Abstract
The signals that control the transcription of osmoregulated genes are not understood satisfactorily. The “turgor control model” suggested that the primary osmoregulatory signal in Enterobacteriaceae is turgor loss, which induces the kdp K+ transport operon and activates the Trk K+ permease. The ensuing increase in cytoplasmic K+ concentration was proposed to be the signal that turns on all secondary responses, including the induction of the proU (proline-glycine betaine transport) operon. The “ionic strength model” proposed that the regulatory signal for all osmotically controlled responses is the increase in the cytoplasmic ionic strength or macromolecular crowding after an osmotic upshift. The assumption in the turgor control model that the induction of kdp is a primary response to osmotic shock predicts that this response should precede all secondary responses. Both models predict that the induction of all osmotically activated responses should be independent of the chemical nature of the solute used to impose osmotic stress. We tested these predictions by quantitative real-time reverse transcription-PCR analysis of the expression of six osmotically regulated genes in Salmonella enterica serovar Typhimurium. After shock with 0.3 M NaCl, proU was induced at 4 min, proP and rpoS were induced at 4 to 6 min, kdp was induced at 8 to 9 min, and otsB and ompC were induced at 10 to 12 min. After an equivalent osmotic shock with 0.6 M sucrose, proU was induced with kinetics similar to those seen with NaCl, but induction of kdp was reduced 150-fold in comparison to induction by NaCl. Our results are inconsistent with both the turgor control and the ionic strength control models.
Organisms respond to changes in external osmolality by accumulating or releasing low-molecular-weight “compatible solutes” that maintain the proper balance between internal and external osmolality. One of the prominent responses of Enterobacteriaceae to high ion concentrations is transcriptional induction a >100-fold of two operons: proU (proVWX), which encodes a transport system for the osmoprotectant compounds glycine betaine and proline, and kdpABC, which specifies the components of a high-affinity K+ transport system.
The signal sensing mechanisms of the osmotic control of transcription of the kdp and proU operons are uncertain. Two osmoregulatory models were proposed for Enterobacteriaceae. The “turgor control model,” which has been very influential in shaping thinking about bacterial osmoregulation, suggests that the fundamental signal is the loss of turgor resulting from the efflux of water after an osmotic upshift (3, 13). This model postulates that the first response of the cells to turgor loss is increased uptake of K+, accomplished by stimulation of the activities of the constitutive, low-affinity K+ transport systems Trk and Kup and by transcriptional induction of the kdpABC operon. The model further proposes that elevated K+ acts as a second messenger to turn on all other osmotically activated responses, including the induction of proU. Finally, the model postulates that when K+ and the counterion glutamate are accumulated to sufficient levels, turgor is restored and the transcription of the kdp operon returns to the prestress basal level. The “ionic strength model” proposes that the regulatory stimulus for the activation of osmotically controlled responses is the increase in the ion concentration of the cytoplasm (35). Because the reduction in volume resulting from plasmolysis increases the concentrations of all cytoplasmic components, from small metabolites to macromolecules, in principle the increase in the concentration of a variety of molecules could provide a signal to set off osmotically activated responses (8). The elevated K+ concentration, together with macromolecular crowding, has been shown to stimulate the activity of the ProP transport system under osmotic stress (10), and it is conceivable that such a signal could be used to regulate the transcription of osmoresponsive genes. Because K+ glutamate is the major determinant of the cytoplasmic ionic strength (6), the turgor control and the ionic strength models are very similar; the main difference between them is that is that the former model suggests that K+ uptake and the induction of the kdp operon are regulated by turgor loss and all other responses are secondary consequences of the accumulation of K+ glutamate, whereas the latter model suggests that all osmotically activated responses, including the induction of the kdp operon, are turned on by the increase in ionic strength.
The two models make a number of testable predictions. It has been proposed in the turgor control model that kdp is induced by low turgor pressure when Trk activity is not sufficient to maintain turgor (3, 13). If the function of the osmotic induction of kdp is to enable the cells to acquire K+ when Trk is inadequate, then it would be expected that the osmotic induction of this operon would precede the secondary responses that are suggested to depend on the accumulation of K+. The fundamental cause for both turgor loss and volume reduction after an osmotic upshift is the decrease in external water activity imposed by impermeant solutes. Thus, both the turgor control and the ionic strength control models imply that the response of every osmotically regulated function would depend on the decrease in the water activity, regardless of the chemical nature of impermeant solutes that are used to impose the osmotic shock. We tested the above predictions by quantitative real-time reverse transcription-PCR (Q-RT-PCR) analysis to determine the kinetics of induction of proU, kdp, and four other osmotically regulated operons in Salmonella enterica serovar Typhimurium.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
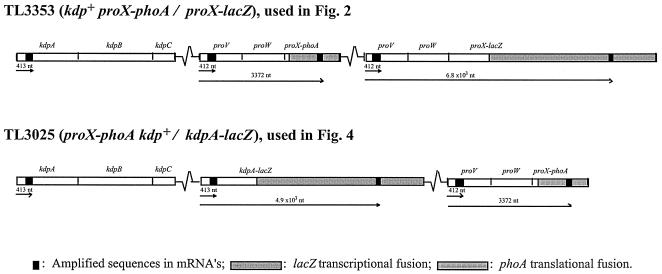
The experiments in this work were done with derivatives of S. enterica serovar Typhimurium LT2. Strain TL3025 (proX1917::TnphoA DUP [kdpA101::Mud1-8::nadA216]/kdp+) is a Kdp+ strain, which carries an intact copy of the kdpABCDE genes and a kdpA-lacZ transcriptional fusion generated by phage Mud1-8 (14) in a chromosomal duplication of the kdpA-nadA segment of the chromosome. Strain TL3353 (DUP [proX(proU)1884::Mud1-8::pheA564]/proX1917::TnphoA) carries a chromosomal duplication of the pheA-to-proU region of the chromosome, in which one copy of the proU operon contains a proX-phoA translational fusion (obtained from E. Eisenstadt via J. Galán) and a second copy carries a proX-lacZ translational fusion (30). The chromosomal duplications in these strains were constructed essentially by the method of Hughes and Roth (20). Both strains are ProU−, but the absence of this transport system has no effect on the transcriptional regulation of the kdp or proU operons (11). The sites of insertion of the proX1917::TnphoA and kdpA101::Mud1-8 fusions are 2,462 and 1,467 nucleotides (nt) downstream of the proU and kdp promoters, respectively, as determined by PCR amplification and DNA sequencing (data not shown). The site of insertion of the proX1884::Mud1-8 is 3.4 kbp downstream of the proU promoter, determined by Southern blot analysis (30). Strain TL3799 (proX1917::TnphoA/F′128 lac+ mhpC31::Tn10dTc) carries the wild-type lac operon of Escherichia coli K-12 on the low-copy-number plasmid F′.
The medium used for the study of the high-osmolality-dependent regulation of the kdp and proU operons was K medium, which has been used previously in other studies of the osmotic control of expression of the kdp and proU operons (1, 11, 16). This medium contains 1 mM KH2PO4, 1 mM (NH4)2SO4, 0.08 mM MgCl2 · 6H2O, 1.8 μM Fe2SO4 · 6H2O, 5 g liter−1 Casamino Acids (Difco), and 10 mM glucose. The osmolality of this medium was increased by 0.3 M NaCl or 0.6 M sucrose, and in each case the pH was adjusted to 7.6 with Tris base. The K+ concentration of the K medium was confirmed with inductively coupled plasma atomic emission spectrometry to be 1 mM. The osmolalities of K medium and K medium containing 0.3 M NaCl or 0.6 M sucrose were 147, 638, and 662 mosM, respectively, measured by a Wescor 5100 osmometer. Growth was with aeration at 37°C. Ampicillin was added at 25 μg ml−1 for maintenance of the chromosomal duplications in strains TL3025 and TL3353, and tetracycline was added at 15 μg ml−1 for the maintenance of the F′128 lac+ mhpC31::Tn10dTc.
RNA extraction and cDNA synthesis.
Cells were inoculated to a density of 5 × 107 ml−1 (optical density at 600 nm = 0.05) in K medium and grown to early exponential phase. After two doublings (optical density at 600 nm = 0.2), the osmolality of the medium was increased by the addition of 0.1 volume 3.0 M NaCl or 0.5 volume of 1.2 M sucrose, dissolved in K medium that was warmed to 37°C. Samples (total, 3 ml) were removed from the cultures at 5 and 2 min before and at various time points after the osmotic upshift: 1 ml, used for β-galactosidase and alkaline phosphatase assays, was mixed rapidly with 100 μg chloramphenicol and kept on ice until the assays; 2 ml was mixed with 4 ml of RNAprotect reagent (QIAGEN) and used for the isolation of RNA with the RNeasy Mini kit (QIAGEN), according to the manufacturer's protocol. Random hexamer-primed cDNAs were obtained from the RNA preparations by using the Superscript First Strand Synthesis system for RT-PCR (Invitrogen).
Amplification primers.
The primers used for the amplification of mRNAs and 16S rRNA were designed with Applied Biosystems Primer Express software (version 1.5). The following primer sequences were used (5′-to-3′ direction), with their nucleotide positions downstream from the translation start site of the relevant gene given in parentheses, unless otherwise indicated: kdpA, GGCGCTACTGACGCTCAATC (nt 192 to 211) and AGGCTTGCCAGTTGGTATTGG (nt 352 to 332); proV, GGATTATCCGGCTCGGGTAA (nt 181 to 200) and GAGCGCAAATGACTGGAAGAC (nt 351 to 331); proP, TGCCTACGCGTTGGGTAAAG (nt 141 to 160) and CCGTATTTATCGCCGAGCAT (nt 281 to 262); rpoS, GTTGGACGCGACTCAGCTTT (nt 160 to 179) and TTTTACCACCAGACGCAGGTT (nt 310 to 290); otsB, TTAACCGTATCCCCCGAACTC (nt 11 to 31) and CCGCGAGACGGTCTAACAAC (nt 146 to 127); ompC, GCGCCGACATCAACGTATTT (nt 882 to 901) and GCCAACAAAGCGCAGAACTT (nt 1042 to 1023); gnd, CAACATCGAAAGCCGTGGTT (nt 58 to 77) and GGCGTTTCGAGGGATTCAA (nt 198 to 180); lacZ, CACCAGCAGCAGTTTTTCCA (nt 3347 to 3366 in Mud1-8) and ATCCAGTGCAGGAGCTCGT (nt 3447 to 3429 in Mud1 to 8); phoA, GCGATGCTGCCTCACTGAAT (nt 751 to 770 in TnphoA) and TTGCGGATTTGGCGTACAG (nt 911 to 893 in TnphoA); and 16S rRNA, ATTGACGTTACCCGCAGAAGA (nt 478 to 498 in rrsA, -B, -C, -E, and -G; nt 477 to 497 in rrsD; nt 468 to 488 in rrsH) and GGGATTTCACATCCGACTTGA (nt 613 to 593 in rrsA, -B, -C, -E, and -G; nt 612 to 592 in rrsD; and nt 603 to 583 in rrsH).
Q-RT-PCR.
Amplification of 8 ng of cDNA was carried out in triplicate with SYBR Green I PCR Master Mix (Applied Biosystems), as described by Balaji et al. (2). The RT-PCR and melting-curve analysis of the amplification products were carried out with an ABI PRISM 7700 Sequence Detector (38).
The mRNA levels for the genes of interest were quantified from the Ct value, which is the PCR cycle number that generated a common signal for each gene in the exponential phase of amplification. The Ct values were converted to template concentrations, using standard curves (15). To correct for sampling errors, the levels of expression of each gene, as determined from their Ct values, were normalized to the level of 16S rRNA, and the data were averaged for the three replicates. Error bars for the proV and the kdpA mRNA levels after shock with 0.3 M NaCl are shown (see Fig. 2A) and for kdpA after shock with 0.6 M sucrose (see Fig. 4B). These errors are similar to those obtained for other genes in all experiments, and the latter are not shown for the ease of visualization of the curves. Induction ratios were obtained by dividing the normalized levels of mRNAs at each time point after the osmotic shock by the average of the normalized levels of those mRNAs before the osmotic shock.
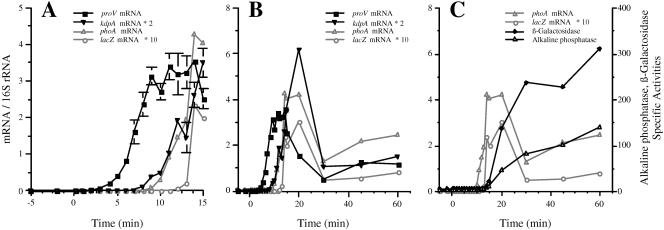
FIG. 2.
Kinetics of induction of the proU and kdp operons by osmotic shock with 0.3 M NaCl. Strain TL3353 (kdp+ proV+ proW+ proX-phoA/proV+pro W+ proX-lacZ) was grown in K medium and shocked with 0.3 M NaCl at 0 min. The accumulation of the kdpA, proV, phoA, and lacZ mRNAs was determined by Q-RT-PCR. The synthesis of alkaline phosphatase and β-galactosidase activities were measured at the indicated time points as described in Materials and Methods. The mRNA levels are normalized to the level of 16S rRNA, and the enzyme activities are in nanomoles of substrate consumed per minute per milligram of protein. (A and B) Normalized mRNA levels displayed for the first 15 min (A) and for 60 min (B); panel A shows an expanded version of the same data as panel B. For the sake of clarity, the mRNA data for kdpA and lacZ are plotted on different scales than for proV and phoA; the normalized mRNA levels were multiplied by 2 for kdpA and by 10 for lacZ. (C) Alkaline phosphatase and β-galactosidase specific activities. The levels of the phoA and lacZ mRNAs from panel B are also included in this panel. The threshold times of induction and times of peak expression were determined by ANOVA, as described in Materials and Methods. The threshold induction times of proV and kdpA were at 4 and 9 min, respectively (P < 0.0001). Mean peak value expression of proV was at 14 min (t = 17.3) and of kdpA was at 20 min 20 min (t = 15.5) (P < 0.0001 for both).
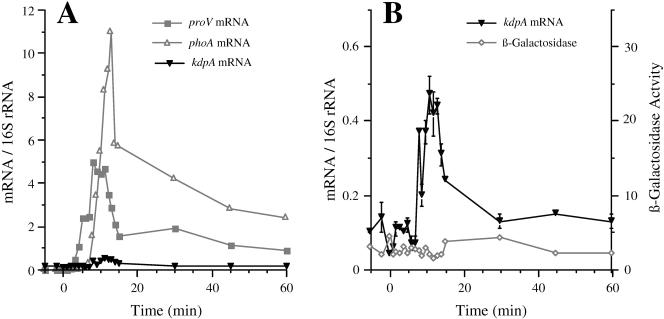
FIG. 4.
The kinetics of induction of the kdp and proU operons after shock with 0.6 M sucrose. Strain TL3025 (kdp+ proX-phoA/kdpA-lacZ) was grown in K medium and exposed to an osmotic shock with 0.6 M sucrose. At the indicated time points, the accumulation of the kdpA, proV, and phoA mRNAs was determined by Q-RT-PCR and the synthesis of alkaline phosphatase and β-galactosidase activities measured as described in Materials and Methods. (A) Normalized kdpA, proV, and phoA mRNA levels. (B) Expanded view of the normalized mRNA levels for the kdpA mRNA and the β-galactosidase activities (in nanomoles per minute per milligram of protein). The induction of alkaline phosphatase from the proX-phoA fusion was also determined. These data, which are not shown, were similar to those obtained with the proX-phoA fusion after shock with 0.3 M NaCl (Fig. 2).
Statistical treatment of data.
The time of induction of each gene was determined by repeated-measures analysis of variance (ANOVA), comparing the preinduction levels of mRNA to the triplicate data obtained at each time point after induction, using a multiple-comparison t test (′diff' option in PROC MIXED software [41a]). The threshold time of induction for each gene was taken as the first time point at which the normalized level of expression of that gene was significantly greater (α = 0.05) than the preinduced value. We tested for differences in the time of peak expression of kdpA versus proV as follows. We calculated the mean value of the proV and kdpA mRNA/16S rRNA levels at each separate time point and then standardized all the individual data points by dividing each by the peak mean value. Standardization was conducted separately for each experiment and each gene product. This standardization transforms the gene expression level to ensure that the curves for both gene products range from ≥0 to 1 and therefore gives us a simple way to compare the relative shapes of these two curves. We then tested all the data collected in each experiment in a single, repeated-measure ANOVA with a standardized expression level as the dependent variable and time and gene (kdpA versus proV) as independent variables. The model included a time × gene interaction term; a significant interaction term indicates that the relative shape of the two curves is different, but it does not indicate where those differences occur. To determine the relative position of the peaks of both of these curves, we used a multiple-comparison t test (′diff' option of PROC MIXED as above) at the time of the peaks of both curves. If the curves were significantly different at these points, we concluded that the relative position of the peaks were not coincident.
Enzyme assays.
β-Galactosidase and alkaline phosphatase assays were carried out as described by Miller (29) and Overdier et al. (31), respectively, with the enzyme specific activities expressed as the number of nanomoles of substrate used per minute per milligram of protein.
Determination of the effect of osmotic upshift on macromolecular synthesis and the induction of the lac operon.
For the determination of the effect of osmotic stress on macromolecular synthesis, strain TL3353 was inoculated at a density of 5 × 107 cells ml−1 in K medium and 10 μCi of [U-14C]glucose (5.5 mCi mmol−1; ICN). After two doublings, the osmolality of the medium was increased by 0.3 M NaCl, as described above for the induction of proU and kdp. At various time points before and after the osmotic shift, 1-ml samples were removed and mixed rapidly with 50 μl of 100-mg ml−1 trichlorocacetic acid at room temperature. The trichlorocacetic acid-insoluble fraction was pelleted by centrifugation, the precipitate was washed three times with 1 ml H2O, and the incorporated radioactivity was determined by liquid scintillation counting.
For determination of the effect of osmotic shock on the induction of the lac operon, strain TL3799 (proX-phoA/F′ lac+) was inoculated at a density of 5 × 107 cells ml−1 into K medium. After two doublings, the lac operon was induced by the addition of 1 mM IPTG (isopropyl-ā-d-thiogalactopyranoside); 15 min later, the osmolality was increased to 0.3 M NaCl, as described above. At various time points before and after the addition of IPTG and NaCl, the alkaline phosphatase and β-galactosidase activities were measured.
RESULTS
Kinetics of induction of the proU and kdp operons after osmotic shock with NaCl.
The kinetics of induction of osmotically regulated genes after shock with 0.3 M NaCl were analyzed in two kdp+ strains (Fig. 1): TL3353, which carried a proV+ proW+ proX-phoA translational fusion and a proV+proW+proX-lacZ transcriptional fusion in a tandem duplication of the proU operon, and TL3025, which in addition to carrying an intact copy of the kdpA+ kdpB+ kdpC+ kdpD+ kdpE+ genes also contained a kdpA-lacZ transcriptional fusion in a duplication of the kdp region of the chromosome. These strains were used because the reporter genes enabled us to monitor the level of expression of the kdp and proU transcripts at widely spaced sites in the mRNAs and to correlate the accumulation of the translation products of the reporters with the levels of the messages. We obtained similar results for the kinetics and magnitude of induction of the kdpA and proV genes in strain TL3353, which carries a single copy of the kdp+ operon and two copies of proU, as in strain TL3025, which carries a kdpA-lacZ fusion in the background of a kdp+ operon and a proX-phoA fusion. We show the data obtained after NaCl shock only for the former strain, but the main conclusions obtained with both strains are summarized below (see Fig. 6).
FIG. 1.
The location of the amplified segments of the proU and kdp mRNAs in the strains used for the Q-RT-PCR analysis of the osmotic control of proU and kdp expression. The arrows indicate the distance from the promoters to the amplified regions in the mRNAs.
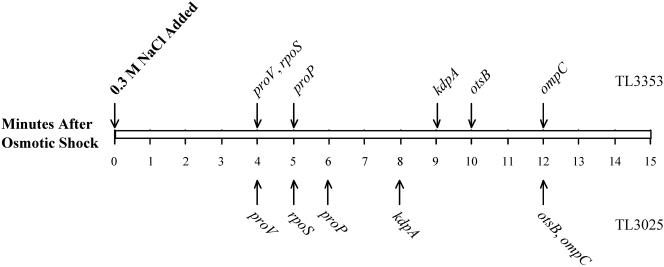
FIG. 6.
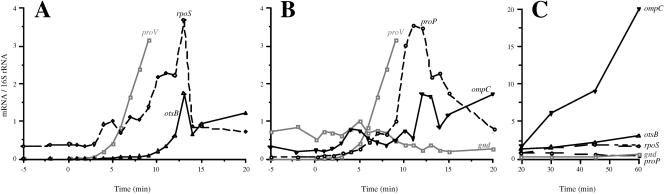
The timeline of induction of osmotically controlled genes. This figure shows the threshold induction times of the proV, kdpA, proP, rpoS, otsB, and ompC genes after shock with 0.3 M NaCl. The threshold induction times of these genes were determined by ANOVA, as described in Materials and Methods. The arrows above the rectangle show the times of induction of each gene in strain TL3353 after shock with 0.3 M NaCl, extracted from the data shown in Fig. 2 and 3, and the arrows below the rectangle show induction times obtained with strain TL3025 in a similar experiment (primary data not shown).
Figure 2 shows the results on the induction of kdpA, proV, proX-phoA, and proX-lacZ mRNAs after shock with 0.3 M NaCl. The primers used in this experiment amplified sequences covering nucleotide positions 253 to 413 in the kdpA mRNA and positions 241 to 412 in the proV mRNA. We also used a pair of primers that amplified sequences at positions 3211 to 3372 downstream of the proU promoter in the proX-phoA fusion and a pair of primers that amplified sequences 6.7 to 6.8 kbp downstream of the proU promoter in the proX-lacZ fusion. Increased accumulation of the 5′ portion of the proV gene was detectable by 4 min after the NaCl shock. The accumulation of this portion of the proV mRNA increased until 14 min after the NaCl shock, at which point it reached a ∼600-fold induction ratio over the preinduced value, and then it decreased to a steady-state level of ∼200-fold induction at 60 min. In contrast, increased accumulation of the 5′ end of the kdpA gene was detectable only 9 min after the shock. Expression of kdpA peaked at a 670-fold induction ratio at 20 min after the shock and decreased to a 170-fold induction ratio at 60 min. The observation that the kdp operon was expressed at a substantial level 1 h after the osmotic upshift has been indicated previously by lacZ reporter gene fusion studies (1, 14). This finding is not consistent with the postulate of the turgor control model that the expression of the kdp operon is shut off after turgor restoration, as proposed by Epstein (13).
We detected induction of the phoA and lacZ mRNAs in the proX fusions 10 and 13 min after the NaCl shock, respectively. The accumulation of the mRNAs of these two genes showed comparable phases of increase and decay that were seen for the proV mRNA, but with about a 6- and 9-min delay, respectively. The temporal order of induction of the amplified portions of the proV, proX-phoA, and proX-lacZ mRNAs was consistent with the physical sequence of these sites in the chromosome. This result validated our conclusion that the 5-min difference in appearance of the proV and kdp mRNAs was due to a difference in the timing of induction of these two genes. We also measured the kinetics of induction of the β-galactosidase and alkaline phosphatase enzymes from the proX-lacZ and proX-phoA fusion (Fig. 2C). Induction of the synthesis of both enzymes was detectable 14 min after the NaCl shock. The delay in the appearance of the products of the phoA and lacZ reporter fusions compared to the appearance of the internal portions of the mRNAs could be due to a combination of factors, including the time required for the completion of the messages, possible inhibitory effects of osmotic shock on translation (see below), or adverse effects of this treatment on the assembly of enzymatically active proteins, which in the case of alkaline phosphatase requires export to the periplasm.
Kinetics of induction of other osmoregulated genes.
We also probed the timing of induction of four other osmotically controlled genes: proP (encoding proline glycine betaine permease), rpoS (encoding stationary-phase σ factor), otsB (encoding trehalose-6-phosphate phosphatase), and ompC (encoding outer membrane porin), after shock with 0.3 M NaCl in strain TL3353. Increase in the expression of the rpoS gene was detectable by 4 min after the NaCl shock (Fig. 3A). This timing was similar to that obtained for proU, although the rpoS gene showed only a maximal 12-fold induction (at 13 min) above the basal level. Increased expression of the otsB gene was detectable by 6 min after the shock. The otsB gene had a transient peak of induction ratio of 370 fold at 13 min (Fig. 3A), after which it fell and then increased again gradually to an induction ratio of 660 at 60 min. The otsBA operon is known to be induced by stationary phase, and it is possible that the increase in the expression of this operon at the later time points was due to the growth-phase-dependent regulation (17).
FIG. 3.
The kinetics of induction of the rpoS, otsB, proP, and ompC genes after shock with 0.3 M NaCl. RNA preparations isolated in the experiment shown in Fig. 2 were used to measure the expression of the rpoS, otsB, proP, and ompC genes at the indicated time points as described in Materials and Methods. (A) Normalized mRNA levels for the rpoS and otsB genes; (B) data for the proP, ompC and gnd genes for the first 20 min after the shock with 0.3 M NaCl. These panels also include the results for the proV gene, copied from Fig. 2. (C) Data for the rpoS, otsB, proP, ompC, and gnd genes for the 20 to 60 min after the osmotic shock.
Increase in the transcription of the proP gene was detectable by 5 min after the NaCl shock and it reached a peak value of ∼60-fold induction by 11 min (Fig. 3B). A previous analysis of the regulation of proP expression conducted with a lacZ fusion indicated that osmotic shock elicited an initial 17-fold induction in the differential rate of synthesis of β-galactosidase (21), but after about 25 min, the proP-lacZ fusion was expressed at only a 2-fold-higher steady-state level in cells that were adapted to high osmolality than in cells growing at low osmolality (12, 21). In E. coli K-12, the proP gene has been shown to be transcribed from two osmotically inducible promoters, one of which is dependent on the RpoS-RNA polymerase (49). The permease activity of the ProP protein is known to be enhanced at high osmolality by a posttranslational mechanism (12, 37). Thus, the initial burst of induction of proP gene could amplify the effect of the posttranslational regulation of the transport system in the accumulation of compatible solutes soon after osmotic upshifts.
The kinetics of induction of ompC were more complicated (Fig. 3B): after an initial increase in expression at 3 to 4 min, there was a reduction to background levels until 11 min, and statistically significant induction was evident only after 12 min. The expression of ompC continued to increase steadily until 60 min after the osmotic shift.
As a control for the quantification of the effect of osmotic shock on transcription, we measured the levels of the gnd (encoding gluconate-6-phosphate dehydrogenase) mRNA, which is subject to a growth rate control but is believed to be independent of other environmental factors (34). There was a gradual reduction in the level of the gnd mRNA after the NaCl shock, resulting in a fourfold decrease at 15 min, which could be a manifestation of the reduction in growth rate after the NaCl shock.
The kdp operon is less sensitive to osmotic induction by sucrose than by NaCl.
Both the turgor control and the ionic strength models predict that the kdp operon should be induced upon plasmolysis to the same extent in response to a given increase in external osmolality by an impermeant solute, regardless of the chemical nature of the solute. However, contrary to this prediction, it was observed with lacZ reporter fusions that permanent induction of the kdp operon could be achieved only with high concentrations of NaCl and ionic solutes but not with nonpolar solutes, such as sucrose (14, 16, 43). Because Q-RT-PCR provides a more sensitive measure of the instantaneous levels of mRNAs than reporter gene fusions, we used the this technique to determine simultaneously the kinetics of induction of the kdp and proU operons after shock with 0.6 M sucrose in a strain that carried a kdpA-lacZ and a proX-phoA fusion. The results are shown in Fig. 4. In contrast to the peak 670-fold induction of the kdpA mRNA that we observed after the shock with 0.3 M NaCl (Fig. 2), the level of this message increased only 4-fold in response to 0.6 M sucrose. This slight induction of the kdp operon was initiated at 8 min after addition of sucrose and peaked at 11 min after the osmotic shock. Another difference in the response of the kdp operon to sucrose and NaCl was that the expression of the operon returned to basal level by 30 min after the addition of sucrose, whereas it was maintained at a 170-fold-induced level at 60 min after the shock with 0.3 M NaCl. We did not detect induction of β-galactosidase from the kdpA-lacZ fusion after shock with sucrose, in accord with the marginal increase of the mRNA accumulation. It should be noted that the osmotic shock experiments with 0.3 NaCl and 0.6 M sucrose were carried out using 1 mM K+; therefore, the K+-dependent component of kdp regulation would have presumably been uniform under both conditions.
The proU operon, however, was induced to similar high levels by 0.6 M sucrose as by 0.3 M NaCl, as measured by the accumulation of the proV and phoA mRNAs and alkaline phosphatase activity. Induction of the 5′ part of the proV mRNA by sucrose was detectable by 3 min after the osmotic upshift and reached a maximum of 590-fold induction at 8 min after shock with sucrose. Induction of the phoA mRNA and alkaline phosphatase activity from the proX-phoA fusion was evident by 6 and 11 min after the shift, respectively. This sequence of responses demonstrates that the osmotic induction of proU also preceded that of kdp after shock with sucrose. The fact that we observed efficient induction of proU with 0.6 M sucrose serves as an important control, demonstrating that this solute imposes an osmotic shock similar to 0.3 M NaCl but did not induce the kdp operon to the same high level.
Osmotic upshift causes a transient inhibition of macromolecular synthesis.
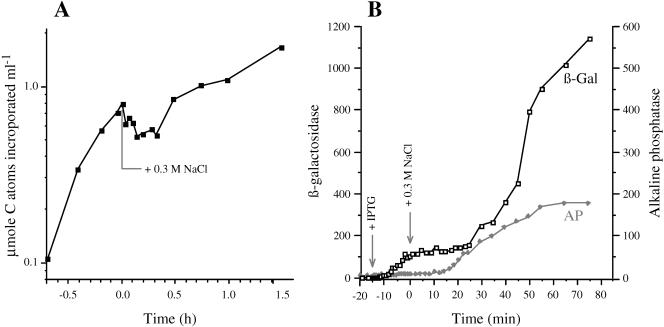
The 4 min required for the induction of the proV mRNA after shock with 0.3 M NaCl (Fig. 2) was considerably longer than the typical response times observed for the transcriptional control of other genes. For example, increased synthesis of enzymatically active araB and lacZ gene products was detectable within 0.7 and 1.5 min after the induction of the ara and lac operons by arabinose or IPTG, respectively (32, 42). The comparatively sluggish response of the proV and other osmotically controlled genes to osmotic induction might be due to an inherent feature of the mechanisms that perceive or relay the regulatory signal to the promoters of these genes. However, osmotic shock has been observed to cause a transient inhibition of nutrient uptake, respiration, and DNA synthesis (19, 27, 28, 40), and the slow induction of the osmotically regulated genes might be due to a general inhibition of metabolism after plasmolysis. To probe the latter possibility, we determined the effect of osmotic shock on the incorporation of radioactivity from [14C]glucose into macromolecules and on the induction of the lac operon under conditions identical to those used for the measurements of RNA levels.
The results of the first experiment are shown in Fig. 5A. Strain TL3353 was grown in the low-osmolar K medium in the presence of [14C]glucose. Initially, there was a rapid incorporation of radioactivity into macromolecules. After two doublings, the osmolality of the culture was increased by 0.3 M NaCl. The osmotic upshift caused an immediate inhibition of the incorporation of 14C. Further incorporation did not resume until ∼0.5 h after the osmotic shock. These results confirm that osmotic shock with 0.3 M NaCl resulted in a transient cessation of overall macromolecular synthesis.
FIG. 5.
Effect of osmotic shock on macromolecular synthesis and induction of the lac operon. (A) Strain TL3353 was grown in K medium containing 10 μCi [U-14C]glucose. At 0 h, the osmolality of the medium was increased with 0.3 M NaCl. Samples were removed from the culture at the indicated time points, and the incorporation of radioactivity into macromolecules was determined as described in Materials and Methods. (B) Strain TL3799 (kdp+ proX-phoA/F′ lac+) was grown in K medium. At −15 min, the lac operon was induced with 1 mM IPTG; at time 0, the osmolality of the culture was increased with 0.3 M NaCl. The accumulation of β-galactosidase and alkaline phosphatase (in nanomoles per minute per milligram of protein) was determined at the indicated time points, as described in Materials and Methods.
In the second experiment, exponential-phase cells of strain TL3799 (proX-phoA/F′ lac+) growing in K medium were treated with IPTG to induce the lac operon; 15 min after the addition of IPTG, the osmolality of the medium was increased to 0.3 M NaCl. The expression of the lac operon was monitored by β-galactosidase assays (Fig. 5B). Using the mathematical treatment of Schleif et al. (42), we were able to detect the formation of newly synthesized β-galactosidase by 1.5 min after the addition of IPTG, in accord with the previously observed kinetics of induction of the lac operon (32, 42). This result confirms that the induction of proV by high osmolality (Fig. 2 and 4) was indeed substantially slower than the induction of lac operon by IPTG. The accumulation of β-galactosidase continued to increase rapidly for 15 min, at which time the cells were exposed to 0.3 M NaCl. The osmotic shock caused a rapid inhibition of β-galactosidase synthesis that persisted for ∼0.5 h and then resumed at a lower rate than before the shock. These results are consistent with the 14C labeling, because both experiments suggest that shock with 0.3 M NaCl inhibited the overall macromolecular synthesis for ∼0.5 h. Thus, this adverse effect of osmotic upshift on macromolecular synthesis could account for the slow induction of proU and the other osmotically regulated genes. As a control, we measured the induction of the alkaline phosphatase from the proX-phoA fusion, which was detectable by 15 min after the osmotic shock, similar to the result reported in Fig. 2.
DISCUSSION
Measurement of mRNA levels after osmotic induction enabled us to construct a temporal sequence of induction of the six osmotically induced operons that we examined. Our time- line of induction of these genes after shock with 0.3 M NaCl in two different strains is shown in Fig. 6. These observations show that the rpoS, proV, and proP genes were induced earliest among the osmotically regulated genes, 4 to 6 min after the shock. It is important to note that the kdp operon was induced only at 8 to 9 min after the shock with 0.3 M NaCl or 4 to 5 min after the induction of proV. Induction of the otsB and ompC genes was observable at 10 to 12 min. This timeline of the regulation of the osmotically controlled operons (Fig. 6) is at odds with the suggestions that the induction of the kdp operon is an early response to an osmotic shift (5, 25) and that most (if not all) osmotically controlled events, including the induction of the proU operon, are secondary consequences of accumulation of K+ (13, 18). Our observation that the induction of the proU operon occurs within 4 min after the osmotic upshift is certainly contrary to an assertion in the literature that the induction of osmoresponsive genes occurs one or more hours after the osmotic upshift (47).
To our knowledge, this is the first work in which induction of genes has been monitored by Q-RT-PCR in bacteria, although this technique been used to follow the kinetics of accumulation of mRNAs in plants after viral or fungal infection (2, 39). The sensitivity of Q-RT-PCR in a bacterial system has been documented by its usefulness in measuring mRNA half-lives at a 0.5-min resolution (41). The osmotic control of gene expression has been studied extensively with lacZ or lux reporter gene fusions (12, 21, 24, 33). Our Q-RT-PCR measurements detected both earlier induction times and higher induction ratios of osmotically controlled genes than were inferred with the reporter gene fusions. Under conditions comparable to ours (K medium; 1 mM K+), Gowrishankar (16), found that 0.3 M NaCl caused a 12-fold induction in steady-state levels of β-galactosidase in a kdp mutant E. coli carrying a kdp-lacZ fusion. One reason why the induction ratio observed by Gowrishankar is substantially less than the one we observed in this work could be that his strain already had an elevated basal level of expression of the kdp-lacZ fusion, due to the kdp mutation (14, 24). Furthermore, it is to be expected that there would be differences in the results of Q-RT-PCR measurements and reporter gene analysis, because the former technique gives the instantaneous level of mRNAs, while the latter technique measures a delayed output, the accumulation of an enzymatic activity integrated over time. Weber and Jung (46) carried a single time point profiling of gene expression in E. coli at 9 min after shock with 0.4 M NaCl and found by Northern analysis that proX, otsA, kdpA, rpoS, and ompC were induced 19.9-, 8.1-, 6.1-, 3.3-, and 1.1-fold, respectively. Because of differences in the species examined and the media and severity of osmotic shock used, our results may not be directly comparable to those of Weber and Jung, but we found that the proV, otsB, kdpA, rpoS, and ompC genes gave induction ratios of 540, 19, 41, 4, and 2, respectively, at 9 min after shock with 0.3 M NaCl. Thus, we measured higher induction ratios for each operon, especially for proU and kdp, with Q-RT-PCR than were detected by Northern analysis (46).
The strains we used for the Q-RT-PCR analysis were wild type with respect to K+ transport, but we examined the effect of K+ transport mutations on the expression kdp and proU operons with lacZ or phoA reporter fusions. Although kdpC::Tn10 and trkA::MudQ mutations increased the basal level of β-galactosidase from the kdpA-lacZ fusion 3- and 36- fold, respectively, over that seen with the wild type in K medium, these K+ transport mutations did not affect the timing of induction of either the kdp or proU operon by 0.3 M NaCl (K. O'Connor and L. N. Csonka, unpublished data). These results suggest that functioning of the Kdp or the TrkA system is not required for the osmotic induction of these two osmotically controlled operons (if at least one of these two K+ permeases is functioning).
Our observed timeline gene induction (Fig. 6) suggests that the osmotically controlled genes can be divided into three groups: rpoS, proV, and proP, which are induced early after the osmotic upshift; kdp, which is induced somewhat later; and otsB and ompC, which are induced last. The fact that these groups of genes have different threshold and peak times of induction suggests that they might respond to different regulatory signals, although we cannot rigorously exclude the possibility that some them could be regulated by a common signal but with different response times. It has been shown by Yan et al. (50) that the accumulation of glutamate is necessary for the maintenance of high concentrations of K+ in osmotically stressed cells. However, glutamate limitation did not impair the high-level expression of proU at high osmolality (9), providing evidence against K glutamate being a regulatory signal for the induction of this operon. The transcriptional control of the kdp operon is mediated by a two-component system, consisting of the membrane-bound signal receptor KdpD and the transcriptional activator KdpE. KdpD has been suggested to sense two periplasmic signals: high osmolality and K+ limitation (22, 43). The synthesis of the RpoS protein is stimulated under a variety of conditions, including starvation or entry into stationary phase, high osmolality, low and high temperatures, and acidic pH (17). The rpoS gene is downstream of the nlpD gene in an operon, and it is transcribed from three promoters, of which one is within nlpD and has a 567-nucleotide-long untranslated region (17) and the other two are upstream of the latter gene. It is not known what the signal is for the osmotic induction of the rpoS gene, but in view of the fact that this gene is up-regulated under a variety of stress conditions that cause growth impairment, it is possible that it may be controlled by a temporary growth rate reduction, such as the one after an osmotic upshift. The otsBA operon is transcribed by the RpoS-RNA polymerase holoenzyme (17); therefore, the observation that the induction of the otsB gene occurs after the induction of rpoS is consistent with this transcriptional activation cascade. The transcription of ompC is controlled by another two-component regulatory system, comprising the membrane signal receptor EnvZ protein, which activates the transcriptional regulatory protein OmpR in response to some periplasmic signal(s) (36). The regulation of transcription of ompC is more complex than that of the other osmotically regulated genes, because in addition to osmolality, this gene (and its paralog, ompF) responds to a variety of other environmental cues, including carbon source, pH, and temperature (36). Superimposed on the transcriptional control, translational control also contributes to the regulation of expression of the ompC gene product, mediated by the micC antisense RNA, which can anneal to the first 22 nucleotides at the 5′ end of the ompC mRNA and inhibit binding of ribosomes to the message (7). The primers that we used for the amplification of the ompC sequences did not overlap micC; therefore, there was no interference from the micC RNA in our measurements of the ompC mRNA levels. It is possible that the long-term increase in the level of the ompC mRNA that we observed may not have been due solely to an osmotic effect but to some other factor, such as growth phase.
In addition to finding that the proU operon was induced before kdp after osmotic shock, our experiments demonstrated that kdp mRNA was induced at 170-fold-higher levels by 0.3 M NaCl than by 0.6 M sucrose. Our direct measurements of mRNA levels confirm data obtained by lacZ reporter gene analysis (14, 16, 43) that in contrast to proU, which responds to low levels of water activity resulting from any impermeant solute, the kdp operon is much more sensitive to ionic solutes as inducers than nonpolar ones and that the induction of the kdp operon can be antagonized by high concentrations of K+. There are contradictory data in the literature about the steady-state regulation of the kdp operon by high osmolality. Laimins et al. (24) reported the finding that constitutes one of the premises of the turgor control model, that the kdp operon was induced only transiently by an osmotic upshift imposed with 0.2 M glucose. However, other researchers noted that osmotic stress imposed by ionic solutes resulted in a high level steady-state induction of the operon, whereas nonpolar solutes did not induce the operon at all, even transiently, as measured with lacZ reporter fusions (14, 16, 43). Our observations may offer a resolution for the discrepancy between the data of Laimins et al. (24) and that of other researchers (14, 16, 43). The mRNA level measurements indicated the proU, kdp, proP, and rpoS genes exhibited a large initial induction soon after the osmotic shock, followed by a drop in their expression. The kdp operon exhibited this pattern of expression after the NaCl and the sucrose shocks, although the magnitude of the response was 2 orders of magnitude less with the nonpolar solute than with the ionic one. Thus, our mRNA data on the regulation of the kdp operon by sucrose, which indicate that the expression of the operon returned to basal level 30 min after the osmotic shock, are consistent with the results that Laimins et al. obtained by kdp-lacZ reporter analyses, although we were not able to detect the small induction of the kdpA-lacZ fusion by β-galactosidase assays after sucrose shock. It should be noted that Laimins et al. used a kdp trk kup mutant strain that was highly defective in K+ uptake, whereas our experiments were conducted with strains that were wild type with respect to K+ accumulation. The triple-mutant strain used by Laimins et al., which required a pathologically high concentration of 50 mM K+ for growth, was near K+ limitation, even at this concentration of the cation. It is possible that the transient induction of the kdp-lacZ fusion that was reported by Laimins et al. after shock may have been magnified in the kdp trk kup triple mutant, compared to that seen by us in the wild-type background.
Regardless of the reasons why the kdp operon responds differently to sucrose and NaCl, critical predictions of both the turgor control and the ionic strength models were not borne out by our data. As discussed in the introduction, the turgor control model entails the prediction that the transcription of the kdp operon should decrease as proU is induced. However, our observation that the transcription of proU was already declining by 20 min after the osmotic shock when the expression of the kdp operon became maximal (Fig. 2) is contrary to this prediction. Osmotic shock with sucrose elicited a much lower induction of the kdp operon than was seen with NaCl, whereas both solutes elicited similar induction of the proU operon. These observations are inconsistent with both the turgor control and the ionic strength models, because these models propose that all impermeant solutes would have similar effects on the regulation of the osmotically controlled genes.
According to one of the assumptions of the turgor control model (13), our result that the kdp operon was induced by 0.3 M NaCl (Fig. 1) would indicate that the constitutive Trk activity and the uninduced levels of Kdp were not sufficient to maintain turgor. If K+ were the second messenger for the induction of the proU operon, as is assumed in the turgor control model, then the fact that the induction of the proU operon begun by 4 min after shock with 0.3 M NaCl would imply that the accumulation of K+ was at least initiated by this time. However, the fact that the kdp operon was not induced until 8 to 9 min after the shock means that turgor has not been restored at this time. Thus, the induction of the kdp operon does not seem to be a rapid mechanism for turgor restoration by K+ accumulation compared to a proposed secondary response, the K+-dependent induction of the proU operon. Moreover, the peak expression of proU occurred at 14 min after shock with 0.3 M NaCl (Fig. 1). Assuming that K+ is the regulatory signal for the expression of proU, this result would imply that the K+ concentration reached a peak level at or before 14 min. However, at this time, kdp expression was still on the increase, suggesting that turgor has not yet been fully restored. Thus, this analysis of our data in light of the turgor control model leads to the seeming paradox that the K+ accumulation peaks before turgor restoration has been achieved.
It is generally accepted that the uptake of K+ is the earliest active osmoregulatory response in Enterobacteriaceae and that organically compatible solutes, such as glycine betaine and proline, are taken up only after K+ has been accumulated to potentially deleterious levels (13, 18, 23, 25, 44, 45, 48). Our observations that proU and proP are induced before kdp suggests that, on the contrary, the cells might be poised to take up organic osmolytes before K+. This conclusion is consistent with the observations that K+ transport mutations did not impair the transport of glycine betaine after an osmotic upshift (28). By accumulating glycine betaine or other organic osmolytes in preference to K+, the cells could avoid the unnecessary energy expenditure for the uptake of K+ and the synthesis of glutamate, which are more deleterious to macromolecular function than the organic compatible solutes.
To obtain a satisfactory understanding of the osmoregulatory responses, it would be necessary to correlate the transcription of osmotically regulated genes over the time course of osmotic adaptation with a variety of relevant parameters, such as turgor pressure, membrane tension, water content, cytoplasmic volume, intracellular K+ concentration, and ionic strength. The water content, cytoplasmic volume internal solute concentrations, and turgor pressure have been determined for nongrowing E. coli cells (6). As our analysis required the use of cells that were able to carry out normal osmotic adaptation, the technology is not available for meaningful determination of these rapidly changing parameters on the necessary time scale.
Analysis of the timing of the induction of the proV, proX-phoA, and proX-lacZ mRNAs (Fig. 2) enables us to estimate the rate of progression of RNA polymerase after osmotic shock. The distances between the amplified portion of the proV mRNA and the amplified portions of the phoA and lacZ mRNAs were 3.0 and 6.8 kb, respectively. Induction of these sequences was detectable 6 and 9 min after the induction of the amplified segment in the proV mRNA, respectively. From these results, we can calculate that the average transit time of the RNA polymerase during minutes 4 to 9 after the osmotic upshift was 0.5 to 0.8 kb min−1. This estimate is considerably less than the RNA polymerase progression rate of 2.6 to 3.3 kb min−1 reported for exponentially growing E. coli (4). The drastic reduction in the rate of RNA elongation rate during the early stages after the osmotic shock could be due to the general inhibition of metabolism documented in Fig. 5.
Although our data provide evidence against predictions of the turgor control and the ionic strength control models, we do not feel that we are in a position to offer satisfactory alternatives for a signal(s) for any of the osmotically controlled genes. A conceptual difficulty in any model of osmoregulation is to account for both the rapid initial increase in transcription of the osmoresponsive operons, such as proU and kdp, and for their continued high steady-state levels of expression after the osmotic adaptation has been completed (8). As plasmolysis has drastic effects on the cell envelope, one could speculate that the rapid induction of these operons might be mediated by a sensor(s) monitoring some physical property of the cell envelope, such as membrane tension, surface area, or turgor pressure. Sugiura et al. (43) proposed that high osmolality and K+ limitation, which are the inducing signals for the kdp operon, are detected by two distinct domains in the membrane-bound KdpD protein. The fact that osmotic shock with sucrose did not elicit high-level induction of kdp while it efficiently induced the proU operon does not support the idea that high osmolality itself is sufficient to activate the KdpD protein. It has been suggested by Sugiura et al. (43) that the ability of the KdpD protein to sense high concentrations of K+ may by perturbed by high concentrations of Na+ and other cations. If this idea is correct, then high concentrations of salts may induce the kdp operon, not because of an osmotic effect but because other cations at elevated concentrations may compete with K+ for binding to the KdpD protein and mimic K+ limitation. An alternative explanation for the higher sensitivity of the kdp operon for ionic solutes than nonpolar ones, proposed by Malli and Epstein (26), was that at intermediate K+ concentrations, ionic solutes cause a limitation of K+ uptake, which is not seen with nonpolar osmolytes.
Our understanding of the regulation of proU is even more unsatisfactory than that of the kdp operon, because thus far no osmotic signal-sensing or signal-transducing proteins that achieve the >600-fold induction of this operon have been identified. The transcriptional regulation of the proU operon is mediated by a negative control mechanism that involves an interaction between the promoter and a downstream silencer (31). Coupled transcription-translation assays demonstrated that the proU promoter was efficient in cell extracts and that it exhibited a response to K+ glutamate concentration that was essentially identical to the response seen with the lacUV5 promoter, which is not osmotically regulated (9). The generalized DNA binding protein Hns plays a role in the repression of the proU operon in media of low osmolality, but this protein is not necessary for the osmotic induction of this operon (25). Conceivably, the transcriptional control of proU might be effected by an ensemble of generalized DNA binding proteins (including Hns), which also contains an undetermined protein that couples the expression of the operon to some osmoregulatory signal. From measurements of the periplasmic and cytoplasmic volumes, water content, and solute levels after plasmolysis, Cayley et al. (6) deduced the turgor pressure in cells that had been grown at different osmolalities. They concluded that the steady-state turgor pressure and the intracellular volume of exponentially growing cells are not equal at low and high osmolalities, but rather that these quantities decrease as a function of external osmolality. Because these parameters change as a function of external osmolality, it is possible that the difference in turgor, volume, water content, or some related quantity (such as cytoplasmic ion concentration, macromolecular crowding, membrane tension, or surface area) could provide a steady-state signal for the maintenance of the high-level expression of the proU operon at high osmolality. The identification of the regulatory signal for the proU operon remains one of the outstanding challenges in the field of osmoregulation.
Acknowledgments
We thank M. Hammer for advice with the RNA isolation and R. LaRossa for helpful discussions.
This work was supported by the NSF Life in Extreme Environments Program (grant MCB-9978253).
REFERENCES
- 1.Asha, H., and J. Gowrishankar. 1993. Regulation of kdp operon expression in Escherichia coli: evidence against turgor as signal for transcription control. J. Bacteriol. 175:4528 to 4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balaji, B., D. B. Bucholtz, and J. M. Anderson. 2003. Barley yellow dwarf virus and cereal yellow dwarf virus quantification by real-time polymerase chain reaction in resistant and susceptible plants. Phytopathology 93:1386-1392. [DOI] [PubMed] [Google Scholar]
- 3.Booth, I. R., and C. F. Higgins. 1990. Enteric bacteria and osmotic stress; intracellular potassium glutamate as a secondary signal of osmotic stress. FEMS Microbiol. Rev. 75:239-246. [DOI] [PubMed] [Google Scholar]
- 4.Bremer, H., and P. P. Dennis. 1996. Modulation of chemical composition and other parameters of the cell by growth rate, p. 1553-1569. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 5.Burg, M. B., E. D. Kwon, and D. Kültz. 1996. Osmotic regulation of gene expression. FASEB J. 10:1598-1606. [DOI] [PubMed] [Google Scholar]
- 6.Cayley, D. S., H. J. Guttman, and M. T. Record, Jr. 2000. Biophysical characterization of changes in amounts and activity of Escherichia coli cell and compartment water and turgor pressure in response to osmotic stress. Biophys. J. 78:1748-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, S., A. Zhan, L. B. Blyn, and G. Storz. 2004. MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli. J. Bacteriol. 186:6689-6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Csonka, L. N., and A. D. Hanson. 1991. Prokaryotic osmoregulation: genetics and physiology. Annu. Rev. Microbiol. 45:569-606. [DOI] [PubMed] [Google Scholar]
- 9.Csonka, L. N., T. P. Ikeda, S. A. Fletcher, and S. Kustu. 1994. The accumulation of glutamate is necessary for optimal growth of Salmonella typhimurium in media of high osmolality but not induction of the proU operon. J. Bacteriol. 176:6324-6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Culham, D. E., J. Henderson, R. A. Crane, and J. M. Wood. 2003. Osmosensor ProP of Escherichia coli responds to the concentration, chemistry, and molecular size of osmolytes in the proteoliposome lumen. Biochemistry 42:410-420. [DOI] [PubMed] [Google Scholar]
- 11.Druger-Liotta, J., V. J. Prange, D. G. Overdier, and L. N. Csonka. 1986. Selection of mutations that alter the osmotic control of transcription of the Salmonella typhimurium proU operon. J. Bacteriol. 169:2449-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunlap, V. J., and L. N. Csonka. 1985. Osmotic regulation of l-proline transport in Salmonella typhimurium. J. Bacteriol. 163:296-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Epstein, W. 1986. Osmoregulation by potassium transport in Escherichia coli. FEMS Microbiol. Rev. 39:73-78. [Google Scholar]
- 14.Frymier, J., T. Reed, S. A. Fletcher, and L. N. Csonka. 1997. Characterization of the transcriptional regulation of the kdp operon of Salmonella typhimurium. J. Bacteriol. 179:3061-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giulietti, A., L. Overbergh, D. Valckx, B. Decallonne, R. Bouillon, and C. Mathieu. 2001. An overview of real-time quantitative PCR: applications to quantify cytokine gene expression. Methods 25:386-401. [DOI] [PubMed] [Google Scholar]
- 16.Gowrishankar, J. 1985. Identification of osmoresponsive genes in Escherichia coli: evidence for participation of potassium and proline transport systems in osmoregulation. J. Bacteriol. 164:434-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the σs (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higgins, C. F., J. Cairney, D. Striling, L. Sutherland, and I. R. Booth. 1987. Osmotic regulation of gene expression: ionic strength as an intracellular signal? Trends Biochem. Sci. 12:339-344. [Google Scholar]
- 19.Houssin, C., N. Eynard, E. Schechter, and A. Ghazi. 1991. Effect of osmotic pressure on membrane energy linked functions in Escherichia coli. Biochim. Biophys. Acta 1056:76-84. [DOI] [PubMed] [Google Scholar]
- 20.Hughes, K. T., and J. R. Roth. 1985. Directed formation of deletions and duplications using Mud(Ap, lac). Genetics 109:263-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jovanovich, S. B., M. Martinell, M. T. Record, Jr., and R. R. Burgess. 1988. Rapid response to osmotic upshift by osmoregulated genes in Escherichia coli and Salmonella typhimurium. J. Bacteriol. 170:534-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung, K., and K. Altendorf. 2002. Towards an understanding of the molecular mechanisms of stimulus perception and signal transduction by the KdpD/KdpE system of Escherichia coli. J. Mol. Microbiol. Biotechnol. 4:223-228. [PubMed] [Google Scholar]
- 23.Kempf, B., and E. Bremer. 1998. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch. Microbiol. 170:319-330. [DOI] [PubMed] [Google Scholar]
- 24.Laimins, L. A., D. B. Rhoads, and W. Epstein. 1981. Osmotic control of kdp operon expression in Escherichia coli. Proc. Natl. Acad. Sci. USA 78:464-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lucht, J. M., and E. Bremer. 1994. Adaptation of Escherichia coli to high osmolality environments: osmoregulation of the high affinity glycine betaine transport system ProU. FEMS Microbiol. Rev. 14:3-20. [DOI] [PubMed] [Google Scholar]
- 26.Malli, R., and W. Epstein. 1998. Expression of the Kdp ATPase is consistent with regulation by turgor pressure. J. Bacteriol. 180:5102-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meury, J. 1988. Glycine betaine reverses the effects of osmotic stress on DNA replication and cellular division in Escherichia coli. Arch. Microbiol. 149:232-239. [DOI] [PubMed] [Google Scholar]
- 28.Meury, J. 1994. Immediate and transient inhibition of the respiration of Escherichia coli under hyperosmotic shock. FEMS Microbiol. Lett. 121:281-286. [DOI] [PubMed] [Google Scholar]
- 29.Miller, J. H. 1991. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Overdier, D. G. 1990. Genetic and molecular analysis of the osmotically regulated proU operon of Salmonella typhimurium. Ph.D. thesis. Purdue University, West Lafayette, IN.
- 31.Overdier, D. G., and L. N. Csonka. 1992. A transcriptional silencer downstream of the promoter in the osmotically controlled proU operon of Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 89:3140-3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmer, D. T., P. H. Blum, and S. W. Artz. 1983. Effects of the hisT mutation of Salmonella typhimurium on translation elongation rate. J. Bacteriol. 153:357-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park, S. F., D. A. Stirling, C. S. J. Hulton, I. R. Booth, C. F. Higgins, and G. S. A. B. Stewart. 1989. A novel, non-invasive promoter probe vector: cloning of the osmoregulated proU promoter of Escherichia coli K12. Mol. Microbiol. 3:1011-1023. [DOI] [PubMed] [Google Scholar]
- 34.Pease, A. J., and R. E. Wolf, Jr. 1994. Determination of the growth rate-regulated steps in expression of the Escherichia coli K-12 gnd gene. J. Bacteriol. 176:115-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poolman, B., P. Blount, J. H. A. Folgering, R. H. E. Friesen, P. C. Moe, and T. van der Heide. 2002. How do membrane proteins sense water stress? Mol. Microbiol. 44:889-902. [DOI] [PubMed] [Google Scholar]
- 36.Pratt, L. A., W. Hsing, K. E. Gibson, and T. E. Silhavy. 1996. From acids to osmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol. Microbiol. 20:911-917. [DOI] [PubMed] [Google Scholar]
- 37.Racher, K. I., R. T. Voegle, E. V. Marshall, D. E. Culham, J. M. Wood, H. Jung, M. Bacon, M. T. Cairns, S. M. Ferguson, W.-J. Liang, P. J. F. Henderson, G. White, and F. R. Hallett. 1999. Purification and reconstitution of an osmosensor: transporter ProP of Escherichia coli senses and responds to osmotic shifts. Biochemistry 38:1676-1684. [DOI] [PubMed] [Google Scholar]
- 38.Rajeevan, M. S., D. G. Ranamukhaarachchi, S. D. Vernon, and E. R. Unger. 2001. Use of real-time quantitative PCR to validate the results of cDNA array and differential display PCR technologies. Methods 25:443-451. [DOI] [PubMed] [Google Scholar]
- 39.Ray, S., J. M. Anderson, F. I. Urmeev, and S. M. Goodwin. 2003. Rapid induction of a protein disulfide isomerase and defense-related genes in wheat in response to the hemibiotrophic fungal pathogen Mycosphaerella graminicola. Plant Mol. Biol. 53:741-754. [DOI] [PubMed] [Google Scholar]
- 40.Roth, W. G., M. P. Lackie, and D. N. Dietzler. 1985. Osmotic stress drastically inhibits active transport of carbohydrates by Escherichia coli. Biochem. Biophys. Res. Commun. 126:434-441. [DOI] [PubMed] [Google Scholar]
- 41.Sabina, J., N. Dover, L. J. Templeton, D. R. Smulski, D. Söll, and R. A. LaRossa. 2003. Interfering with different steps of protein synthesis explored by transcriptional profiling of Escherichia coli K-12. J. Bacteriol. 185:6158-6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a.SAS Institute. 1994. SAS/STAT software, release 6.09 and release 6.08: maintenance enhancement for PROC Mixed. SAS Institute, Inc., Cary, N.C.
- 42.Schleif, R. 1987. The L-arabinose operon, p. 1473-1481. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 43.Sugiura, A., K. Hirokawa, K. Nakashima, and T. Mizuno. 1994. Signal-sensing mechanisms of the putative osmosensor KdpD in Escherichia coli. Mol. Microbiol. 14:929-938. [DOI] [PubMed] [Google Scholar]
- 44.Sugiura, A., K. Nakashima, K. Tanaka, and T. Mizuno. 1992. Clarification of the structural and functional features of the osmoregulated kdp operon of Escherichia coli. Mol. Microbiol. 6:1769-1776. [DOI] [PubMed] [Google Scholar]
- 45.Sutherland, L., J. Cairney, M. J. Elmore, I. R. Booth, and C. F. Higgins. 1986. Osmotic regulation of transcription: induction of the proU betaine transport gene is dependent on the intracellular potassium. J. Bacteriol. 168:805-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weber, A., and K. Jung. 2002. Profiling early osmostress-dependent gene expression in Escherichia coli using DNA microarrays. J. Bacteriol. 184:5502-5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wood, J. M. 1999. Osmosensing by bacteria: signals and membrane-based sensors. Microbiol. Mol. Biol. Rev. 63:230-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wood, J. M., E. Bremer, L. N. Csonka, R. Kraemer, B. Poolman, T. van der Heide, and L. T. Smith. 2001. Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comp. Biochem. Physiol. A 130:437-460. [DOI] [PubMed] [Google Scholar]
- 49.Xu, J., and R. C. Johnson. 1995. Fis activates the RpoS-dependent stationary-phase expression of proP in Escherichia coli. J. Bacteriol. 177:5222-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan, D., T. P. Ikeda, A. E. Shauger, and S. Kustu. 1996. Glutamate is required to maintain the steady-state potassium pool in Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 93:6527-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]