Abstract
To assess links between the diversity of nitrite-oxidizing bacteria (NOB) in agricultural grassland soils and inorganic N fertilizer management, NOB communities in fertilized and unfertilized soils were characterized by analysis of clone libraries and denaturing gradient gel electrophoresis (DGGE) of 16S rRNA gene fragments. Previously uncharacterized Nitrospira-like sequences were isolated from both long-term-fertilized and unfertilized soils, but DGGE migration patterns indicated the presence of additional sequence types in the fertilized soils. Detailed phylogenetic analysis of Nitrospira-like sequences suggests the existence of one newly described evolutionary group and of subclusters within previously described sublineages, potentially representing different ecotypes; the new group may represent a lineage of noncharacterized Nitrospira species. Clone libraries of Nitrobacter-like sequences generated from soils under different long-term N management regimes were dominated by sequences with high similarity to the rhizoplane isolate Nitrobacter sp. strain PJN1. However, the diversity of Nitrobacter communities did not differ significantly between the two soil types. This is the first cultivation-independent study of nitrite-oxidizing bacteria in soil demonstrating that nitrogen management practices influence the diversity of this bacterial functional group.
Nitrogen fertilization is the most important management strategy for the improvement of agricultural crops. However, up to 60% of N fertilizer applied can be lost through leaching of mobile N compounds (NO3−) and transformation into N2O and N2 (53), leading to water pollution and contributing to global warming. The key process during natural N cycling is microbial chemolithoautotrophic nitrification, the conversion of NH4+ to NO3− via NO2−. Autotrophic nitrification is carried out by two distinct groups, ammonia-oxidizing bacteria (AOB) and nitrite-oxidizing bacteria (NOB), and it is generally assumed that the first step is rate-limiting (50). This assumption is based mainly on observations that NO2− concentrations are low or negligible in most natural systems. Consequently, with the exception of high-N-load sewage-processing systems, where nitrite concentrations are high (NO2−-N up to 70 mg liter−1) (17, 22) and nitrite washout is of concern (41, 63), research into community composition, activity, and environmental responses of nitrifying bacteria has focused on AOB (37). These studies have demonstrated the influence of land use management regimes on the diversity and activity of AOB, as well as links to changes in ammonia oxidation rates (4, 9, 11, 49, 70), but a comprehensive understanding of soil nitrification also requires characterization of NOB.
Analysis of NOB in natural environments has been limited by reliance on traditional, cultivation-based methods. In contrast to AOB, for which most 16S rRNA gene sequences of cultured organisms and related environmental clones fall within a monophyletic group (51, 66), NOB are classified into four genera, based traditionally on cell morphology and on phylogenetic analysis of 16S rRNA gene sequences (66). The most intensively studied NOB genus, Nitrobacter, falls within the Alphaproteobacteria and contains several characterized cultured representatives (7, 46, 62). The Nitrospira genus has traditionally been placed within the Proteobacteria, but 16S rRNA gene analysis suggests that it forms a separate phylum (66). With the exception of a few recent investigations of Nitrospira-like sequences in freshwater systems (1, 30, 52), environmental studies have focused on wastewater treatment systems (17, 58, 59, 61). Two further NOB groups have been characterized containing two cultured representatives of Nitrospina gracilis (Deltaproteobacteria) and one of Nitrococcus mobilis (Gammaproteobacteria) (69) isolated from marine environments. Nothing is known of their environmental significance.
Recently, several genus-specific 16S rRNA gene probes have been developed that allow the detection and characterization of Nitrobacter- and Nitrospira-like sequences in environmental samples by cultivation-independent methods (13, 16, 26, 30). These have enabled assessment of their distribution in sewage processing and freshwater systems by fluorescence in situ hybridization (FISH) (16, 24, 32, 33, 58, 60, 61) and by analysis of clone libraries (12, 30, 58). The results indicate dominance of Nitrospira spp. and suggest that Nitrobacter spp. may be adapted to high nitrite and oxygen concentrations (16, 59), potentially reducing their importance in natural environments. Phylogenetic analysis of clone sequences further suggests that the phylum Nitrospira forms an extended and highly diverse group (16), with several clusters containing 16S rRNA gene sequences isolated from soil environments. Analysis of the evolutionary relationships of AOB based on 16S rRNA gene sequence similarities has demonstrated that different ecosystem types are represented in distinct phylogenetic groups or clusters (36, 37). Comparable analysis of NOB, based mainly on analysis of clone libraries generated from sewage systems, suggested the separation of Nitrospira into several sublineages probably representing different ecotypes (16), but little is known about Nitrobacter. The aim of this study was to investigate how agricultural management practices influence NOB diversity by examining communities of Nitrospira and Nitrobacter in grassland soils. 16S rRNA genes were amplified from extracted RNA using reverse transcriptase PCR (RT-PCR), which provides greater sensitivity than DNA-targeted methods due to higher intracellular target copy numbers. Amplification products were analyzed by denaturing gradient gel electrophoresis (DGGE) and by cloning and sequencing, for detailed phylogenetic analysis. Public-domain 16S rRNA gene databases were analyzed to assess evolutionary relationships of clone sequences not previously associated with NOB and to design primers encompassing NOB and closely related lineages.
MATERIALS AND METHODS
Sample location and collection.
Soils were sampled from the Rowden long-term experimental site at the Institute of Grassland and Environmental Research (IGER), North Wyke, United Kingdom (latitude, 50°46′29"N; longitude, 3°55′23"W). These were grazed swards under long-term pasture for the previous 50 years and under the current management regime since 1982. All plots had received a spring dressing of potassium (50 kg ha−1 year−1) and phosphorus (25 kg ha−1 year−1) and an application of lime every 4 to 5 years to maintain soil pH above 6.0. The soil is a poorly drained silty clay loam of the Hallsworth series. Long-term-unfertilized plots were dominated by Holcus lanatus, Lolium perenne, Agrostis stolonifera, and Phleum pratense, whereas those soils under long-term inorganic N fertilizer management (200 kg N ha−1 year−1) were dominated by Holcus lanatus. The N dynamics of the fertilized and unfertilized plots have been studied in detail previously (28) and demonstrated NH4+ concentrations of 5.9 and 3.7 mg of N · kg of dry soil−1 and significantly different (P = 0.09) nitrification rates of 1.6 and 1.0 μg of N g−1 day−1 for the long-term-fertilized and unfertilized plots, respectively. For this study, 10 2-m-by-3-m subplots were constructed separately within a long-term-fertilized plot and an unfertilized plot. Within each of the background long-term-managed plots, five subplots were randomly selected and maintained under the previous management regime, and the remaining five plots were managed under the alternative regime. That is, inorganic N fertilization was discontinued in long-term-fertilized plots, and inorganic N fertilization of previously unfertilized plots commenced at the rate used for long-term management, i.e., four applications per year of approximately 50 kg of N ha−1 in each dressing. Subplots were cut 3 times during the growing season. Five soil cores (2.5 by 2.5 by 7 cm) were collected from each subplot at the end of the first year of the experimental management regime. Soil samples were broken down, and plant roots were removed, sieved (3.35-mm mesh), and bulked to generate one soil sample per subplot. Soil samples were frozen at −20°C immediately after sieving until further processing.
RNA extraction.
Nucleic acids were extracted from 0.5 g of bulked soil by disruption with a Hybaid Ribolyser Cell Disruptor (Thermo Hybaid, Ltd., Ashford, United Kingdom) (speed 4, twice for 10 s each time) in 2-ml screw-cap Blue Matrix Ribolyser tubes (Hybaid) according to Griffiths et al. (25). Extracted nucleic acids were resuspended in 50 μl of RNase-free sterile water for the preparation of RT-PCR templates.
Development of 16S rRNA gene PCR assays.
The phylogeny of 16S rRNA gene sequences of recognized nitrobacters, nitrospirae, and related clone sequences was initially explored using the 59 and 21 prealigned 1.2-kb sequences annotated as Nitrospira (genus) and Nitrobacter (genus), respectively, within RDP II (15). Individual sequences were compared with the GenBank database by using the Bl2Seq algorithm as implemented in BLASTn (2). Closest match sequences of >1 kb that were not included within the initial alignment were added using ClustalX 1.81 (EMBL, Heidelberg, Germany), with sequences of close phylogenetic neighbors serving as outgroups. Alignments were trimmed and manually refined, and distance analysis between pairs of 16S rRNA gene sequences was carried out using the Jukes-Cantor correction and the neighbor-joining tree method, as implemented in PAUP 4.010. Sequences or groups of sequences flanking nearest phylogenetic neighbors or representing isolated clades were again compared with the GenBank database, and additional closest match sequences were added to the alignment. This process was iterated until database searches did not retrieve new sequences.
For primer design, a subset of sequences was chosen from these alignments that represented all clades within the monophyletic boundaries of the target groups. Primrose 1.7 (3) was used to search for sites conserved within Nitrospira, excluding all other bacteria, and, for Nitrobacter, to search for sites conserved within Nitrobacter excluding all other bacteria and against a subset of sequences representing only the closest phylogenetic neighbors Bradyrhizobium and Afipia (71). Suggested probes were manually refined to ensure bias toward the target group, and the specificity of probe sequences was confirmed by a BLASTn search and by testing for specific amplification against a set of type strains, including the following common soil bacteria: Rhodobacter sp. strain DSMZ12077, Nocardioides plantarum NCIMB12834, Rhizobium radiobacter ATCC 6466, Novosphingobium capsulatum ATCC 14666, Acinetobacter hebeiensis DSMZ586, Arthrobacter globiformis NCIMB8602, Flexibacter ruber NCIMB1436, Stenotrophomonas maltophilia NCIMB9204, Planctomyces maris NCIMB2232, Escherichia coli K-12 NCBI10218, Nocardia asteroides ATCC 19247, Paracoccus alcaliphilus ATCC 51199, Arthrobacter globiformis ATCC 4336, Clostridium sphenoides NCIMB10627, Mesorhizobium loti ATCC 33669, Nitrosomonas europaea ATCC 25978, Pseudomonas pseudoalcaligenes NCIMB9867, Nitrosococcus oceanus ATTC 19707, Nitrospina gracilis (kindly provided by F. W. Valois, Woods Hole Oceanographic Institution, MA), Nitrococcus mobilis ATCC 25380, and Bradyrhizobium japonicum ATCC 10324. Annealing temperatures during PCR were optimized by gradient PCR, selecting the highest possible temperature at which sufficient product yield was generated from target DNA, while amplifying no product from nontarget templates. Target templates used were Nitrobacter hamburgensis sp. strain X14 NCBI13809, Nitrobacter vulgaris sp. strain Z NCBI13808, and Nitrospira moscoviensis DSMZ10035.
RT-PCR amplification of environmental 16S rRNA gene fragments.
RT-PCR amplification followed the protocol of Griffiths et al. (25) with previously described modifications (23), using SuperScript II RT RNase H reverse transcriptase (Gibco BRL, MI) and the conserved bacterial 16S rRNA gene PCR primer 1492r (40), at a raised transcription temperature of 52°C to ensure complete denaturation of rRNA secondary structure. Following transcription of 16S rRNA gene templates into cDNA, cDNA was amplified by nested PCR for cloning and DGGE analysis by using the 16S rRNA gene primers Nbacter-1050r (5′-CACCTGTGCTCCATGCTCCG-3′) and Nspira-705r (5′-GGCCTTCYTCCCGAT-3′) in conjunction with the conserved bacterial primer 27f (40). For DGGE, PCR products were reamplified using the general bacterial primers 357f-GC-518r (45). Prior to secondary amplification, PCR amplification products were diluted 1:30 to prevent amplification of nontarget sequences. Individual reagents and their concentrations were as follows: 1× PCR buffer (Bioline Ltd., London, United Kingdom), 1 U Biotaq DNA polymerase (Bioline), 1.5 mM MgCl2, each primer at a concentration of 0.2 μM, and each deoxynucleoside triphosphate (Bioline) at a concentration of 250 μM. Amplification was performed with a total volume of 50 μl by using an Omn-E Express thermal cycler (Thermo Hybaid) and applying the thermal cycle conditions shown in Table 1. PCR-amplified fragments were resolved by electrophoresis on 1% (wt/vol) horizontal agarose gels with visualization of DNA by ethidium bromide fluorescence and using Bioline Hyperladder DNA fragment size markers (Bioline).
TABLE 1.
Primers and PCR amplification conditions
| Primer set | Target group | Thermocycling programb,c
|
||
|---|---|---|---|---|
| Initial denaturation period (min) | Annealing temp (°C) | Extension time (s) | ||
| 27f-Nbac-1050r | Nitrobacter (genus) | 5 | 65.2 | 80 |
| 27f-Nspira-705r | Nitrospira (genus) | 5 | 63 | 50 |
| 357f-GC-518ra | Eubacteria | 2 | 55 | 30 |
A GC clamp was attached to the 5′ end of primer 357f-GC (45).
The general thermocycling program used was as follows: 5 min at 94°C; 10 cycles of 30 s at 94°C, 30 s at the specified annealing temperature, and the specified extension time at 72°C; 25 cycles of 30 s at 92°C, 30 s at the specified annealing temperature, and the specified extension time at 72°C (increasing by 1 s per cycle); and a 10-min final extension at 72°C.
Nitrobacter- and Nitrospira-specific amplifications were performed with a “hot start” by adding the Taq polymerase after the initial denaturation step. All nested second-stage nonspecific amplifications were started by placing cooled tubes containing the PCR mix into a preheated (94°C) PCR block.
DGGE analysis.
DGGE was carried out as described previously (38) using the DCode Universal Mutation Detection System (Bio-Rad Laboratories, Hemel Hempstead, United Kingdom) and 1.5-mm, 8% polyacrylamide gels containing denaturant gradients of 40 to 70%, for analysis of 357f-GC-518r PCR products. Gels were silver stained (44) for 15 min and destained for 10 min in distilled H2O prior to digital imaging.
Cloning and sequence analysis.
For Nitrospira, one clone library was created using PCR amplicons generated with the Nitrospira 27f-Nspira-705r PCR assay. Three PCR amplicons generated from soil samples of each of the two long-term background management regimes (i.e., long-term-fertilized and unfertilized) were pooled to compensate for PCR drift (68) and to ensure that dominant sequences were represented in the clone library. PCR fragments were purified by standard agarose electrophoresis, and excised DNA bands were cleaned using QiaQuick columns (QIAGEN, Hilden, Germany), ligated into the pGEM T-vector system (Promega Ltd., Southampton, United Kingdom), and transformed into XL1-Blue MRF Kan supercompetent E. coli cells (Stratagene Inc., Cambridge, United Kingdom) according to the manufacturer's instructions. Transformed colonies were screened for inserts of the correct size by PCR amplification with the Nitrospira PCR assay, as described above, and 100 clones with inserts of the correct fragment size were subjected to nested PCR with the 357f-GC-518r primers (45). PCR products were screened by DGGE for sequence differences within the hypervariable V3 region. Clone libraries selected for full-length sequencing of the entire insert represented at least two replicate clones with DGGE migration patterns identical to each individual DGGE band in amplicons of environmental samples. Additional replicate clones of the recognized dominant sequences on the DGGE gels were also selected for sequencing. For Nitrobacter, cloning inserts were generated with the Nitrobacter 27f-Nbac-1050r PCR assay as described above. Four different clone libraries from at least three pooled amplicons of each of the four different soil management regimes were produced to ensure recovery of dominant sequence types. Ligations and transformations were performed as described above, and transformed colonies were screened for inserts of the correct size by PCR amplification with the Nitrobacter PCR assay. Forty-three clones with the correct fragment size insert of each library were subjected to nested PCR and screening by DGGE as described above. The 1-kb 27f-Nbac-1050r PCR amplification products of clones showing DGGE migration patterns identical to those of pure culture amplicons generated from Nitrobacter winogradskyi, N. vulgaris, and N. hamburgensis were subjected to restriction fragment length polymorphism (RFLP) analysis with HinfI (G↓ANTC) and BglII (A↓GATCT) (Promega) according to the manufacturer's instructions, to discriminate between highly similar B. japonicum- and Nitrobacter-like sequences. At least 10 clones from each library were selected for sequencing according to their DGGE migration and RFLP patterns. Additional clones representing the observed diverging patterns were also selected for sequencing. Representatives of Nitrospira and Nitrobacter clone libraries were amplified with vector primers M13f-M13r (Promega), and M13 PCR products were purified by standard preparative agarose electrophoresis as described above. To ensure sequence reads without ambiguities, M13 vector products were sequenced with sufficient overlap of sequencing reads using the SP6 and T7 vector primers (Promega) for the Nitrospira assay and, additionally, with the conserved internal 16S rRNA gene primers 357f and 518r (45) for the Nitrobacter assay. Sequencing reactions were performed using the BigDye Terminator Cycle Sequencing kit (PE Biosystems, Warrington, United Kingdom), and the cycle sequencing products were analyzed with a model ABI377 automated sequencer (PE Biosystems).
For final sequence assembly, clone sequences were added to compiled alignments and subjected to iterative database searches and phylogenetic analysis, as described above. For final phylogenetic analysis, 0.7- and 1.3-kb alignments for Nitrospira and Nitrobacter, respectively, were trimmed to 0.65 and 1.0 kb, respectively, and refined by eye. Single-base sequence inserts that were present only in individual sequences at otherwise conserved positions were deleted, and sequences outside the core alignment of the clone sequences generated in this study were omitted from the final phylogenetic analysis. To exclude chimeric 16S rRNA gene structures of clone sequences prior to phylogenetic analysis, sequences were analyzed using the Bellerophon server (http://foo.maths.uq.edu.au/∼huber/bellerophon.pl) (31). Phylogenetic relationships between pairs of 16S rRNA gene sequences were calculated using maximum likelihood, maximum parsimony, and distance analysis as implemented in PAUP 4.010 (64). Analysis was limited to variable positions estimated using maximum likelihood. LogDet/paralinear distances were calculated using the minimal evolution criterion, and data were bootstrapped 1,000 times. Phylogenetic trees were constructed from distance analysis using the neighbor-joining method (39, 43). Additional alignments containing 16S rRNA gene sequences of maximum length (1.4 kb) only were analyzed as described above, and tree topologies were compared for consistency. Clone sequences and sequences generated from DGGE bands were aligned for analysis of mismatches using Sequencer 4.1 (Genes Codes Corporation, Ann Arbor, MI) and BioEdit (freeware by Tom Hall, Department of Microbiology, University of North Carolina). DNA-DNA sequence similarity percentages were calculated using the sequence similarity matrix function as implemented in BioEdit. Nitrobacter and Bradyrhizobium sequences were analyzed for restriction enzyme recognition sites discriminating between B. japonicum and Nitrobacter spp., using the restriction mapping facility incorporated into BioEdit and the virtual digest tool available at http://tools.neb.com/REBsites/index.php3.
Nucleotide sequence accession numbers.
The partial clone sequences determined in this study have been deposited in the GenBank database under accession numbers AY876601 to AY876630.
RESULTS
Primer design.
Two primers specific for Nitrospira and three for Nitrobacter suggested by the Primrose 1.7 program were evaluated for their specificity against the intended target groups by a PCR-DGGE cloning approach on nucleic acid extracts from the IGER grassland soil samples and by comparison with the final alignments used for analyzing the phylogenetic relations of Nitrospira spp. and Nitrobacter spp. Nitrospira primers yielded similar DGGE banding patterns, with major bands identified as associated with the phylum Nitrospira. For Nitrospira, equally good amplification results were achieved with primers Nspira-705r (5′-GGCCTTCYTCCCGAT-3′) and Nspira-710r (5′-CTTCGCCGGCCTTCYTC-3′). The degeneracy at E. coli position 712 was introduced to incorporate a G-to-A substitution evident for marine sublineage IV (Fig. 1). Both primers showed one mismatch against clone sequences AY627547 and AY493911 (Fig. 1) and three and two mismatches against E. coli, respectively. Primer Nspira-710r showed two additional mismatches against clone sequences AY221079 and AF317762 and was therefore excluded during further analysis. All Nitrobacter primer sequences suggested by Primrose 1.7 generated PCR products from pure-culture Nitrobacter templates. BLASTn comparisons of all 16S rRNA gene sequences deposited in GenBank with the primer generated with bias against all bacteria (primer A, 1421r-5′-CGCACCGCCTTCAGGTAAAA-3′) retrieved only Nitrobacter-associated sequences at the 100% identity level. However, DGGE analysis of PCR products amplified from grassland nucleic acids with primer A demonstrated multiple banding patterns and generated clone sequences that were associated exclusively with Bradyrhizobium spp. and Methylosinus spp. Primer A was therefore excluded from further analysis. Sequence fragments that comigrated with Nitrobacter sequences during DGGE analysis were retrieved from grassland soil nucleic acids with primers created with biases against Afipia spp. (primer B, 828r-5′-TGAGMAACCCACTAACGGC-3′) and Bradyrhizobium spp. (primer Nbac-1050r, 5′-CACCTGTGCTCCATGCTCCG-3). BLASTn analysis and alignment of primer B with all Bradyrhizobium-related 16S rRNA gene sequences demonstrated numerous mis-hits within this group. Primer B was therefore excluded from further analysis. Alignment of the Nbac-1050r primer sequence with all Bradyrhizobiaceae revealed a minimum of one mismatch; Nbac-1050r also showed a minimum of one mismatch when aligned with Afipia felis and as many as five mismatches against Afipia massiliensis/broomeae-related sequences. Only sequences associated with Nitrobacter were retrieved by BLASTn at the 100% identity level when Nbac-1050r was compared against all sequences deposited at GenBank. Alignment of the Nbac-1050r sequence with all known nitrobacters showed insertion of one base within the primer target area in the sequence deposited for N. winogradskyi strain IFO 14297. Nbac-1050r also demonstrated good suitability as a primer in a combined DGGE approach.
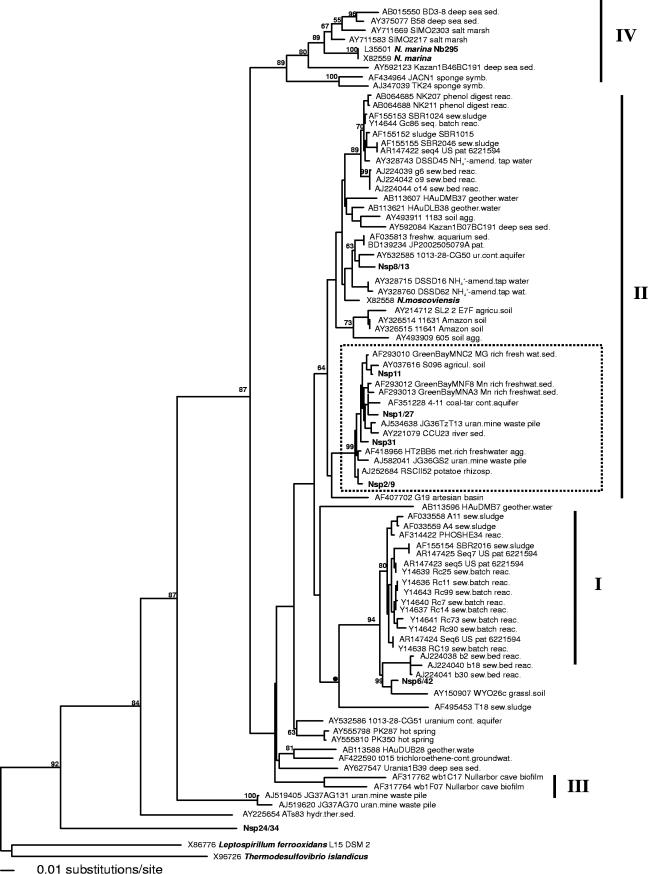
FIG. 1.
Evolutionary distance dendrogram showing the positions of environmental 16S rRNA gene 0.65-kb clone sequences generated with the 27f-Nspira-705r primer assay from North Wyke grassland soils (boldfaced) in relation to pure Nitrospira culture sequences (boldfaced) and associated clone sequences. With the exception of clone Nspira31, sequences generated in this study were included in the analysis only when represented by at least two replicate clones (100% sequence similarity). The tree is based on results of a neighbor-joining analysis of LogDet paralinear distances (Pin. var. 0.3545; 356 parsimony informative characters). Bootstrap values are given at nodes when they exceed 50% of replicates (1,000 resamplings). Bar indicates an estimated 0.01 change per nucleotide position. The cluster marked within sublineage II was recovered in all inferred tree topologies. Sublineage designations are given according to Daims et al. (17). Clone sequences with accession numbers AF293012 and AF293013 are putative chimeras between Nitrospira and gammaproteobacterium-associated sequences with chimeric breakpoints at E. coli positions 1005 and 930, respectively (http://foo.maths.uq.edu.au/∼huber/doc/chimerasMar03.nds).
RT-PCR amplification.
Crude nucleic acid extracts of the clay-loam soil from the IGER North Wyke experimental field site showed little indication of contamination by humic acids, and RT-PCR amplification was successful for 19 of the 20 samples. Direct PCR amplification of cDNA generated with the general 1492r primer was most successful when the highest possible denaturation temperature (52°C) was used during transcription of extracted rRNA, generating almost full length 16S rRNA transcription products. cDNA was usually of sufficient quality to generate amplicons of high yield in Nitrobacter- and Nitrospira-specific PCR assays without nested PCR.
16S rRNA DGGE analysis of Nitrospira communities.
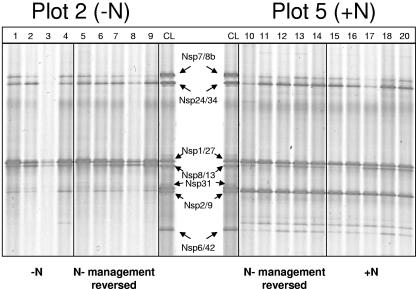
DGGE analysis of the 27f-Nspira-705r 16S rRNA PCR products nested with 357f-GC-518r, spanning the hypervariable V3 region only, produced identical patterns for replicate subplots within each of the two swards, demonstrating the presence of at least seven sequence types with different migration patterns and band intensities. DGGE migration patterns of the long-term-fertilized soil samples showed the presence of three additional bands that were absent or present only as weak bands in the long-term-unfertilized soil samples (Fig. 2), indicating changes in relative abundance of the different sequence types in Nitrospira communities. One of the dominant bands (Nsp2/9), present in all of the background long-term-fertilized samples (i.e., those currently long-term fertilized, and those where application was discontinued), was absent from, or present only as a weak band in, all of the background unfertilized samples. Band Nsp6/42 and one unidentified band migrating to a position above Nsp6/42 were present in all of the background long-term-fertilized subplots as weak bands but absent from almost all of the long-term-unfertilized subplots. Band Nsp1/27, migrating close to band Nsp8/13, appeared more consistently and with greater density throughout each of the four management regimens. One additional faint band (Nsp31) was present in two of the long-term-unfertilized subplots only, but, with this exception, DGGE profiles of subplots subjected to a reversed N treatment within the background long-term-managed pastures showed little difference from remaining replicate subplots with the continued management regime.
FIG. 2.
DGGE analysis of nested 160-bp Nitrospira-like 16S rRNA gene sequences amplified by RT-PCR from rRNA extracted from agricultural grassland soils at the IGER North Wyke experimental field station. Amplification products from soils were obtained using RT transcription primer 1492r and PCR primer 27f (40) in conjunction with Nspira-705r, nested with 357f-GC and 518r (45). Clone library (CL) PCR products were assembled from RT-27f-Nspira-705r amplicons of North Wyke grassland soils and reamplified with 357f-GC and 518r for DGGE analysis. (Left panel) Long-term-unfertilized pasture. Lanes 1 to 4, replicate subplots from long-term-unfertilized pasture; lanes 5 to 9, subplots from the long-term-unfertilized pasture after application of inorganic N fertilizer for one season. (Right panel) Long-term N-fertilized pasture. Lanes 10 to 14, replicate subplots from long-term N-managed pasture with fertilizer addition discontinued for one season; lanes 15 to 20, long-term N-fertilized subplots. Clone sequences shown in lanes CL are (from the top) Nsp7/8b, Nsp24/34, Nsp1/27, Nsp8/13, Nsp31, Nsp2/9, and Nsp6/42, as marked by arrows indicating comigrating sequences in soil samples. No 27f-Nspira-705r RT-PCR product could be generated from one long-term-unfertilized subplot.
16S rRNA DGGE analysis of Nitrobacter communities.
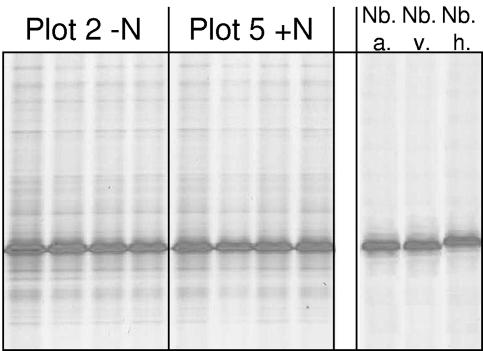
A single dominant comigrating sequence type was obtained by DGGE analysis of environmental 16S rRNA gene PCR products and of PCR products amplified from pure cultures of N. vulgaris and Nitrobacter alkalicus generated with the Nitrobacter PCR assay and nested with 357f-GC-518. The nested amplicon generated from N. hamburgensis migrated to a position marginally above the main soil N. vulgaris/N. alkalicus DGGE band. Amplicons generated from soil samples showed the additional presence of several very faint bands. DGGE migration patterns were identical for all soil samples analyzed (Fig. 3).
FIG. 3.
DGGE analysis of nested 160-bp Nitrobacter-like 16S rRNA gene sequences amplified by RT-PCR from rRNA extracted from agricultural grassland soils at the IGER North Wyke experimental field station. Amplification products from soils were obtained using RT transcription primer 1492r and PCR primer 27f (40) in conjunction with Nbac-1050r, nested with 357f-GC and 518r (45). (Left panels) Replicate subplots from long-term-unfertilized soils and replicate subplots from long-term N-fertilized soils. (Right panel) Amplicons generated from pure culture extracts.: Nb. a., Nitrobacter alkalicus; Nb. v., Nitrobacter vulgaris; Nb. h., Nitrobacter hamburgensis.
Analysis of Nitrospira clone sequences.
BLASTn GenBank database searches using the 0.7-kb clone sequences generated by the Nitrospira PCR assay retrieved clone sequences that, with two exceptions (clones Nsp24/34 and Nsp7/8b), matched most closely with sequences associated with the phylum Nitrospira in previous studies (16) (Table 2). Closest matches with sequence identities of >95% were found between almost all clone groups generated in this study and clone sequences retrieved from soil or freshwater sediments. Clone group Nsp24/34 was associated with relatively low sequence identity (79.7%) with a single clone retrieved from hydrothermal vent sediment, and the closest match of clone group Nsp7/8b was an unassociated soil clone (88.4% identity; clone 353). This group also showed the lowest 16S rRNA gene sequence identity to a pure-culture Nitrospira strain (68.9% identity to Nitrospira marina; NCBI L35501 and X82559). Clone group Nsp8/13 (94.9%) showed the highest sequence identities to the recognized NOB strain Nitrospira moscoviensis (X82558), but similarities to the recognized strain N. marina (L35501 and X82559) were only 84.5%.
TABLE 2.
Similarities of 0.7-kb Nitrospira-like 16S rRNA gene clone sequences with closest match sequences retrieved from GenBank
| Nitrospira-like clone | Closest matcha (origin) | Similarity (%) |
|---|---|---|
| Nsp7/8b | AJ582056; KCMC5; (metal-contaminated soil) | 95.9 |
| Nsp24/34 | AY225654; ATs83 (hydrothermal sediment) | 79.7 |
| Nsp1/27 | AJ534638; JG36TzT13 (contaminated soil) | 98.7 |
| Nsp8/13 | AF035813; (freshwater aquarium sediment) | 97.3 |
| Nsp31 | AJ252684; RSCII52 (rhizosphere soil) | 98.7 |
| Nsp2/9 | AF293010; GreenBayMNC2 (freshwater sediment) | 99.0 |
| Nsp6/42 | AY150907; WYO26c (grassland soil) | 97.8 |
GenBank clone sequence accession number; clone designation.
Initial phylogenetic analysis suggested the association of clone group Nsp7/8b with a distinct group containing soil clone sequences and one isolate recently suggested as a representative of the new phylum Gemmatimonadetes (73), which was therefore excluded from final analysis. The final sequence alignment produced by inferring the relationships of clone sequences in this study by iterative BLASTn searches and phylogenetic analysis contained 85 clone and pure-culture sequences. Only three sequences, however, were derived from recognized NOB strains (L35501, Nitrospira marina Nb295; X82559, Nitrospira marina; X82558, Nitrospira moscoviensis). Inferring the phylum Nitrospira-associated tree topology based on 0.65-kb 16S rRNA gene sequences with maximum likelihood, maximum parsimony, and Log/Det paralinear distance analysis suggested the placement of sequences into several discriminated clusters (Fig. 1). In general, cluster groupings within the genus Nitrospira reflected the division into sublineages proposed by Daims et al. (16). Clone sequences generated in this study were mainly associated with sublineages I and II (Fig. 1). Clone sequences Nsp2/9, Nsp11, Nsp31, and Nsp1/27s were associated with a well-supported cluster within sublineage II, exclusively containing clone sequences retrieved from soil and freshwater sediments (Fig. 1). Clone group Nsp8/13 was also placed within sublineage II but showed no association to a cluster with sufficient statistical support. Clone group Nsp6/42 was placed together with its closest BLASTn match within sublineage I, which is otherwise dominated by sequences retrieved from sewage samples. Placement of clone group Nsp24/34 at an intermediate position between the Leptospirillum and Nitrospira genera suggested the existence of an additional, deep-branching, previously unrecognized lineage. Inferring the tree topology, based on 356 parsimony informative positions with maximum likelihood, maximum parsimony, and LogDet paralinear distance analysis, gave consistent results for the major sublineage grouping, but bootstrap support for cluster grouping beyond the sublineage level was generally low.
Analysis of Nitrobacter clone sequences.
The initial sequence alignment, produced by iterative BLASTn GenBank database searches and phylogenetic analysis, suggested placement of recognized Nitrobacter sequences as a monophyletic group within the Alphaproteobacteria with high similarity to Bradyrhizobium spp. and Afipia spp. Further phylogenetic analysis, however, separated Bradyrhizobium spp. into two separate clades with high bootstrap support (95%): one containing most Bradyrhizobium spp. (Bradyrhizobium elkanii group) and a second comprising Afipia broomeae-related sequences as an outgroup and B. japonicum-associated sequences, positioned as nearest neighbor to Nitrobacter spp. in all phylogenetic analysis methods used (48, 67). Subsequent database and phylogenetic analyses therefore focused on B. japonicum/Nitrobacter-associated sequences, producing a final alignment containing 42 sequences retrieved from GenBank.
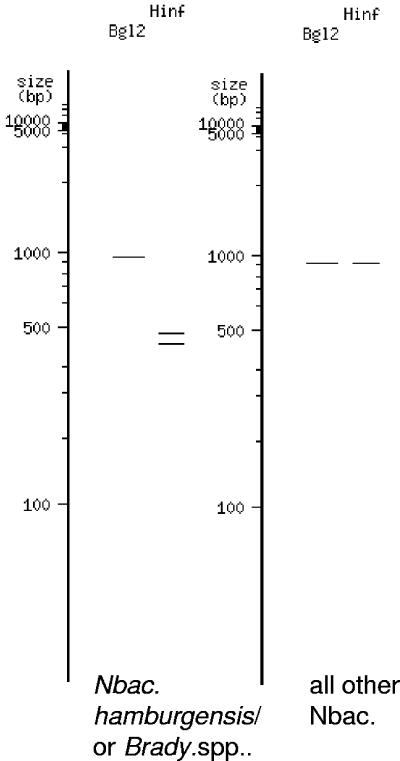
Analyzing the B. japonicum/Nitrobacter sp. sequence alignment revealed a 1-bp mismatch of the Nbacter3-1050r primer sequence with B. japonicum-associated sequences. Consensus sequences spanning the hypervariable 160-bp V3 region used in DGGE analysis of clone libraries and soil samples were identical for the Nitrobacter group and B. japonicum group A (Fig. 4) and with two G/C transversions for B. japonicum group B (Fig. 4). V3 region comparisons of recognized Nitrobacter sequences from previous studies discriminated different Nitrobacter strains by one A/C transversion for the N. hamburgensis group and several substitutions for the Nitrobacter KB212-KB215 group. High sequence identities within the V3 region were reflected by high average identities of full-length 16S rRNA gene sequences. The B. japonicum and Nitrobacter groups were on average 97.5% and 97.8% identical, and the two groups showed 98.1% consensus identity. In summary, the 1-bp mismatch in the primer sequence suggested the likely presence of B. japonicum-related sequences in soil clone libraries, and 100% consensus sequence identity of the hypervariable V3 region indicated the need for screening of clones in addition to DGGE analysis. Restriction enzyme digestion of the 27f-Nbac-1050r amplicon with HinfI discriminated all recognized nitrobacters from B. japonicum, with the exception of the N. hamburgensis group (Fig. 5). A HinfI recognition site for the B. japonicum and N. hamburgensis groups was located at E. coli position 558. Additional discrimination was achieved by restriction digestion of the 27f-Nbac-1050r amplicon with BglII, discriminating B. japonicum groups A and B, consistent with a recognition site at E. coli position 648 (Fig. 5).
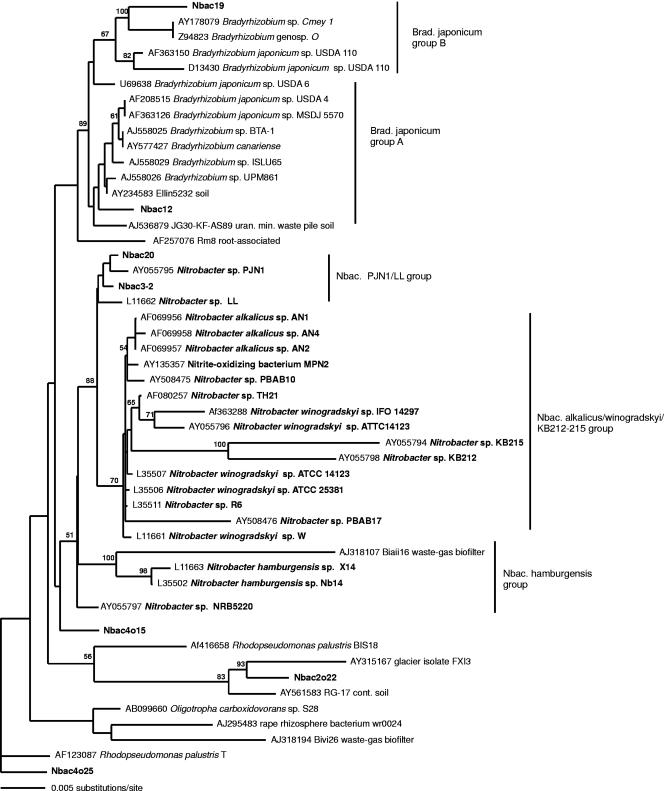
FIG. 4.
Evolutionary distance dendrogram showing the positions of environmental 16S rRNA gene 0.91-kb clone sequences generated with the 27f-Nbac-1050r primer assay from North Wyke grassland soils (boldfaced) in relation to pure Nitrobacter culture sequences (boldfaced) and associated clone and culture sequences. Sequences generated in this study were included in the analysis only when represented by at least two replicate clones (100% sequence similarity). The tree is based on results of a neighbor-joining analysis of LogDet paralinear distances (Pin. var. 0.7331; 81 parsimony informative characters). Bootstrap values are indicated at nodes when they exceed 50% of replicates (1,000 resamplings). Bar indicates an estimated 0.005 change per nucleotide position.
FIG. 5.
Restriction fragment length polymorphism pattern for 1-kb Nitrobacter sp. and closely related Bradyrhizobium japonicum sp. 16S rRNA gene sequences derived from the virtual digest tool available at http://tools.neb.com/REBsites/index.php3.
Of the four clone libraries, 83% (long-term N fertilized), 63% (long-term fertilized, N application discontinued), 65% (long-term unfertilized, N currently applied), and 52% (long-term unfertilized) were identified as Nitrobacter-like sequences. Closest matches of all clones selected for sequencing according to DGGE and RFLP analyses were Nitrobacter spp. and B. japonicum. Additional clones with divergent DGGE and RFLP patterns that were excluded from the final phylogenetic analysis were predominantly associated with the B. elkanii group or with an uncharacterized alphaproteobacterial clone group, wr0197 (34). Overall, the combination of clone screening by DGGE and RFLP led to a significant increase in the proportion of sequenced Nitrobacter clones.
Inferring the final Nitrobacter-associated tree topology based on 0.95-kb 16S rRNA gene sequences with maximum-likelihood analysis and LogDet paralinear distance analysis suggested the predominant placement of sequences generated from the four different clone libraries as closest neighbors to Nitrobacter sp. strain PJN1 and, additionally, placement of a minor proportion (approximately 2 out of 10 clones per library) as closest neighbors to Nitrobacter sp. strain LL (Fig. 4). All sequenced clones within the libraries that were associated with the main DGGE migration band, and with a corresponding HinfI recognition site, either fell within B. japonicum group A (Nbac12 group) or were isolated in an intermediate topological position between B. japonicum and Nitrobacter spp. (Nbac4o15 group). Two additional clone sequences containing the HinfI recognition site were associated with the N. hamburgensis group (Nbac21 group). Closely related clone sequences with diverging DGGE migration patterns were associated with Rhodopseudomonas palustris (Nbac4o25) and a glacier isolate (Nbac2o22). Clone sequences with a Nitrobacter/B. japonicum matching DGGE migration pattern, but with a HinfI and a BglII recognition site (Nbac19 group), were associated with B. japonicum group B.
Separation of clades associated with Nitrobacter strain PJN1/Ll, N. alcalicus/winogradskyi/KB212-KB215 (Fig. 4), and B.japonicum was achieved by all phylogenetic analysis methods employed. However, discrimination of sequences of N. hamburgensis from B. japonicum was possible only by using maximum-likelihood and LogDet paralinear distance analysis, and statistical bootstrap support for nodes separating B. japonicum- and Nitrobacter sp.-associated clades was below 50%. Almost identical tree topologies were achieved by analyzing 1.4-kb sequences only.
DISCUSSION
Investigation of soil nitrifiers has focused on AOB (4, 49, 70), and little is known of the diversity and activity of soil NOB. Analysis of sewage treatment systems indicates, however, that the diversity of NOB communities may be related to nitrite concentration, partial oxygen pressure, and the availability of simple organic substrates, which are utilized by many mixotrophic NOB (16, 54). Furthermore, the response of such systems to changes in nitrite concentration, and the consequent removal of toxic nitrite, may depend on NOB activity and species composition (16, 56, 57, 59). In agricultural systems, fertilization regimes usually involve several applications of inorganic and organic nitrogen annually. The onset of nitrification, while depending on soil type, moisture, and temperature, is generally rapid, and nitrite rarely accumulates. However, Webster et al. (70) recently demonstrated that lags in nitrification in grassland soils, following input of high levels of ammonia, are related to the relative abundance of particular AOB phylotypes (Nitrosospira clusters 3a and 3b). This study provides the first evidence for the influence of agricultural N management regimes on the diversity of nitrite-oxidizing bacteria.
Soil Nitrospira communities.
Community analysis of soil NOB posed contrasting problems for Nitrospira and Nitrobacter. Few nitrospirae have been cultured, but numerous 16S rRNA gene sequences, retrieved from a variety of environments, provide evidence for a large and highly diverse group (16). Highly divergent 16S rRNA gene sequences within the phylum Nitrospira enabled the application of DGGE as a fast screening method for the analysis of Nitrospira grassland soil communities. Confirmation that all Nitrospira-like sequences within this monophyletic group are derived from NOB awaits characterization of cultured strains, but there is some support from studies combining FISH and nitrification process measurements or microautoradiography (16, 56, 57, 59). Probes designed by Schramm (58) and Daims (16) have been successfully applied for detection of Nitrospira in wastewater treatment systems and freshwater environments (56), but application of FISH to soil environments is rarely feasible. The FISH probe S-Ntspa-0712-a-A-21 (16) and primer Nspira705r target the same motif but yielded multiple DGGE banding patterns when used as PCR primers, indicating nonspecific amplification. Exploitation of the Nitrospira-specific 16S rRNA gene motif with the PCR assay developed in this study offers an additional approach to this FISH probe and, with DGGE or clone screening, enables characterization of Nitrospira communities and detailed phylogenetic analysis. The high sequence diversity within the genus Nitrospira and the newly added clone sequence groups necessitated a PCR primer assay that was not exclusive of currently undiscovered groups but had sufficient specificity to target preferentially all Nitrospira sequences. Alignment of the Nspira-705r primer sequence with sequences from the phylum Nitrospira indicates an increasing number of mismatches against the Leptospirillum and Thermodesulfovibrio groups. Clone sequences associated with the newly proposed phylum Gemmatimonadetes (73) mostly showed two or three mismatches within the primer target sequence, although some showed one mismatch.
Phylogenetic analysis placed soil clone group Nsp2/9 in a cluster of sequences previously assigned with Nitrospira sublineage II (Fig. 1) (16), which constituted one of the dominant DGGE bands in the long-term N-fertilized plot but was absent or at low relative abundance in long-term-unfertilized soils. Soil clone groups Nsp11, Nsp31, and Nsp1/27, when analyzed with additional sequences retrieved from soil, freshwater, and groundwater environments, also fell into the same cluster within the highly diverse sublineage II (Fig. 1). This cluster was retrieved with good bootstrap support in all inferred tree topologies and may indicate a previously unrecognized ecotype. Additionally, the concurrent placement of soil clone groups Nsp6/42 and Nsp8/13 with single sequences isolated from similar habitats within sublineage I and within the main cluster of sublineage II, respectively, which are otherwise dominated by sewage clone sequences, may indicate further phylogenetic discrimination with the inclusion of more environmental sequences. However, with the exception of the cluster within sublineage II, bootstrap support for discrimination of sublineages I, III, and IV into different clusters was low, and more-detailed correlation of ecophysiology and phylogeny requires inclusion of additional sequences. Nitrospira sequences in current public databases are dominated by those from wastewater treatment plants and sewage reactors, and phylogenetic analysis of these sequences may be biased toward evolutionary groups adapted to high NO2− concentrations.
Soil Nitrobacter communities.
In contrast to Nitrospira, public databases contain only two clone sequences associated with recognized nitrobacters. All other sequences within the monophyletic boundaries of Nitrobacter are derived from a relatively small number of pure NOB cultures. Previous studies (18, 47, 48) indicate close evolutionary relationships between Nitrobacter, Bradyrhizobiaceae, and Afipia, and the likely abundance of Bradyrhizobiaceae in soil therefore necessitated design of Nitrobacter primers discriminating against these groups. The PCR assay developed here achieved the required selectivity, but close evolutionary relationships within all recognized Nitrobacter strains and between B. japonicum and Nitrobacter clades required thorough analysis of almost full length 16S rRNA gene sequences for discrimination and prevented the use of DGGE as a screening method for environmental Nitrobacter communities. For example, the average full-length 16S rRNA gene sequence identity of all nitrobacters was 98%, and that between Nitrobacter and B. japonicum clades was also 98%. The genus Nitrobacter therefore represents a very narrow group of highly similar 16S rRNA gene phylogeny, closely related to phylogenetic neighbors with very different characteristics. Detection and identification of Nitrobacter in soil environments has previously involved enrichment of NOB on selective media prior to PCR amplification or serotyping (6, 18, 20, 26). Enumeration of Nitrobacter in environmental samples by most-probable-number-PCR used the Nitrobacter specific PCR primer FGPS1269 with a general bacterial primer (14, 19-21). In assessing the specificity of this primer set within a highly competitive soil environment, clone sequences that were mainly closely related to Bradyrhizobium spp., Methylosinus spp., and the undescribed clone group wr0197 (34) were retrieved. No Nitrobacter-associated clone sequence was generated from grassland soils in this study with the Nitrobacter PCR primer FGPS1269 (data not shown). However, the combination of a highly specific Nitrobacter PCR assay with clone screening by DGGE and RFLP allowed direct PCR amplification of Nitrobacter-like sequences without the potential bias of laboratory enrichment. As suggested by Grundmann et al. (26), the close evolutionary relationships of all Nitrobacter strains indicate that alternative, more-discriminatory target genes may be required for detection of different ecotypes (for example, genes encoding the nitrite oxidoreductase alpha and beta subunits [norA and norB]).
Influence of management regime on nitrite oxidizer communities.
Differences in NO2− oxidation rates have been observed previously using soil slurries from long-term-fertilized and unfertilized pastures investigated in this study (27). The successful amplification of Nitrospira-like sequences from soil indicates a sufficient degree of specificity of the PCR assay, while amplification of non-Nitrospira sequences suggests inclusion of currently closely related uncharacterized groups. Nitrospira-like sequences have been documented in numerous clone surveys generated with general bacterial PCR primers (5, 8, 10, 29, 35, 42, 65, 72). This may result from a bias of bacterial primers toward the phylum Nitrospira but also suggests the ubiquity and high relative abundance of these organisms in natural communities, indicating a role in other ecosystem processes, in addition to nitrite oxidation, which may not be sufficient to support such high population levels.
Sequence information derived from the 16S rRNA Nitrobacter PCR assay indicated the dominance of strains related to Nitrobacter sp. strain PJN1, which was recently isolated from barley rhizoplane samples (55) in media supplemented with root exudates, suggesting adaptation to the rhizosphere. Its high optimum substrate concentration for growth (11.6 mM NO2−N), compared to 0.35 mM for Nitrospira moscoviensis, may suggest adaptation to high nitrite concentrations, as with other Nitrobacter spp. (50). The presence of Nitrobacter sp. strain PJN1 sequence types in both fertilized and unfertilized soil thus suggests the presence of microenvironments within the rhizosphere with temporary or constant high nitrite concentrations that may provide the ecological niche for Nitrobacter spp. within an otherwise low-substrate habitat. The similar proportions of Nitrobacter sequence types and the low number of clone sequences did not enable confident discrimination of the different Nitrobacter communities. Nevertheless, association of the soil clone groups with Nitrobacter sp. strain PJN1, Nitrobacter sp. strain LL, or N. hamburgensis suggests sufficient resolution to distinguish Nitrobacter soil communities with respect to recognized Nitrobacter strains. DGGE migration patterns of Nitrospira communities and ratios of relative abundances of Nitrobacter and B. japonicum suggest different NOB communities in the long-term-managed pastures. The long-term differences in the N fertilization regimes may have favored Nitrospira strains of the Nsp2/9, and to a lesser extent the Nsp6/42, sequence type in the long-term N-fertilized plot and may have also increased the cell numbers of Nitrobacter in relation to B. japonicum. However, there was little evidence of a change in community structure within 1 year of reversal of the treatment regimes.
In conclusion, this study provides methods for the molecular detection of Nitrobacter and Nitrospira in environmental samples. These methods have been used for the first molecular characterization of soil NOB communities. The results indicate a low diversity of Nitrobacter strains but extend evidence that Nitrospira sequences are diverse and abundant in natural environments and suggest a broad ecosystem function for this group.
Acknowledgments
This study was supported by the Biotechnology and Biological Sciences Research Council, grant 1/D15955.
We acknowledge H. Daims and M. Wagner for comments on a draft of the manuscript.
REFERENCES
- 1.Altmann, D., P. Stief, R. Amann, D. de Beer, and A. Schramm. 2003. In situ distribution and activity of nitrifying bacteria in freshwater sediment. Environ. Microbiol. 5:798-803. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Ashelford, K. E., A. J. Weightman, and J. C. Fry. 2002. PRIMROSE: a computer program for generating and estimating the phylogenetic range of 16S rRNA oligonucleotide probes and primers in conjunction with the RDP-II database. Nucleic Acids Res. 30:3481-3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avrahami, S., W. Liesack, and R. Conrad. 2003. Effects of temperature and fertilizer on activity and community structure of soil ammonia oxidizers. Environ. Microbiol. 5:691-705. [DOI] [PubMed] [Google Scholar]
- 5.Bakermans, C., and E. L. Madsen. 2002. Diversity of 16S rDNA and naphthalene dioxygenase genes from coal-tar-waste-contaminated aquifer waters. Microb. Ecol. 44:95-106. [DOI] [PubMed] [Google Scholar]
- 6.Bartosch, S., C. Hartwig, E. Spieck, and E. Bock. 2002. Immunological detection of Nitrospira-like bacteria in various soils. Microb. Ecol. 43:26-33. [DOI] [PubMed] [Google Scholar]
- 7.Bock, E., H. P. Koops, U. C. Moller, and M. Rudert. 1990. A new facultatively nitrite oxidizing bacterium, Nitrobacter vulgaris sp nov. Arch. Microbiol. 153:105-110. [Google Scholar]
- 8.Bond, P. L., R. Erhart, M. Wagner, J. Keller, and L. L. Blackall. 1999. Identification of some of the major groups of bacteria in efficient and nonefficient biological phosphorus removal activated sludge systems. Appl. Environ. Microbiol. 65:4077-4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Briones, A. M., S. Okabe, Y. Umemiya, N. B. Ramsing, W. Reichardt, and H. Okuyama. 2002. Influence of different cultivars on populations of ammonia-oxidizing bacteria in the root environment of rice. Appl. Environ. Microbiol. 68:3067-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brofft, J. E., J. V. McArthur, and L. J. Shimkets. 2002. Recovery of novel bacterial diversity from a forested wetland impacted by reject coal. Environ. Microbiol. 4:764-769. [DOI] [PubMed] [Google Scholar]
- 11.Bruns, M. A., J. R. Stephen, G. A. Kowalchuk, J. I. Prosser, and E. A. Paul. 1999. Comparative diversity of ammonia oxidizer 16S rRNA gene sequences in native, tilled, and successional soils. Appl. Environ. Microbiol. 65:2994-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burrell, P. C., J. Keller, and L. L. Blackall. 1998. Microbiology of a nitrite-oxidizing bioreactor. Appl. Environ. Microbiol. 64:1878-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burrell, P. C., C. M. Phalen, and T. A. Hovanec. 2001. Identification of bacteria responsible for ammonia oxidation in freshwater aquaria. Appl. Environ. Microbiol. 67:5791-5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cébron, A., T. Berthe, and J. Garnier. 2003. Nitrification and nitrifying bacteria in the lower Seine River and estuary (France). Appl. Environ. Microbiol. 69:7091-7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daims, H., J. L. Nielsen, P. H. Nielsen, K. H. Schleifer, and M. Wagner. 2001. In situ characterization of Nitrospira-like nitrite oxidizing bacteria active in wastewater treatment plants. Appl. Environ. Microbiol. 67:5273-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daims, H., P. H. Nielsen, J. L. Nielsen, S. Juretschko, and M. Wagner. 2000. Novel Nitrospira-like bacteria as dominant nitrite-oxidizers in biofilms from wastewater treatment plants: diversity and in situ physiology. Water Sci. Technol. 41:85-90. [Google Scholar]
- 18.Degrange, V., and R. Bardin. 1995. Detection and counting of Nitrobacter populations in soil by PCR. Appl. Environ. Microbiol. 61:2093-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Degrange, V., R. Lensi, and R. Bardin. 1997. Activity, size and structure of a Nitrobacter community as affected by organic carbon and nitrite in sterile soil. FEMS Microbiol. Ecol. 24:173-180. [Google Scholar]
- 20.Degrange, V., M. M. Couteaux, J. M. Anderson, M. P. Berg, and R. Lensi. 1998. Nitrification and occurrence of Nitrobacter by MPN-PCR in low and high nitrifying coniferous forest soils. Plant Soil 198:201-208. [Google Scholar]
- 21.Feray, C., B. Volat, V. Degrange, A. Clays-Josserand, and B. Montuelle. 1999. Assessment of three methods for detection and quantification of nitrite-oxidizing bacteria and Nitrobacter in freshwater sediments (MPN-PCR, MPN-Griess, immunofluorescence). Microb. Ecol. 37:208-217. [DOI] [PubMed] [Google Scholar]
- 22.Freitag, T. E., and J. I. Prosser. 2004. Differences between betaproteobacterial ammonia-oxidizing communities in marine sediments and those in overlying water. Appl. Environ. Microbiol. 70:3789-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freitag, T. E., and J. I. Prosser. 2003. Community structure of ammonia-oxidizing bacteria within anoxic marine sediments. Appl. Environ. Microbiol. 69:1359-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gieseke, A., P. Arnz, R. Amann, and A. Schramm. 2002. Simultaneous P and N removal in a sequencing batch biofilm reactor: insights from reactor- and microscale investigations. Water Res. 36:501-509. [DOI] [PubMed] [Google Scholar]
- 25.Griffiths, R. I., A. S. Whiteley, A. G. O'Donnell, and M. J. Bailey. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grundmann, G. L., M. Neyra, and P. Normand. 2000. High-resolution phylogenetic analysis of NO2-oxidizing Nitrobacter species using the rrs-rrl IGS sequence and rrl genes. Int. J. Syst. Evol. Microbiol. 50:1893-1898. [DOI] [PubMed] [Google Scholar]
- 27.Hatch, D. J., S. C. Jarvis, and R. J. Parkinson. 1996. Nitrification in grassland soils, p. 239-241. In Transactions of the 9th Nitrogen Workshop. Technische Universitaet, Braunschweig, Braunschweig, Germany.
- 28.Hatch, D. J., S. C. Jarvis, R. J. Parkinson, and R. D. Lovell. 2000. Combining field incubation with nitrogen-15 labelling to examine nitrogen transformations in low to high intensity grassland management systems. Biol. Fertil. Soils 30:492-499. [Google Scholar]
- 29.Holmes, A. J., N. A. Tujula, M. Holley, A. Contos, J. M. James, P. Rogers, and M. R. Gillings. 2001. Phylogenetic structure of unusual aquatic microbial formations in Nullarbor caves, Australia. Environ. Microbiol. 3:256-264. [DOI] [PubMed] [Google Scholar]
- 30.Hovanec, T. A., L. T. Taylor, A. Blakis, and E. F. Delong. 1998. Nitrospira-like bacteria associated with nitrite oxidation in freshwater aquaria. Appl. Environ. Microbiol. 64:258-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 32.Juretschko, S., A. Loy, A. Lehner, and M. Wagner. 2002. The microbial community composition of a nitrifying-denitrifying activated sludge from an industrial sewage treatment plant analyzed by the full-cycle rRNA approach. Syst. Appl. Microbiol. 25:84-99. [DOI] [PubMed] [Google Scholar]
- 33.Juretschko, S., G. Timmermann, M. Schmid, K. H. Schleifer, A. Pommerening-Röser, H. P. Koops, and M. Wagner. 1998. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl. Environ. Microbiol. 64:3042-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaiser, O., A. Puhler, and W. Selbitschka. 2001. Phylogenetic analysis of microbial diversity in the rhizoplane of oilseed rape (Brassica napus cv. Westar) employing cultivation-dependent and cultivation-independent approaches. Microb. Ecol. 42:136-149. [DOI] [PubMed] [Google Scholar]
- 35.Kindaichi, T., T. Ito, and S. Okabe. 2004. Ecophysiological interaction between nitrifying bacteria and heterotrophic bacteria in autotrophic nitrifying biofilms as determined by microautoradiography-fluorescence in situ hybridization. Appl. Environ. Microbiol. 70:1641-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koops, H. P., and A. Pommerening-Röser. 2001. Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol. Ecol. 37:1-9. [Google Scholar]
- 37.Kowalchuk, G. A., and J. R. Stephen. 2001. Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu. Rev. Microbiol. 55:485-529. [DOI] [PubMed] [Google Scholar]
- 38.Kowalchuk, G. A., P. L. E. Bodelier, G. H. J. Heilig, J. R. Stephen, and H. J. Laanbroek. 1998. Community analysis of ammonia-oxidising bacteria, in relation to oxygen availability in soils and root-oxygenated sediments, using PCR, DGGE and oligonucleotide probe hybridisation. FEMS Microbiol. Ecol. 27:339-350. [Google Scholar]
- 39.Lake, J. A. 1994. Reconstructing evolutionary trees from DNA and protein sequences—paralinear distances. Proc. Natl. Acad. Sci. USA 91:1455-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Academic Press, Chichester, England.
- 41.Lewis, W. M., and D. P. Morris. 1986. Toxicity of nitrite to fish: a review. Am. Fish. Soc. 115:183-195. [Google Scholar]
- 42.Li, L. N., C. Kato, and K. Horikoshi. 1999. Bacterial diversity in deep-sea sediments from different depths. Biodivers. Conserv. 8:659-677. [Google Scholar]
- 43.Lockhart, P. J., A. W. D. Larkum, M. A. Steel, P. J. Waddell, and D. Penny. 1996. Evolution of chlorophyll and bacteriochlorophyll: the problem of invariant sites in sequence analysis. Proc. Natl. Acad. Sci. USA 93:1930-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCaig, A. E., L. A. Glover, and J. I. Prosser. 2001. Numerical analysis of grassland bacterial community structure under different land management regimens by using 16S ribosomal DNA sequence data and denaturing gradient gel electrophoresis banding patterns. Appl. Environ. Microbiol. 67:4554-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S ribosomal RNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Navarro, E., M. P. Fernandez, F. Grimont, A. Claysjosserand, and R. Bardin. 1992. Genomic heterogeneity of the genus Nitrobacter. Int. J. Syst. Bacteriol. 42:554-560. [Google Scholar]
- 47.Orso, S., M. Gouy, E. Navarro, and P. Normand. 1994. Molecular phylogenetic analysis of Nitrobacter spp. Int. J. Syst. Bacteriol. 44:83-86. [DOI] [PubMed] [Google Scholar]
- 48.Parker, M. A., B. Lafay, J. J. Burdon, and P. van Berkum. 2002. Conflicting phylogeographic patterns in rRNA and nifD indicate regionally restricted gene transfer in Bradyrhizobium. Microbiology 148:2557-2565. [DOI] [PubMed] [Google Scholar]
- 49.Phillips, C. J., D. Harris, S. L. Dollhopf, K. L. Gross, J. I. Prosser, and E. A. Paul. 2000. Effects of agronomic treatments on structure and function of ammonia-oxidizing communities. Appl. Environ. Microbiol. 66:5410-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prosser, J. I. 1989. Autotrophic nitrification in bacteria. Adv. Microb. Physiol. 30:125-181. [DOI] [PubMed] [Google Scholar]
- 51.Purkhold, U., M. Wagner, G. Timmermann, A. Pommerening-Röser, and H. P. Koops. 2003. 16S rRNA and amoA-based phylogeny of 12 novel betaproteobacterial ammonia-oxidizing isolates: extension of the dataset and proposal of a new lineage within the nitrosomonads. Int. J. Syst. Evol. Microbiol. 53:1485-1494. [DOI] [PubMed] [Google Scholar]
- 52.Regan, J. M., G. W. Harrington, and D. R. Noguera. 2002. Ammonia- and nitrite-oxidizing bacterial communities in a pilot-scale chloraminated drinking water distribution system. Appl. Environ. Microbiol. 68:73-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robertson, G. P. 1997. Nitrogen use efficiency in row-crop agriculture: crop nitrogen use and soil nitrogen loss, p. 347-365. In L. E. Jackson (ed.), Ecology in agriculture. Academic Press, San Diego, Calif.
- 54.Satoh, H., S. Okabe, Y. Yamaguchi, and Y. Watanabe. 2003. Evaluation of the impact of bioaugmentation and biostimulation by in situ hybridization and microelectrode. Water Res. 37:2206-2216. [DOI] [PubMed] [Google Scholar]
- 55.Satoh, K., T. Yanagida, K. Isobe, H. Tomiyama, R. Takahashi, H. Iwano, and T. Tokuyama. 2003. Effect of root exudates on growth of newly isolated nitrifying bacteria from barley rhizoplane. Soil Sci. Plant Nutr. 49:757-762. [Google Scholar]
- 56.Schramm, A. 2003. In situ analysis of structure and activity of the nitrifying community in biofilms, aggregates, and sediments. Geomicrobiol. J. 20:313-333. [Google Scholar]
- 57.Schramm, A., D. De Beer, A. Gieseke, and R. Amann. 2000. Microenvironments and distribution of nitrifying bacteria in a membrane-bound biofilm. Environ. Microbiol. 2:680-686. [DOI] [PubMed] [Google Scholar]
- 58.Schramm, A., D. De Beer, M. Wagner, and R. Amann. 1998. Identification and activities in situ of Nitrosospira and Nitrospira spp. as dominant populations in a nitrifying fluidized bed reactor. Appl. Environ. Microbiol. 64:3480-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schramm, A., D. de Beer, J. C. van den Heuvel, S. Ottengraf, and R. Amann. 1999. Microscale distribution of populations and activities of Nitrosospira and Nitrospira spp. along a macroscale gradient in a nitrifying bioreactor: quantification by in situ hybridization and the use of microsensors. Appl. Environ. Microbiol. 65:3690-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schramm, A., D. De Beer, H. van den Heuvel, S. Ottengraf, and R. Amann. 1998. In situ structure/function studies in wastewater treatment systems. Water Sci. Technol. 37:413-416. [Google Scholar]
- 61.Schramm, A., L. H. Larsen, N. P. Revsbech, N. B. Ramsing, R. Amann, and K. H. Schleifer. 1996. Structure and function of a nitrifying biofilm as determined by in situ hybridization and the use of microelectrodes. Appl. Environ. Microbiol. 62:4641-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sorokin, D. Y., G. Muyzer, T. Brinkhoff, J. G. Kuenen, and M. S. M. Jetten. 1998. Isolation and characterization of a novel facultatively alkaliphilic Nitrobacter species, N. alkalicus sp. nov. Arch. Microbiol. 170:345-352. [DOI] [PubMed] [Google Scholar]
- 63.Stein, L. Y., and D. J. Arp. 1998. Loss of ammonia monooxygenase activity in Nitrosomonas europaea upon exposure to nitrite. Appl. Environ. Microbiol. 64:4098-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Swofford, D. L. PAUP*;: phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland, Mass.
- 65.Tarre, S., and M. Green. 2004. High-rate nitrification at low pH in suspended- and attached-biomass reactors. Appl. Environ. Microbiol. 70:6481-6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Teske, A., E. Alm, J. M. Regan, S. Toze, B. E. Rittmann, and D. A. Stahl. 1994. Evolutionary relationships among ammonia- and nitrite-oxidizing bacteria. J. Bacteriol. 176:6623-6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Berkum, P., Z. Terefework, L. Paulin, S. Suomalainen, K. Lindstrom, and B. D. Eardly. 2003. Discordant phylogenies within the rrn loci of rhizobia. J. Bacteriol. 185:2988-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wagner, A., N. Blackstone, P. Cartwright, M. Dick, B. Misof, P. Snow, G. P. Wagner, J. Bartels, M. Murtha, and J. Pendleton. 1994. Surveys of gene families using polymerase chain-reaction—PCR selection and PCR drift. Syst. Biol. 43:250-261. [Google Scholar]
- 69.Watson, S. W., and J. B. Waterbury. 1971. Characteristics of two marine nitrite-oxidizing bacteria, Nitrospina gracilis nov. gen. nov. sp. and Nitrococcus mobilis nov. gen. nov. sp. Arch. Microbiol. 77:203-230. [Google Scholar]
- 70.Webster, G., T. M. Embley, T. E. Freitag, Z. Smith, and J. I. Prosser. 2005. Links between ammonia oxidiser species composition, functional diversity and nitrification kinetics in grassland soils. Environ. Microbiol. 7:676-684. [DOI] [PubMed] [Google Scholar]
- 71.Willems, A., and M. D. Collins. 1992. Evidence for a close genealogical relationship between Afipia (the causal organism of cat scratch disease), Bradyrhizobium japonicum and Blastobacter denitrificans. FEMS Microbiol. Lett. 96:241-246. [DOI] [PubMed] [Google Scholar]
- 72.Williams, M. M., J. W. S. Domingo, M. C. Meckes, C. A. Kelty, and H. S. Rochon. 2004. Phylogenetic diversity of drinking water bacteria in a distribution system simulator. J. Appl. Microbiol. 96:954-964. [DOI] [PubMed] [Google Scholar]
- 73.Zhang, D., D. M. Zhang, Y. P. Liu, W. W. Cao, and G. X. Chen. 2004. Community analysis of ammonia oxidizer in the oxygen-limited nitritation stage of OLAND system by DGGE of PCR amplified 16S rDNA fragments and FISH. J. Environ. Sci. 16:838-842. [PubMed] [Google Scholar]