Abstract
Fungus-growing termites efficiently decompose plant litter through their symbiotic relationship with basidiomycete fungi of the genus Termitomyces. Here, we investigated phenol-oxidizing enzymes in symbiotic fungi and fungus combs (a substrate used to cultivate symbiotic fungi) from termites belonging to the genera Macrotermes, Odontotermes, and Microtermes in Thailand, because these enzymes are potentially involved in the degradation of phenolic compounds during fungus comb aging. Laccase activity was detected in all the fungus combs examined as well as in the culture supernatants of isolated symbiotic fungi. Conversely, no peroxidase activity was detected in any of the fungus combs or the symbiotic fungal cultures. The laccase cDNA fragments were amplified directly from RNA extracted from fungus combs of five termite species and a fungal isolate using degenerate primers targeting conserved copper binding domains of basidiomycete laccases, resulting in a total of 13 putative laccase cDNA sequences being identified. The full-length sequences of the laccase cDNA and the corresponding gene, lcc1-2, were identified from the fungus comb of Macrotermes gilvus and a Termitomyces strain isolated from the same fungus comb, respectively. Partial purification of laccase from the fungus comb showed that the lcc1-2 gene product was a dominant laccase in the fungus comb. These findings indicate that the symbiotic fungus secretes laccase to the fungus comb. In addition to laccase, we report novel genes that showed a significant similarity with fungal laccases, but the gene product lacked laccase activity. Interestingly, these genes were highly expressed in symbiotic fungi of all the termite hosts examined.
Fungus-growing termites (subfamily, Macrotermitinae) are distributed throughout tropical Africa and Asia, where they are the dominant soil invertebrates (1, 41). The Macrotermitinae have a highly efficient system for digesting plant litter due to their symbiotic relationship with basidiomycete fungi of the genus Termitomyces (order, Agaricales; family, Tricholomataceae). These termites have a great impact on plant litter decomposition and carbon cycling in tropical ecosystems (42, 43). For example, Buxton (5) demonstrated that fungus-growing termites consumed 90% of the dry woody litter in an arid tropical area of Kenya.
Because of their unique characteristics, such as symbiosis with Termitomyces fungi and sophisticated division of labor, the Macrotermitinae have been the subject of extensive research (for reviews, see references 8, 31, and 42). Fungus-growing termites cultivate symbiotic fungi in their nest on a special substrate composed of dead plant material known as the fungus comb or fungus garden. In most Macrotermes species, termite workers ingest dead plant material and deposit undigested or partially digested feces on the top rim of the fungus comb. Thus, there is an age gradient within the fungus comb. As the fungus comb ages, decolorization from the top to the bottom of the fungus comb is observed. After a certain period, the aged part of the fungus comb is eaten by the host termites (42). Several roles of symbiotic fungi have been proposed, including the provision of glycosyl hydrolases (26), enrichment of nitrogen (6, 27), and lignin degradation (14, 19, 30), with the significance of each role apparently varying in importance among host termite species (20, 31).
Previously, we demonstrated that water-soluble phenolic compounds in the fungus comb of Macrotermes gilvus were degraded during fungus comb aging (21). Higher plants synthesize and accumulate a variety of phenolic compounds as secondary metabolites. Although physiological functions of plant phenolic compounds are not yet fully understood, it is thought that they contribute to plant defenses against pests and pathogens, and therefore, they also influence the decomposition of plant litter by microorganisms in the detritus food chain (33). Consequently, phenol degradation in the fungus comb is considered to be important for improving palatability of termite food, especially that containing high phenol content such as fallen leaves and bark.
The ability of white rot basidiomycetes to degrade a variety of aromatic compounds, such as lignin (23) and aromatic pollutants (29), has been extensively studied. It has been shown that the extracellular phenol-oxidizing enzymes lignin peroxidase (EC 1.11.1.14), manganese peroxidase (EC 1.11.1.13), and laccase (EC 1.10.3.2) are responsible for the depolymerization of lignin (13, 23). Lignin peroxidase catalyzes the oxidation of various aromatic compounds to form aryl cation radicals (23) while manganese peroxidase oxidizes Mn(II) to Mn(III), which diffuses from the enzyme and oxidizes various phenolic compounds. These enzymes require hydrogen peroxide for their activities. Laccase also catalyzes the oxidation of various phenolic compounds and aromatic amines, but this reaction is coupled with the reduction of molecular oxygen to water. Because of their broad substrate specificities, they can be also involved in the degradation of a variety of plant phenols. Although Mora and Lattaud (28) reported the presence of the oxidation activity of 2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) and syringaldazine in the fungus combs of several fungus-growing termites in Africa, and although those authors considered that these reactions were catalyzed by laccase, there is no molecular evidence for the presence of laccase in the fungus comb, and there is no information on the other phenol-oxidizing enzymes such as the peroxidases described above. To better understand phenol degradation in the fungus comb and the contribution of symbiotic fungi to its degradation process, further studies on phenol-oxidizing enzymes in the fungus comb are required.
In this study, we investigated phenol-oxidizing enzymes in fungus combs of fungus-growing termites in Thailand. Laccase was the sole detectable phenol-oxidizing enzyme, and laccase cDNA sequences were identified directly from the fungus combs, showing the distribution and diversity of laccase genes in symbiotic fungi. We also found laccase-like sequences that did not encode laccase and that were highly expressed in fungus combs.
MATERIALS AND METHODS
Fungus combs, microbial strains, and culture conditions.
The fungus combs of five fungus-growing termites were collected from May 2000 to October 2001 in Thailand. The termite hosts and locations sampled in this study were M. gilvus, Microtermes sp. and Odontotermes sp. from the Prachinburi Province, Odontotermes longignathus and Hypotermes sp. from the Saraburi Province, and M. gilvus from the Pathum Thani Province. The sampling locations were separated from each other by at least 50 km. The fungus combs were stored at −80°C until use. Termitomyces strains described previously (34) (JCM accession no. 11082, 11086, 11088, 11089, 11091 to 11094, 11096 to 11098, 11100 to 11106, 11110, 11153, and 11157) and strain NS/Mg (JCM accession no. 13351), which was newly isolated from a fungus comb of M. gilvus in the Pathum Thani Province, were maintained on a potato dextrose agar medium (0.4% potato extract, 2% glucose; Nissui) at 30°C in our laboratory (a complete list of the strains examined in this study is given in Table S1 of the supplemental material). The partial rRNA gene sequence of strain NS/Mg was determined as described previously (34) and submitted to the DDBJ database (accession no. AB202123). The partial large-subunit rRNA gene sequence (1.2 kbp) of this strain was closely related to that of Termitomyces sp. group 1 (accession no. AB073514), showing 94% nucleotide identity. For determination of enzyme activity, Termitomyces spp. strains were also cultured in KB liquid medium (1% glucose, 1.2 mM [low nitrogen {LN}] or 12 mM [high nitrogen {HN}] ammonium tartrate, and 100 ml of Kirk's basal III mineral medium [37] per liter). Four mycelial plugs (4 mm) from each of the Termitomyces strains grown on potato dextrose agar medium were transferred to a 500-ml flask containing 120 ml of KB liquid medium under LN and HN conditions and cultured with agitation (120 rpm) at 28°C.
Plate assay of phenol-oxidizing enzyme.
Termitomyces spp. strains were incubated at 30°C for 4 weeks on KB agar medium containing 0.01% guaiacol under HN and LN conditions. A brownish pigment was observed if the fungi produced a phenol-oxidizing enzyme(s).
Enzyme activity.
The activities of phenol-oxidizing enzymes in fungus combs and in a liquid culture of Termitomyces spp. were examined. The fungus comb was gently ground using a mortar and pestle. Approximately 0.1 g of the ground comb was added to a 1.5-ml polypropylene tube containing 1 ml MiliQ water and subsequently mixed. The supernatant was recovered by centrifugation (10,000 × g for 5 min at 4°C), and enzyme activities were determined. Lignin peroxidase and manganese peroxidase activities were measured as described previously (37, 39) using 3,4-dimethoxybenzyl alcohol and Mn(II) as substrates, respectively. Laccase activity was measured spectrophotometrically by following the oxidation of 2,6-dimethoxyphenol (DMP) or ABTS in sodium citrate at pH 5.0 and 4.5, respectively. The oxidation rates of DMP and ABTS were determined using a Δɛ469 of 49.6 mM−1 cm−1 and a Δɛ415 of 36.0 mM−1 cm−1, respectively (39, 40).
RNA extraction.
Poly(A)+ RNA was extracted from the fungus comb that was ground into a fine powder with a mortar and pestle under liquid nitrogen. Approximately 0.1 g of this fungus comb powder was then transferred to a polypropylene tube containing 0.2 ml of extraction buffer (4 M guanidine thiocyanate, 0.1 M Tris-HCl, pH 7.5, 1% 1,4-dithiothreitol, 0.5% lauroylsarcosine). After mixing thoroughly, 0.6 ml of dilution buffer (0.1 M Tris-HCl, pH 7.5, 0.4 M LiCl, and 20 mM EDTA) was added. The supernatant containing RNA was separated from the debris by centrifugation (17,000 × g). Poly(A)+ RNA was isolated from the supernatant using an Oligotex mRNA kit according to the manufacturer's protocol (Takara). Total RNA was extracted from a 23-day-old Termitomyces culture (strain KU418) according to the method described previously by Han et al. (15), and poly(A)+ RNA was extracted with an Oligotex mRNA kit. First-strand cDNA was synthesized from 0.1 μg (fungus comb samples) or 1 μg (strain KU418) of poly(A)+ RNA using Superscript II (Invitrogen) and poly(T) primer 5′-TTTACCTCTTCAGC(T)19-3′ at 42°C for 50 min and subjected to PCR.
Isolation of the laccase genes and cDNA.
Degenerate primers, primer 1 (5′-GGMACSTTCTGGTAYCAY-3′) and primer 2 (5′-CCRTGCARRTGGAAKGGRTG-3′), targeting a copper binding domain of basidiomycete laccases, were designed to amplify laccase-like sequences from the fungus comb and Termitomyces sp. strain KU418. PCR was carried out using a PTC-200 thermocycler (MJ Research) and Ex-Taq (Takara). Thermal cycling consisted of 35 cycles with an initial denaturation step at 94°C for 30 s, an annealing step at 50°C for 30 s, and an extension step at 72°C for 1 min. PCR products (ca. 900 bp) were purified from 1% agarose gel using a Minelute gel extraction kit (QIAGEN) before being ligated into the pGEM-T vector (Promega) to construct laccase cDNA libraries from five fungus combs and Termitomyces sp. strain KU418. Approximately 30 clones of each library were randomly selected, and their insert DNA sequences were amplified by PCR using a universal primer set of the vector and sorted into groups by restriction fragment length polymorphism (RFLP) using either HaeIII or HhaI. The DNA sequences of representative clones from each library were determined using M13 forward and reverse primers on a DNA sequencer (ABI model 377) and a BigDye Terminator cycle sequencing kit (Applied Biosystems).
Full-length cDNA sequences of lcc1-2 and lcc2-5 were determined using rapid amplification of cDNA ends (RACE)-PCR. Primer 3 (5′-GGTGCAAAGCAGCCTCAAGATCT-3′) and primer 4 (5′-TGGAACATCAATAATGTTTCGTAC-3′) were used for amplification of the 3′-end sequences of lcc1-2 and lcc2-5, respectively. The PCR conditions for lcc1-2 were 35 cycles of 94°C for 30 s followed by 65°C for 1.5 min. The PCR conditions for lcc2-5 were 35 cycles of 94°C for 30 s, 58°C for 1 min, and 72°C for 1 min. For the amplification of the 5′ end, reverse transcription reactions for lcc1-2 and lcc2-5 were performed with primer 5 (5′-TACCAGTCGGCAAGAG-3′) and primer 6 (5′-ATGCTTGAGTTGTGCC-3′), respectively, using a 5′ RACE-PCR kit version 2 (Gibco BRL). The PCR conditions were 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min. PCR products from the 3′ and 5′ RACE-PCR were cloned and sequenced as described above.
The coding regions of the lcc1-2 and lcc2-5 genes were amplified from genomic DNA extracted from the Termitomyces sp. strain NS/Mg using an Isoplant kit (Nippon Gene). The 5′- and 3′-flanking regions of the lcc1-2 and lcc2-5 genes were obtained using inverse PCR (38) with primer 7 (5′-CCTCAAGATCTCCTTCCTTCT-3′) and primer 8 (5′-AGATCCTTCAGAGGATCGTTC-3′) for lcc1-2 and primer 9 (5′-TCTGTCATTGAGGTCGACTTC-3′) and primer 10 (5′-TAAATGAGCCGAAGGATCTTCG-3′) for lcc2-5. Genomic DNA (0.5 μg) from Termitomyces sp. strain NS/Mg was digested with either PstI or EcoRI for the lcc1-2 and lcc2-5 amplifications, respectively. PCR conditions were 30 cycles of 94°C for 20 s and 67°C for 8 min or 5 min for lcc1-2 and lcc2-5, respectively. PCR products were purified using a PCR purification kit (QIAGEN) and analyzed by direct sequencing.
Semiquantitative analysis of gene expression.
In order to estimate expression levels of laccase genes semiquantitatively, we optimized PCR conditions by changing parameters related to the PCR cycle, annealing temperature, and extension time (reference 17 and references therein). The concentrations of the first-strand cDNA and primers 1 and 2 were 0.1 ng/μl and 0.5 μM, respectively. Thermal cycling consisted of an initial denaturation step at 95°C for 2 min followed by 18 or 20 cycles of 95°C for 30 s, 50°C for 1 min, and 72°C for 3 min and, subsequently, a final extension step at 72°C for 10 min. Concentrations of PCR products were estimated by electrophoresis on an agarose gel stained with SYBR green I using Molecular Imager FX (Bio-Rad). Approximately 0.2 to 0.3 nM of PCR products (ca. 900 bp) was obtained. PCR products were purified using a Minelute PCR purification kit (QIAGEN) and cloned into a pCR 2.1 vector using a TOPO TA cloning kit (Invitrogen). Ninety-six randomly chosen clones from the library were sorted by comparing RFLP. Expression levels were expressed as the clone abundance (percent) of each RFLP group.
Phylogenetic analysis.
Taking PCR error into consideration, clones isolated from a sample with more than 99% nucleotide sequence identity were considered sufficiently similar and were grouped together. Representative sequences were used for phylogenetic analysis. Based on sequence alignment and the “GT-AG” rule, some sequences appeared to contain introns. In these cases, putative introns were manually removed and subsequently used for further analyses. All of the sequences were checked for the presence of chimera using the Bellerophon server (http://foo.maths.uq.edu.au/∼huber/bellerophon.pl) (18). No chimera sequences were found. A similarity search was conducted by using BLASTX (2) against the nonredundant protein database (May 2005). The protein sequences were aligned using ClustalX version 1.8 (36). A neighbor-joining tree was constructed using the MEGA package with a PAM matrix (24). The sampling variance of the distance values was estimated from 1,000 bootstrap resamplings of the alignment columns. The signal peptide sequence was predicted using the SignalP program (3). GENETYX version 10.1 (Software Development) was used to calculate sequence identity.
Partial purification of laccase from the fungus comb.
Approximately 100 g of M. gilvus fungus comb from Pathum Thani was gently ground with 1.0 liter of distilled water by using a mortar and pestle. Laccase was precipitated by ammonium sulfate at 65% saturation and collected by centrifugation (10,000 × g, 30 min). The precipitate was dissolved in approximately 60 ml of MilliQ water. The fungus comb of M. gilvus contains considerable amounts of dissolved organic matter (DOM) (21). DOM appeared to contain acidic macromolecular compounds and interfered with enzyme purification. To remove the DOM, 0.8 g of DEAE-Sephadex A50 powder (Amersham) was added to the crude fungus comb extract. Most of the laccase did not bind to the DEAE-Sephadex powder under these conditions. The solution separated from Sephadex was dialyzed against 10 mM sodium acetate (pH 4.0). After 16 h, the solution was adjusted to pH 6.0 and subsequently applied to a column (1.5 by 10 cm) of DEAE-Toyopearl (Tosoh) equilibrated with 10 mM phosphate (pH 6.0). The column was washed with the starting buffer until all of the DOM had been removed. Elution with 20 mM sodium acetate (pH 4.0) resulted in 77% laccase recovery in this step. The enzyme solution was then applied to a Superdex 200HR 10/30 column (Amersham) with a 50 mM sodium acetate-0.15 M ammonium sulfate buffer (pH 4.0) at a flow rate of 0.25 ml/min. The enzyme was further purified using a HiTrap-phenyl Sepharose HP column (Amersham). A sample containing 2 M ammonium sulfate was applied to the column and eluted using a linear gradient of 1.5 to 0.7 M ammonium sulfate in 100 mM acetate (pH 4.0). The fractions exhibiting laccase activity were pooled and reapplied to the same column a total of four times. The N-terminal amino acid sequences were determined using an automated sequencer (ABI model 494cLC).
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in the DDBJ database under the following accession numbers: AB201126 to AB201165.
RESULTS
Phenol-oxidizing enzyme activity in the fungus comb.
We examined a series of phenol-oxidizing enzymes, laccase, lignin, and manganese peroxidases in the fungus combs of Microtermes sp., Odontotermes sp., and M. gilvus. Of these enzymes, only laccase activity was detected in all of the fungus combs examined (Table 1). The laccase activity monitored by DMP oxidation was not affected by the addition of H2O2 (data not shown), indicating the absence of horseradish peroxidase type enzyme in the fungus comb because horseradish peroxidase can oxidize DMP in the presence of H2O2. Inhibitory effects of several chemicals on the laccase activity in the fungus comb of M. gilvus were similar to those on laccases from known basidiomycete isolates (Table 2). The laccase activity was also significantly inhibited in a nitrogen atmosphere (data not shown), indicating that oxygen is required for this activity. The optimum pH for DMP oxidation was pH 5.0.
TABLE 1.
Laccase activity in fungus combs
| Termite species (location) | Activity (μkat/g of comb)a |
|---|---|
| M. gilvus (Prachinburi) | 32.6 |
| M. gilvus (Pathum Thani) | 33.3 |
| Odontotermes sp. (Prachinburi) | 9.58 |
| Microtermes sp. (Prachinburi) | 5.15 |
The data shown are mean values of duplicate experiments.
TABLE 2.
Inhibitory effect of chemicals on laccase activity from M. gilvus comb in Pathum Thania
| Inhibitor | Concn (mM) | Inhibition (%)b
|
||
|---|---|---|---|---|
| Fungus comb | P. cinnabarinusc | C. hirsutusc | ||
| DTTd | 1.0 | 100 | 100 | 100 |
| NaN3 | 0.1 | 97 | 100 | 100 |
| EDTA | 5.0 | 11 | 0e | 0e |
DMP was used as a substrate.
Zero inhibition refers to 16 nkat of laccase activity.
Laccases from Picnoporus cinnabarinus and Coriorus hirsutus (12).
DTT, dithiothreitol.
EDTA, 4.0 mM.
Laccase activity from Termitomyces strains.
The ability of the symbiotic fungi to produce phenol-oxidizing enzymes was examined in 22 Termitomyces strains from fungus combs of various termite species (34) using a plate assay method. Phenol-oxidizing activity was observed under both HN and LN conditions in most of the strains assayed (data not shown), except for two strains from Hypotermes sp. and one strain from O. longignathus that only exhibited the activity under LN conditions. No activity was observed from one Odontotermes sp. strain under any condition. The strains that exhibited higher activity were cultured in KB liquid medium, and the series of phenol-oxidizing enzymes were examined. Only laccase activity was found, and no lignin or manganese peroxidase activities were detected in any strain. As shown in Fig. 1, significant laccase activity was detected under LN conditions in strain KU418 that was isolated from the fungus comb of M. gilvus. Weak laccase activities (less than 6.7 nkat/ml) were also detected in other strains isolated from the fungus combs of Hypotermes sp. (strain KU428), Microtermes sp. (strain KU430), Odontotermes sp. (strain KU432), and O. longignathus sp. (strain KU426).
FIG. 1.
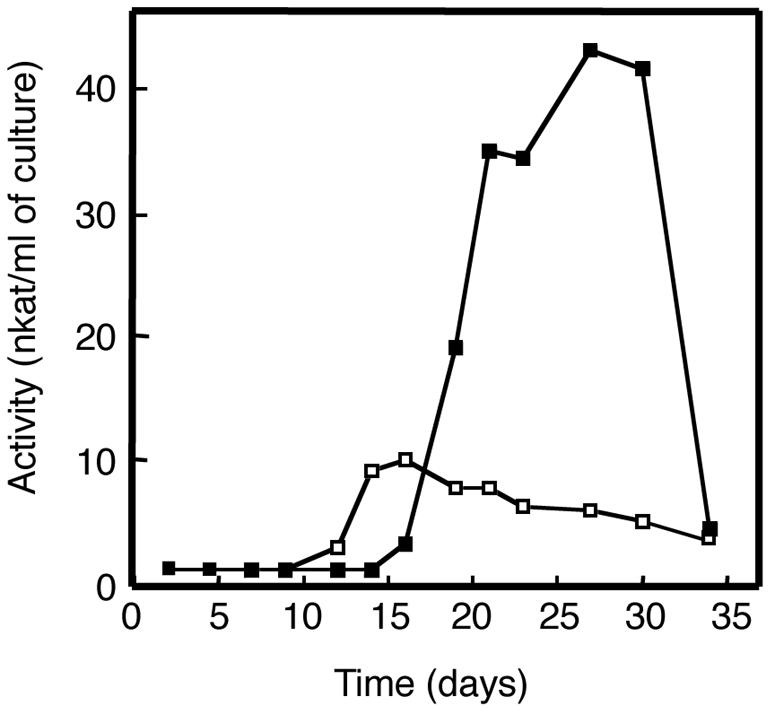
Laccase activity in the culture supernatant of Termitomyces sp. strain KU418 isolated from M. gilvus under low-nitrogen (closed squares) and high-nitrogen (open squares) conditions. The reaction mixture contained 0.5 mM DMP and 100 μl of culture supernatant in 50 mM sodium citrate, pH 5.0.
Cloning and characterization of putative laccase genes.
In order to identify laccase genes expressed in the fungus comb, reverse transcription (RT)-PCR was carried out on RNA extracted from the fungus comb of M. gilvus from Pathum Thani with degenerate primers targeting the highly conserved copper binding domains II and III (Fig. 2). RT-PCR products of an appropriate size (ca. 900 bp) were obtained and cloned. The sequences of the clones showed a significant similarity with fungal laccases. The full-length cDNAs of two representative clones (lcc1-2 and lcc2-5) were isolated from the fungus comb RNA. The corresponding gene sequences were also identified from genomic DNA of the symbiotic fungal strain NS/Mg isolated from the same fungus comb. Comparison between the genomic and cDNA sequences revealed that the coding regions of genomic lcc1-2 and lcc2-5 were interrupted by 23 and 21 introns and encoded 524 and 534 amino acids, respectively. Deduced amino acid sequences from the lcc1-2 and lcc2-5 cDNAs contained 6 and 10 potential N-glycosylation sites (N-X-S/T), respectively. Putative signal peptides were detected from the first Met to Ala 18 of LCC1-2 and to Gly 21 of LCC2-5, respectively. The LCC1-2 amino acid sequence contained all of the amino acid residues that are essential for copper ion binding in laccase, whereas the copper binding domains in LCC2-5 were incomplete (Fig. 2). The LCC2-5 amino acid sequence lacked 3 of 11 amino acid residues involved in copper ion binding. Two disulfide bonds, Cys 103-Cys 505 and Cys 135-Cys 222, were found in the crystal structure of laccase 1 from Coprinus cinerea (9). The equivalent four Cys residues were also conserved in the LCC1-2 sequence, while one set of Cys residues was present in the LCC2-5 sequence. A BLAST search showed that best hits for LCC1-2 and LCC2-5 amino acid sequences were laccase 1 (Q12541) and laccase 2 (Q12542), respectively, from the basidiomycete Agaricus bisporus. Amino acid identities and similarities between LCC1-2 and laccase 1 were 66 and 76%, while those between LCC2-5 and laccase 2 were 47 and 62%, respectively.
FIG. 2.
Sequence comparison among C. cinerea laccase 1 (CCLCC1) (DDBJ accession no. AAD30964), A. bisporus laccase 2 (ABLCC2) (accession no. Q12542), LCC1-2, and LCC2-5. Identical and similar amino acids are shaded. The conserved amino acid residues potentially involved in copper ion binding are marked by asterisks, and position 129 of A. bisporus laccase 2 is also marked with an arrow. Numbers under the His and Cys residues indicate types of copper ions that bind to each residue. The amino acid sequences that are used to design the degenerate primers are underlined. The Cys residues that form disulfide bonds in C. cinerea laccase 1 are marked with closed (Cys 103-Cys 505) and open (Cys 135-Cys 222) inverted triangles (9). A closed circle indicates the Leu residue that influences a laccase redox potential. Experimentally determined N-terminal amino acid sequences of LCC1-2 and LCC2-5 are italicized.
Laccase cDNA fragments from various fungus combs.
We have thus far demonstrated that our primer set designed here was capable of amplifying laccase cDNA from the fungus comb of M. gilvus. Therefore, laccase cDNAs in the fungus comb of different termites were analyzed using the same method. RT-PCR products with expected sizes were obtained from all the fungus comb samples. A total of 71 sequences were identified from the fungus combs of five termite species and from strain KU418 grown in KB liquid culture (Fig. 1). Of these, 69 sequences showed significant similarities to basidiomycete laccases (BLAST scores and E values ranged from 441 to 234 and 1e-122 to 2e-60, respectively). Termination codons were found in putative open reading frames of two sequences from the fungus comb of Odontotermes sp. and one sequence from strain KU418, and these were not analyzed further. In addition to the lcc1-2 and lcc2-5 genes, the numbers of distinct cDNA sequences eventually identified were six from M. gilvus, three from Odontotermes sp., and six from Microtermes sp. in Prachinburi; seven from Hypotermes sp. and five from O. longignathus in Saraburi; one from M. gilvus in Pathum Thani; and nine from strain KU418.
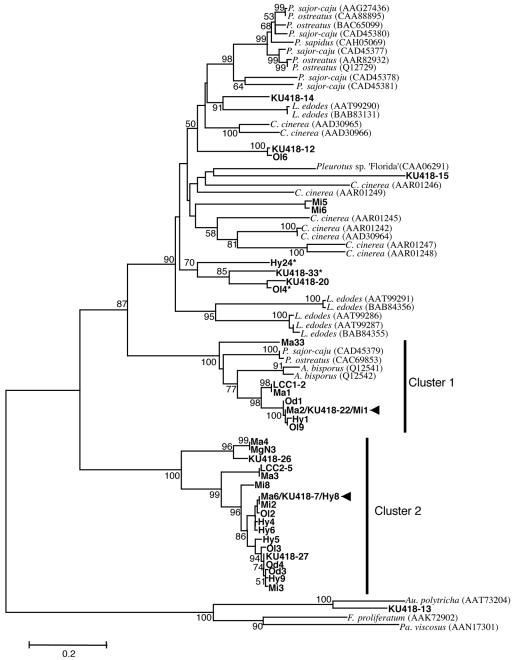
Phylogenetic analysis placed most of the putative laccase amino acid sequences into two major clusters (clusters 1 and 2) (Fig. 3). All of the sequences from the M. gilvus fungus combs from Prachinburi and Pathum Thani were found in these clusters, while five sequences identified from Microtermes sp., Hypotermes sp., and O. longignathus (Mi5, Mi6, Hy24, Ol4, and Ol6) were placed outside these clusters and appeared more closely related to the laccases of known basidiomycetes. All of the sequences in cluster 2 lacked the His residue that is essential for type 3 copper binding (position 129 in A. bisporus LCC2 [shown in Fig. 2]), while other clones possessed His at that position. Among 10 sequences from strain KU418, 3 sequences were found in cluster 2. Sequences that were identical to those of KU418-22 and KU418-7 were identified from the fungus combs (Fig. 3). The phylogenetic position of sequence KU418-13 was close to that of the ascomycete laccases, although a laccase from the basidiomycete Auricularia polytricha also clustered in the same group as the closest relative.
FIG. 3.
Neighbor-joining tree for amino acid sequences of putative laccases identified from the fungus combs and Termitomyces sp. strain KU418 and related proteins. The scale bar represents 0.2 amino acid substitutions per position. Accession numbers are shown after the names of organisms. Bootstrap confidence values greater than 50 are indicated at the nodes. Branches containing identical sequences are indicated with arrowheads. Clone designations and genus abbreviations are as follows: MaX, M. gilvus in Prachinburi; MgN3, M. gilvus in Pathum Thani; OdX, Odontotermes sp.; MiX, Microtermes sp.; OlX, O. longignathus; Hy, Hypotermes sp.; KU418-X, Termitomyces sp. strain KU418, where X means number; P., Pleurotus; L., Lentinula; C., Coprinopsis; A., Agaricus; Au., Auricularia; F., Fusarium; Pa., Panorbis. Asterisks indicate the clones that contain a putative intron(s).
The relative abundance of putative laccase sequences in each clone library was analyzed semiquantitatively to estimate expression levels of laccase genes in the fungus comb (Fig. 4). The most abundant clones found in all the three libraries were grouped together in cluster 2. Except for the sequences in cluster 2, Ma1, lcc1-2, and Hy1 were the most abundant sequences from M. gilvus in Prachinburi, M. gilvus in Pathum Thani, and Hypotermes sp. in Saraburi.
FIG. 4.
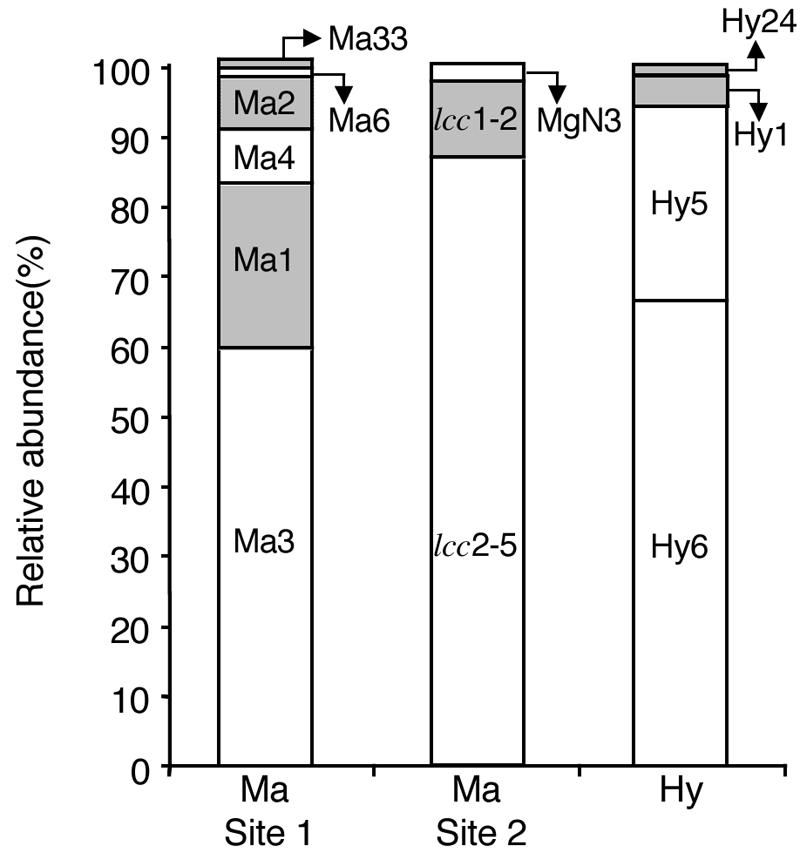
Relative abundance of cDNA clones from the fungus combs of M. gilvus in Prachinburi (site 1), M. gilvus in Pathum Thani (site 2), and Hypotermes sp. in the Saraburi Province. Clone designations are shown in the legend of Fig. 3. The sequences possessing a conserved His residue at position 129 of A. bisporus laccase 2 are shaded.
Partial purification of laccase from the fungus comb.
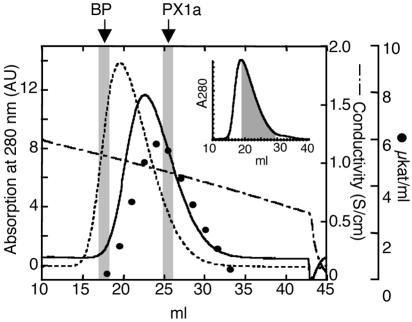
Crude enzyme from the fungus comb of M. gilvus from Pathum Thani was separated into two fractions containing laccase activity using a DEAE-Toyopearl column. One fraction (PX1) that was eluted with 20 mM sodium acetate (pH 4.0) contained laccase activity that was three times higher than that of the other fraction that was eluted with a salt gradient. Although sodium dodecyl sulfate-polyacrylamide gel electrophoresis of PX1 gave a single band with a molecular mass of 75 kDa, analysis of the N-terminal amino acid sequence indicated that this band contained two proteins. PX1 was further purified using gel filtration and subsequent hydrophobic-interaction chromatography. When PX1 was subjected to a Phenyl Sepharose column, a broad single peak appeared, where laccase activity was found in the shaded part of the peak (Fig. 5, inset), indicating that PX1 consisted of laccase and nonlaccase proteins. The fractions with laccase activity were pooled, and the chromatographic separation with the same column was repeated. Figure 5 shows the chromatograms of the separated laccase and nonlaccase fractions (designated PX1a and BP, respectively) after chromatographic runs were performed four times. The purified BP fraction did not exhibit the oxidation activities for ABTS, DMP, potassium ferrocyanide, and o- and p-catechol under the laccase assay conditions. The N-terminal amino acid sequence of BP was determined to be NLKEITLDIVNA. Since the PX1a fraction still contained BP protein, the N-terminal amino acid sequence of the protein with laccase activity was estimated to be AVRHFDIPXTVT by subtracting the BP amino acid sequence. This protein is tentatively called PX1a in this paper. The N-terminal amino acid sequences of PX1a and BP were found in the deduced amino acid sequences of LCC1-2 and LCC2-5, respectively (Fig. 2). Thus, it is highly likely that the gene lcc1-2 encodes laccase PX1a and that lcc2-5 is not a laccase gene.
FIG. 5.
Phenyl Sepharose chromatogram of BP (broken line) and PX1a (solid line). The fractions indicated with shaded squares were subjected to N-terminal amino acid sequencing. The closed circles indicate laccase activity of PX1a fractions. The inset shows a typical chromatogram of PX1 on Phenyl Sepharose. The fractions with laccase activity are shaded. Further details are described in Results. AU, absorbance units.
DISCUSSION
A number of phenol-oxidizing enzymes have been characterized in basidiomycete fungi by culturing them in the laboratory. However, to our knowledge, this is the first detailed molecular study of such an enzyme characterized directly in a natural environment without cultivation. We consider the naturally occurring condition of Termitomyces very important in order to understand the real nature of the symbiotic relationship with the host termite and their efficient decomposition of plant litter. Here, we clearly demonstrated that laccase was the sole detectable phenol-oxidizing enzyme in the fungus combs of Microtermes sp., Odontotermes sp., and M. gilvus. No peroxidase activity was detected in either the fungus comb or culture supernatants of Termitomyces spp. strains, although many white rot basidiomycetes produce extracellular peroxidases. A detailed analysis of the fungus comb of M. gilvus from Pathum Thani and its symbiotic fungus revealed that the lcc1-2 gene of Termitomyces sp. strain NS/Mg encodes laccase PX1a and that this is the dominant isozyme under symbiotic conditions. The LCC1-2 amino acid sequence contains all of the conserved His and Cys residues required for copper ion binding and an additional conserved Leu residue affecting the redox potential of laccase (Fig. 2). According to the classification of Eggert et al. (10), LCC1-2 belongs to the class 2 laccases that have a moderate redox potential (0.71 to 0.47 V) (25). A Met or Phe residue is located at the same position in the class 1 or 3 laccase sequence. All the results found in this study indicated that laccase in the fungus comb of M. gilvus had the common characteristics of laccase in its catalytic properties and primary structure.
In addition to the lcc1-2 laccase gene, we identified a novel gene, lcc2-5, and its gene product, BP, from the fungus comb of M. gilvus. The discovery of the BP protein was attributed to its chromatographic behavior, which was markedly similar to that of laccase PX1a. The LCC2-5 amino acid sequence showed significant similarity with that of A. bisporus laccase 2, but it lacked three His residues required for copper ion binding in the deduced amino acid sequence (Fig. 2), and the gene product, BP, also lacked laccase activity. The laccase molecule has four copper ions distributed among three sites, each of which is defined according to its spectroscopic properties (32). The T1 site contains the type 1 blue copper that is responsible for absorption at around 600 nm. The T2 site contains a type 2 copper with a characteristic electron paramagnetic resonance. In the T3 site, the pair of strongly coupled type 3 coppers is electron paramagnetic resonance silent in the presence of dioxygen. The mononuclear T1 site extracts an electron from a reducing substrate and mediates its transfer to the trinuclear T2/T3 center where molecular oxygen is reduced. However, as shown in Fig. 2, while LCC2-5 (BP protein) possesses the necessary residues for type 1 copper binding, absorption at around 600 nm was not observed (data not shown), suggesting a distortion of the tertiary structure of a potential T1 site in LCC2-5.
Putative cDNA fragments of laccase were amplified using the degenerate PCR primers from the fungus combs of various termite hosts. Their deduced amino acid sequences showed similarity with those of fungal laccases, but some of the sequences had Glu or Gln residues at the site of the conserved His residue corresponding to His 129 of A. bisporus laccase 2. Phylogenetic analysis showed that all of these sequences were placed in cluster 2 and that they formed a lineage distinct from those of basidiomycete laccases (Fig. 3). LCC2-5 was also found in cluster 2, suggesting that while the sequences in cluster 2 are closely related to laccase genes, they do not most certainly encode “true” laccase. Interestingly, these pseudolaccase cDNA sequences were identified from all of the fungus combs tested in this study. Amino acid identities among these pseudolaccases ranged from 61 to 100%. Also, the transcription levels of the pseudolaccase genes were much higher than those of the putative laccase genes (Fig. 4). Expressed sequence tag analysis for symbiotic fungus of M. gilvus is in progress in our laboratory, and the sequence corresponding to lcc2-5 was one of the abundant sequences (T. Johjima, unpublished data). These findings suggest that the pseudolaccase genes are nonetheless essential for either the symbiotic fungi themselves or symbiosis with the host termites. Although laccase gene sequences from plants and bacteria have been deposited in public databases, their phylogenetic positions were distinct from those of fungal laccase clusters (data not shown), suggesting that the putative laccase and laccase-like cDNA sequences from the fungus combs were certainly of fungal origin.
Multiple laccase genes are often found in single organisms. The saprophytic fungus C. cinerea, for example, has eight different laccase genes that were reported previously (16) and one additional gene in the public databases. Termitomyces sp. strain KU418 expressed seven putative laccase genes when the fungus was cultured in KB liquid medium under LN conditions. These sequences showed higher diversity than those found in either the C. cinerea laccases or the sequences from the M. gilvus fungus comb that were found only in cluster 1 (Fig. 3). In M. gilvus from Prachinburi, only one identical sequence set (Ma2 and KU418-22) was retrieved from the clone library of the fungus comb and strain KU418, although four times more clones were sequenced from the KU418 clone library than from the fungus comb library. This finding could possibly be attributed to differences in culture conditions between the fungus comb and KB medium, because the differential expression of fungal laccase genes is often found depending upon nutritional conditions, copper ion concentrations (references 4 and 7 and references therein), and the presence of various aromatic compounds (35). It is unlikely that more than one species of symbiotic fungi are associated with a fungus comb, and consequently, the multiple laccase genes were identified, since Katoh et al. (22) demonstrated no genetic variation in symbiotic fungus from a single large nest of the termite Odontotermes formosanus. We have also analyzed fungal community structures in the fungus combs of Odontotermes sp., Microtermes sp., and M. gilvus from Prachinburi, showing that single or closely related Termitomyces species almost exclusively grew on each fungus comb (S. Moriya, unpublished data). These findings support the idea that Termitomyces fungi have multiple laccase genes.
Putative functional laccase cDNA fragments that clustered outside cluster 2 were identified from the fungus combs of all the termite hosts examined in this study, indicating that laccase is widely distributed among the symbiotic fungi of fungus-growing termites. Phylogenetic analysis showed that the laccase sequences from the fungus comb and Termitomyces sp. strain KU418 were closely related to the laccases from A. bisporus, L. edodes, Pleurotus spp., and C. cinerea. Like the Termitomyces fungi, these fungi belong to the order Agaricales and mostly produce class 2 laccases. Previously, we analyzed phylogenetic relationships among Termitomyces fungi used in this study, except for strain NS/Mg (34), and showed that fungal strains isolated from O. longignathus (Saraburi Province), Odontotermes sp., and M. gilvus (Prachinburi Province) were closely related and shared more than 99% nucleotide identity in partial sequences (1.2 to 1.4 kbp) of the large-subunit rRNA genes. In the laccase phylogeny, specific relationships were not found among laccase homologs from those symbiotic fungi. It is uncertain whether this is due to differential expression of multiple laccase genes in each fungus comb as seen in the M. gilvus fungus comb and strain KU418 or each symbiotic fungus possessing individual laccase genes. Comprehensive analyses of laccase genes in those symbiotic fungi would be required to answer the question.
Hyodo et al. (20) analyzed the chemical composition of fungus combs from Odontotermes spp., Hypotermes makhamensis, Ancistrotermes pakistanicus, Pseudacanthotermes militaris, and four Macrotermes spp. and found that lignin was preferentially degraded compared to carbohydrates in only Macrotermes fungus combs. Those authors noted that the Macrotermes species examined in their study tended to use leaf litter for construction of the fungus comb, while the other genera, with the exception of P. militaris, appeared to use predominantly wood, and therefore, the different material of the fungus comb possibly affects lignin degradation in fungus combs. In this study, laccase activity was detected and higher laccase activity was found in the fungus combs of M. gilvus than in Odontotermes sp. and Microtermes sp. (Table 1). Does the higher laccase activity promote preferential lignin degradation in this genus? This study provides insufficient information on lignin degradation because laccase is basically incapable of oxidation of nonphenolic lignin moieties that comprise up to 85% of the lignin polymer because of the low redox potential of laccase. However, it is known that laccase can oxidize nonphenolic compounds and degrade lignin in the presence of a laccase mediator such as 3-hydroxyanthranilate (11). Thus, laccase mediators must be studied to estimate the contribution of laccase to lignin degradation in the fungus comb of the genus Macrotermes. Further functional studies on laccase and laccase-like protein from the symbiotic fungi are necessary to clarify the importance of these enzymes for efficient decomposition of plant material by fungus-growing termites in tropical ecosystems.
Supplementary Material
Acknowledgments
This work was partially supported by grants for the Bioarchitect Research Program and the Eco Molecular Science Research Program from RIKEN.
We are grateful to the Biomolecular Characterization Laboratory in RIKEN for amino acid sequence analysis and H. Yuzawa, C. Disyen, K. Sirihongsuwan, and C. Boontong for assistance. We are also grateful to S. Hattori for helping with the anaerobic experiment and the National Research Council of Thailand for permission to research in Thailand.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abe, T., and T. Matsumoto. 1979. Studies on the distribution and ecological role of termites in a lowland rain forest of west Malaysia. 3. Distribution and abundance of termites in Pasoh Forest Reserve. Jpn. J. Ecol. 29:337-351. [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 4.Burke, R. M., and J. W. Cairney. 2002. Laccases and other polyphenol oxidases in ecto- and ericoid mycorrhizal fungi. Mycorrhiza 12:105-116. [DOI] [PubMed] [Google Scholar]
- 5.Buxton, R. D. 1981. Termites and the turnover of dead wood in an arid tropical environment. Oecologia 51:379-384. [DOI] [PubMed] [Google Scholar]
- 6.Collins, N. M. 1983. The utilization of nitrogen resources by termites (Isoptera), p. 381-412. In J. A. Lee, S. McNeill, and I. H. Rorison (ed.), Nitrogen as an ecological factor. Blackwell Scientific, Oxford, United Kingdom.
- 7.Collins, P. J., and A. D. W. Dobson. 1997. Regulation of laccase gene transcription in Trametes versicolor. Appl. Environ. Microbiol. 63:3444-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darlington, J. P. E. C. 1994. Nutrition and evolution in fungus-growing termites, p. 105-130. In J. H. Hunt and C. A. Nalepa (ed.), Nourishment and evolution in insect societies. Westview Press, Boulder, Colo.
- 9.Ducros, V., A. M. Brzozowski, K. S. Wilson, S. H. Brown, P. Ostergaard, P. Schneider, D. S. Yaver, A. H. Pedersen, and G. J. Davies. 1998. Crystal structure of the type-2 Cu depleted laccase from Coprinus cinereus at 2.2 Å resolution. Nat. Struct. Biol. 5:310-316. [DOI] [PubMed] [Google Scholar]
- 10.Eggert, C., P. R. LaFayette, U. Temp, K. E. Eriksson, and J. F. Dean. 1998. Molecular analysis of a laccase gene from the white rot fungus Pycnoporus cinnabarinus. Appl. Environ. Microbiol. 64:1766-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eggert, C., U. Temp, J. F. Dean, and K. E. Eriksson. 1996. A fungal metabolite mediates degradation of non-phenolic lignin structures and synthetic lignin by laccase. FEBS Lett. 391:144-148. [DOI] [PubMed] [Google Scholar]
- 12.Eggert, C., U. Temp, and K. E. Eriksson. 1996. The ligninolytic system of the white rot fungus Pycnoporus cinnabarinus: purification and characterization of the laccase. Appl. Environ. Microbiol. 62:1151-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gold, M. H., H. Wariishi, and K. Valli. 1989. Extracellular peroxidase involved in lignin degradation by the white rot basidiomycete Phanerochaete chrysosporium, p. 127-140. In J. R. Whitaker and P. E. Sonnet (ed.), Biocatalysis in agricultural biotechnology. Proceedings of the ACS Symposium. ACS, Washington, D.C.
- 14.Grassé, P.-P., and C. Noirot. 1958. Le meule des termites champignonnistes et sa signification symbiotique. Ann. Sci. Nat. Zool. Biol. Anim. 20:113-128. [Google Scholar]
- 15.Han, J. H., C. Stratowa, and W. J. Rutter. 1987. Isolation of full-length putative rat lysophospholipase cDNA using improved methods for mRNA isolation and cDNA cloning. Biochemistry 26:1617-1625. [DOI] [PubMed] [Google Scholar]
- 16.Hoegger, P. J., M. Navarro-Gonzalez, S. Kilaru, M. Hoffmann, E. D. Westbrook, and U. Kues. 2004. The laccase gene family in Coprinopsis cinerea (Coprinus cinereus). Curr. Genet. 45:9-18. [DOI] [PubMed] [Google Scholar]
- 17.Hongoh, Y., M. Ohkuma, and T. Kudo. 2003. Molecular analysis of bacterial microbiota in the gut of the termite Reticulitermes speratus (Isoptera; Rhinotermitidae). FEMS Microbiol. Ecol. 44:231-242. [DOI] [PubMed] [Google Scholar]
- 18.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 19.Hyodo, F., T. Inoue, J.-I. Azuma, I. Tayasu, and T. Abe. 2000. Role of the mutualistic fungus in lignin degradation in the fungus-growing termite Macrotermes gilvus (Isoptera; Macrotermitinae). Soil Biol. Biochem. 32:653-658. [Google Scholar]
- 20.Hyodo, F., I. Tayasu, T. Inoue, J.-I. Azuma, T. Kudo, and T. Abe. 2003. Differential role of symbiotic fungi in lignin degradation and food provision for fungus-growing termites (Macrotermitinae: Isoptera). Func. Ecol. 17:186-193. [Google Scholar]
- 21.Johjima, T., T. Inoue, M. Ohkuma, N. Noparatnaraporn, and T. Kudo. 2003. Chemical analysis of food processing by the fungus-growing termite Macrotermes gilvus. Sociobiology 42:815-824. [Google Scholar]
- 22.Katoh, H., T. Miura, K. Maekawa, N. Shinzato, and T. Matsumoto. 2002. Genetic variation of symbiotic fungi cultivated by the macrotermitine termite Odontotermes formosanus (Isoptera: Termitidae) in the Ryukyu Archipelago. Mol. Ecol. 11:1565-1572. [DOI] [PubMed] [Google Scholar]
- 23.Kirk, T. K., and R. L. Farrell. 1987. Enzymatic “combustion”: the microbial degradation of lignin. Annu. Rev. Microbiol. 41:465-505. [DOI] [PubMed] [Google Scholar]
- 24.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 25.Kumar, S. V., P. S. Phale, S. Durani, and P. P. Wangikar. 2003. Combined sequence and structure analysis of the fungal laccase family. Biotechnol. Bioeng. 83:386-394. [DOI] [PubMed] [Google Scholar]
- 26.Martin, M. M., and J. S. Martin. 1978. Cellulose digestion in the midgut of the fungus-growing termites Macrotermes natalensis: the role of acquired digestive enzymes. Science 199:1453-1455. [DOI] [PubMed] [Google Scholar]
- 27.Matsumoto, T. 1976. The role of termites in an equatorial rain forest ecosystem of west Malaysia. I. Population density, biomass, carbon, nitrogen and calorific content and respiration rate. Oecologia 22:153-178. [DOI] [PubMed] [Google Scholar]
- 28.Mora, P., and C. Lattaud. 1999. Screening termite species for laccase: role of symbiotic fungi. Insect Sci. Applic. 19:51-55. [Google Scholar]
- 29.Pointing, S. B. 2001. Feasibility of bioremediation by white-rot fungi. Appl. Microbiol. Biotechnol. 57:20-33. [DOI] [PubMed] [Google Scholar]
- 30.Rohrmann, G. F. 1978. The origin, structure, and nutritional importance of the comb in two species of Macrotermitinae. Pedobiologia 18:89-98. [Google Scholar]
- 31.Rouland-Lefèvre, C. 2000. Symbiosis with fungi, p. 289-306. In T. Abe, D. E. Bignell, and M. Higashi (ed.), Termites: evolution, society, symbioses, ecology. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 32.Solomon, E. I., U. M. Sundaram, and T. E. Machonkin. 1996. Multicopper oxidases and oxygenases. Chem. Rev. 96:2563-2606. [DOI] [PubMed] [Google Scholar]
- 33.Swift, M. J., O. W. Heal, and J. M. Anderson. 1979. Decomposition in terrestrial ecosystems. Blackwell Scientific, Oxford, United Kingdom.
- 34.Taprab, Y., M. Ohkuma, T. Johjima, Y. Maeda, S. Moriya, T. Inoue, P. Suwanarit, N. Noparatnaraporn, and T. Kudo. 2002. Molecular phylogeny of symbiotic basidiomycetes of fungus-growing termites in Thailand and their relationship with the host. Biosci. Biotechnol. Biochem. 66:1159-1163. [DOI] [PubMed] [Google Scholar]
- 35.Terron, M. C., T. Gonzalez, J. M. Carbajo, S. Yague, A. Arana-Cuenca, A. Tellez, A. D. Dobson, and A. E. Gonzalez. 2004. Structural close-related aromatic compounds have different effects on laccase activity and on lcc gene expression in the ligninolytic fungus Trametes sp. I-62. Fungal Genet. Biol. 41:954-962. [DOI] [PubMed] [Google Scholar]
- 36.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tien, M., and T. K. Kirk. 1988. Lignin peroxidase of Phanerochaete chrysosporium. Methods Enzymol. 161:238-249. [Google Scholar]
- 38.Triglia, T., M. G. Peterson, and D. J. Kemp. 1988. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 16:8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wariishi, H., K. Valli, and M. H. Gold. 1992. Manganese(II) oxidation by manganese peroxidase from the basidiomycete Phanerochaete chrysosporium. Kinetic mechanism and role of chelators. J. Biol. Chem. 267:23688-23695. [PubMed] [Google Scholar]
- 40.Wolfenden, B. S., and R. L. Willson. 1982. Radical-cations as reference chromogens in kinetic studies of one-electron transfer-reactions. Pulse radiolysis studies of 2,2′-azinobis-(3-ethylbenzthiazoline-6-sulphonate). J. Chem. Soc. Perkin. Trans. II:805-812. [Google Scholar]
- 41.Wood, T. G., and W. A. Sands. 1978. The role of termites in ecosystems, p. 245-292. In M. V. Brian (ed.), Production ecology of ants and termites. Cambridge University Press, Cambridge, United Kingdom.
- 42.Wood, T. G., and R. J. Thomas. 1989. The mutualistic association between Macrotermitinae and Termitomyces, p. 69-92. In N. Wilding, N. M. Collins, P. M. Hammond, and J. F. Webber (ed.), Insect-fungus interaction. Academic Press, London, United Kingdom.
- 43.Yamada, A., T. Inoue, D. Wiwatwitaya, M. Ohkuma, T. Kudo, T. Abe, and A. Sugimoto. 2005. Carbon mineralization by termites in tropical forests, with emphasis on fungus-combs. Ecol. Res. 20:453-460.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.