading, while residues in the conservative hydrophobic core are indicated by yellow shading. There are four conserved segments in GOOX and related flavoproteins (motifs A to D), and the conserved residues are indicated by cyan shading. The flavinylation sites, Y384 in PpPCMH, H422 in PsVAO, and the consensus histidine in motif A, such as H105 in ZmCKX and H121 in BsCOX2, are indicated by red type and by asterisks. DNRW, MITR, RUBI, and MCRA are involved in the biosynthesis of antitumor antibiotics in Streptomyces; however, their substrates are still not known. Alignment was performed by manual editing based on the structural information and secondary structure prediction. This alignment suggests that the sugar oxidases, including GOOX, CcHEOX, NlNEC5, and HaCHOX, apparently evolved so that they have similar residues for FAD recognition but dissimilar residues for carbohydrate binding.
To predict the structural fold of GOOX, a sequence similarity search was performed with the structural databases, which revealed that this enzyme displayed putative homology to the Zea mays cytokinin dehydrogenase (ZmCKX) (Fig. 2). The crystal structures of ZmCKX (17) and its structural homologues, such as Brevibacterium sterolicum cholesterol oxidase 2 (BsCOX2) (14), Pseudomonas putida p-cresol methylhydroxylase (PpPCMH) (3), and Penicillium simplicissimum vanillyl-alcohol oxidase (PsVAO) (7), display similar two-domain topologies and thus are defined as the PCMH superfamily because the PpPCMH structure was the first reported structure (18). These flavoenzymes share a conserved FAD-binding domain but have diverse substrate-binding domains due to similar FAD recognition mechanisms but distinct substrate interactions.
There are four conserved motifs (motifs A to D) in the FAD-binding domain (Fig. 2). Motifs A to C are located in theN-terminal portion, whereas motif D is located in the C-terminal tail. Motif A has features similar to those of the “P-loop,” which is highly conserved in nearly all nucleotide-binding proteins and is responsible for binding to the pyrophosphate group of the cofactor nucleotide diphosphate moiety (1). Residues in motif B interact with the isoalloxazine ring, the ribitol, and the pyrophosphate group of the cofactor FAD. On the other hand, the last two motifs are near theFAD-binding pocket and form conserved salt bridges, hydrogen bonds, and hydrophobic contacts for structural stabilization. Thus, it is possible to build a structural model for the FAD-binding domain of GOOX based on the ZmCKX structure.
To date, most of the known sugar oxidases and dehydrogenases catalyze carbohydrate oxidation with FAD assistance. One interesting feature of these flavoenzymes is that enzyme function does not correlate with folding topology. For example, GOOX, NlNEC5, HaCHOX, and CcHEOX belong to the PCMH superfamily, whereas GOX, CDH, and POX belong to the glutathione reductase superfamily. Thus, these sugar oxidases provide elegant examples of convergent evolution, where starting from various ancestral folds, the same FAD-assisted carbohydrate oxidation was obtained at distinct active sites. In addition, it is worth noting that even starting from similar structural folds, these sugar oxidases apparently evolved so that they have dissimilar residues for interaction with the common substrates. For example, although GOOX, CcHEOX (9), and HaCHOX (4) all can catalyze the oxidation of glucose, maltose, lactose, and cellobiose and have similar FAD-binding domains, their substrate-binding domains apparently lack conserved carbohydrate-interacting residues (Fig. 2). Thus, it would be difficult to predict the specific carbohydrate-binding residues unless the three-dimensional structures are available.
Distinct flavinylation.
Five different types of autocatalytic flavinylation have been identified: four types via the 8α-methyl group of the isoalloxazine ring cross-linking to His, Tyr, and Cys and one type via the C-6 atom to Cys (20). The crystal structures of ZmCKX and BsCOX2 provide a structure base for covalent attachment of the 8α-methyl group to the Nδ1 atom of the consensus histidine in motif A (8α-N1-histidyl FAD) (14, 17). In addition, mutational studies have demonstrated that replacement of this histidine with alanine, threnine, or tyrosine in CsTHCAS (H114A), EcBBE (H104T), and AoHDNO (H71Y) can abolish the flavin incorporation, as well as the enzyme activity (13, 19, 24). Unexpectedly, mutation of the corresponding histidine residue in GOOX, H70, to alanine, serine, cysteine, or tyrosine did not eliminate the covalent FAD linkage and had little effect on the Km (Fig. 3 and Table 4).Instead, the H70 mutants displayed kcat values that were 50- to 600-fold lower. The flavin absorption spectrum of wild-type GOOX has two maxima, at 380 nm and 444 nm. When the spectra of the mutants were compared, the maximum wavelength was shifted from 380 nm to 400 nm in the H70 mutants (Fig. 1). These findings indicate that H70 is crucial for efficient redox catalysis, perhaps through modulation of the oxidative power of the FAD cofactor.
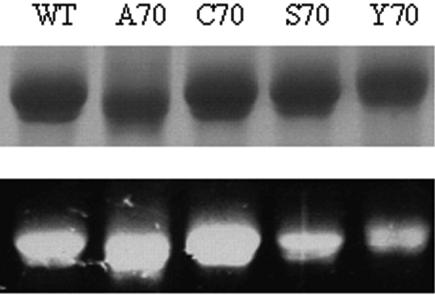
FIG. 3.
SDS-PAGE analysis of purified wild-type (WT) and mutant enzymes. The gel was visualized by transillumination at 312 nm (bottom) and then stained with Coomassie brilliant blue R-250 (top). This analysis demonstrated that the FAD cofactor is covalently bound to the H70 variants.
The H70 mutants suggest that this histidine residue is not the flavinylation site or it is not the only FAD attachment site. The binding type of the cofactor FAD/flavin mononucleotide (covalent versus noncovalent) is one of the least conserved properties among flavoproteins, and the flavinylation sites are often located in poorly conserved regions (5, 8). For example, even though PsVAO and PpPCMH exhibit 30% sequence identity and share some substrate specificity, they have different flavinylation types (3, 7). The FAD cofactor is cross-linked to PsVAO and PpPCMH via the 8α-methyl group at nonconserved residues H422 (8α-N3-histidyl FAD) and Y384 (8α-O-tyrosyl FAD), respectively (Fig. 1). Thus, GOOX might have a distinct covalent FAD linkage. Attempts to identify the FAD attachment site using mass spectrometry have not been successful so far.
ADDENDUM
While the manuscript was under review, we solved the crystal structure of the recombinant GOOX, and the results revealed the first known double-attachment flavinylation, 6-S-cysteinyl, 8α-N1-histidyl FAD. The FAD cofactor is cross-linked to the enzyme via the C-6 atom and the 8α-methyl group of the isoalloxazine ring with C130 and H70, respectively.
Acknowledgments
This study was supported by National Science Council grants NSC93-2311B010-009 and NSC93-2320-B227-004.
REFERENCES
- 1.Aravind, L., L. M. Iyer, D. D. Leipe, and E. V. Koonin. 2004. A novel family of P-loop NTPases with an unusual phyletic distribution and transmembrane segments inserted within the NTPase domain. Genome Biol. 5:R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carter, C. J., and R. W. Thornburg. 2004. Tobacco nectarin V is a flavin-containing berberine bridge enzyme-like protein with glucose oxidase activity. Plant Physiol. 134:460-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cunane, L. M., Z. W. Chen, W. S. McIntire, and F. S. Mathews. 2005. p-Cresol methylhydroxylase: alteration of the structure of the flavoprotein subunit upon its binding to the cytochrome subunit. Biochemistry 44:2963-2973. [DOI] [PubMed] [Google Scholar]
- 4.Custers, J. H., S. J. Harrison, M. B. Sela-Buurlage, E. van Deventer, W. Lageweg, P. W. Howe, P. J. van der Meijs, A. S. Ponstein, B. H. Simons, L. S. Melchers, and M. H. Stuiver. 2004. Isolation and characterisation of a class of carbohydrate oxidases from higher plants, with a role in active defence. Plant J. 39:147-160. [DOI] [PubMed] [Google Scholar]
- 5.Dym, O., and D. Eisenberg. 2001. Sequence-structure analysis of FAD-containing proteins. Protein Sci. 10:1712-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fitzpatrick, P. F. 2004. Carbanion versus hydride transfer mechanisms in flavoprotein-catalyzed dehydrogenations. Bioorg. Chem. 32:125-139. [DOI] [PubMed] [Google Scholar]
- 7.Fraaije, M. W., R. H. H. van den Heuvel, W. J. H. van Berkel, and A. Mattevi. 1999. Covalent flavinylation is essential for efficient redox catalysis in vanillyl-alcohol oxidase. J. Biol. Chem. 274:35514-35520. [DOI] [PubMed] [Google Scholar]
- 8.Fraaije, M. W., and A. Mattevi. 2000. Flavoenzymes: diverse catalysts with recurrent features. Trends Biochem. Sci. 25:126-132. [DOI] [PubMed] [Google Scholar]
- 9.Groen, B. W., S. De Vries, and J. A. Duine. 1997. Characterization of hexose oxidase from the red seaweed Chondrus crispus. Eur. J. Biochem. 244:858-861. [DOI] [PubMed] [Google Scholar]
- 10.Hallberg, B. M., G. Henriksson, G. Pettersson, A. Vasella, and C. Divne. 2003. Mechanism of the reductive half-reaction in cellobiose dehydrogenase. J. Biol. Chem. 278:7160-7166. [DOI] [PubMed] [Google Scholar]
- 11.Hallberg, B. M., C. Leitner, D. Haltrich, and C. Divne. 2004. Crystal structure of the 270 kDa homotetrameric lignin-degrading enzyme pyranose 2-oxidase. J. Mol. Biol. 341:781-796. [DOI] [PubMed] [Google Scholar]
- 12.Johansson, T., and P. O. Nyman. 1996. A cluster of genes encoding major isozymes of lignin peroxidase and manganese peroxidase from the white-rot fungus Trametes versicolor. Gene 170:31-38. [DOI] [PubMed] [Google Scholar]
- 13.Kutchan, T. M., and H. Dittrich. 1995. Characterization and mechanism of the berberine bridge enzyme, a covalently flavinylated oxidase of benzophenanthridine alkaloid biosynthesis in plants. J. Biol. Chem. 270:24475-24481. [DOI] [PubMed] [Google Scholar]
- 14.Lario, P. I., N. Sampson, and A. Vrielink. 2003. Sub-atomic resolution crystal structure of cholesterol oxidase: what atomic resolution crystallography reveals about enzyme mechanism and the role of the FAD cofactor in redox activity. J. Mol. Biol. 326:1635-1650. [DOI] [PubMed] [Google Scholar]
- 15.Liaw, S., D. Y. Lee, L. P. Chow, G. X. Lau, and S. N. Su. 2001. Structural characterization of the 60-kDa bermuda grass pollen isoallergens, a covalent flavoprotein. Biochem. Biophys. Res. Commun. 280:738-743. [DOI] [PubMed] [Google Scholar]
- 16.Lin, S. F., T. Y. Yang, T. Inukai, M. Yamasaki, and Y. C. Tsai. 1991. Purification and characterization of a novel glucooligosaccharide oxidase from Acremonium strictum T1. Biochim. Biophys. Acta 1118:41-47. [DOI] [PubMed] [Google Scholar]
- 17.Malito, E., A. Coda, K. D. Bilyeu, M. W. Fraaije, and A. Mattevi. 2004. Structures of Michaelis and product complexes of plant cytokinin dehydrogenase: implications for flavoenzyme catalysis. J. Mol. Biol. 341:1237-1249. [DOI] [PubMed] [Google Scholar]
- 18.Mathews, F. S., Z. W. Chen, H. D. Bellamy, and W. S. McIntire. 1991. Three-dimensional structure of p-cresol methylhydroxylase (flavocytochrome c) from Pseudomonas putida at 3.0-A resolution. Biochemistry 30:238-247. [DOI] [PubMed] [Google Scholar]
- 19.Mauch, L., V. Bichler, and R. Brandsch. 1989. Site-directed mutagenesis of the FAD-binding histidine of 6-hydroxy-D-nicotine oxidase. Consequences on flavinylation and enzyme activity. FEBS Lett. 257:86-88. [DOI] [PubMed] [Google Scholar]
- 20.Mewies, M., W. S. McIntire, and N. S. Scrutton. 1998. Covalent attachment of flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN) to enzymes: the current state of affairs. Protein Sci. 7:7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer, A. S., and A. Isaksen. 1995. Application of enzymes as food antioxidants. Trends Food Sci. Technol. 6:300-304. [Google Scholar]
- 22.Moschou, E. A., B. V. Sharma, S. K. Deo, and S. Daunert. 2004. Fluorescence glucose detection: advances toward the ideal in vivo biosensor. J. Fluoresc. 14:535-547. [DOI] [PubMed] [Google Scholar]
- 23.Schomburg, I., A. Chang, C. Ebeling, M. Gremse, C. Heldt, G. Huhn, and D. Schomburg. 2004. BRENDA, the enzyme database: updates and major new developments. Nucleic Acids Res. 32:D431-D433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sirikantaramas, S., S. Morimoto, Y. Shoyama, Y. Ishikawa, Y. Wada, and F. Taura. 2004. The gene controlling marijuana psychoactivity: molecular cloning and heterologous expression of Δ1-tetrahydrocannabinolic acid synthase from Cannabis sativa L. J. Biol. Chem. 279:39767-39774. [DOI] [PubMed] [Google Scholar]
- 25.Wohlfahrt, G., S. Witt, J. Hendle, D. Schomburg, H. M. Kalisz, and H. J. Hecht. 1999. 1.8 and 1.9 A resolution structures of the Penicillium amagasakiense and Aspergillus niger glucose oxidases as a basis for modelling substrate complexes. Acta Crystallogr. D Biol. Crystallogr. 55:969-977. [DOI] [PubMed] [Google Scholar]