Abstract
The overexpression of serine acetyltransferase from the Ni-hyperaccumulating plant Thlaspi goesingense causes enhanced nickel and cobalt resistance in Escherichia coli. Furthermore, overexpression of T. goesingense serine acetyltransferase results in enhanced sensitivity to cadmium and has no significant effect on resistance to zinc. Enhanced nickel resistance is directly related to the constitutive overactivation of sulfur assimilation and glutathione biosynthesis, driven by the overproduction of O-acetyl-l-serine, the product of serine acetyltransferase and a positive regulator of the cysteine regulon. Nickel in the serine acetyltransferase-overexpressing strains is not detoxified by coordination or precipitation with sulfur, suggesting that glutathione is involved in reducing the oxidative damage imposed by nickel.
Heavy-metal resistance mechanisms in bacteria have been shown to exist in various species (15). Soft metals, such as Cd, Hg, and Ag, have a chemical preference for thiol ligands (9), deactivating enzymes that contain thiols at their active sites. In addition, metals interact with other important trace elements in cells, inhibiting their normal physiological functions (15). Redox-active metals, like Cu, Fe, and Mn, have direct oxidizing capacity, and other metals, like Ni, Co, and Zn, cause indirect oxidative stress through uncoupling of electron transport in both respiration and photosynthesis and depletion of glutathione (GSH), leading to the accumulation of reactive oxygen species. The intracellular generation of superoxide by Cd, Ni, and Co is toxic in Escherichia coli (6), and superoxide dismutase is involved in protection against this metal-induced oxidative stress (6).
While the generation of oxidative stress in bacteria is catalyzed by certain metals, oxyanions like selenate and arsenate cause the disruption of normal metabolism involved with chemically related nonmetals, such as sulfate and phosphate. At present, there are three major mechanisms of heavy-metal tolerance thought to exist naturally in bacteria. First, heavy-metal concentrations in cells are restricted through efflux mechanisms. Second, metals that prefer to bind to sulfur are segregated into complexes with thiol-containing compounds. Third, the oxidation states of specific metal ions can be altered to reduce their toxicity. For many different metals, tolerance and homeostasis in bacteria involve a combination of these basic mechanisms (15).
Metal tolerance mechanisms have also been extensively documented in plants (4). Plants are known that can tolerate elevated concentrations of various metals by either exclusion or hyperaccumulation of the metal. Numerous plants have been identified that, when growing in there natural habitats, can contain up to 3% (30,000 μg g−1) of their shoot dry weight as either Ni or Zn (19). Such unique plants have been termed metal hyperaccumulators (3). We previously observed that constitutive activation of serine acetyltransferase (SAT), driving accumulation of GSH in shoot tissue, is strongly correlated with the ability to hyperaccumulate Ni in a diverse group of Thlaspi species in the family Brassicaceae, including Thlaspi goesingense (5). In addition, overexpression of sat in the nonaccumulating model species Arabidopsis thaliana causes increased GSH accumulation, which drives enhanced resistance to Ni (5).
Here, we report on a similar finding, that overexpression of T. goesingense sat in E. coli enhanced resistance to both Ni and Co and paradoxically increased sensitivity to Cd. Overexpression of sat, the bottleneck in sulfur assimilation (8), would be expected to enhance both sulfur assimilation and GSH biosynthesis through the action of O-acetyl-l-serine (OAS), the product of SAT, which acts as a positive regulator of the cysteine regulon in E. coli (12). Enhanced biosynthesis of GSH has previously been observed in plants overexpressing sat (2, 5, 8, 25). Interestingly, the recent overexpression of both an E. coli SAT (cysE) and a γ-glutamylcysteine synthetase (gshA) gene resulted in a greater accumulation of γ-glutamylcysteine. These two genes, in conjunction with a phytochelatin synthase from Schizosaccharomyces pombe, resulted in overproduction of phytochelatins in E. coli and increased Cd accumulation, but not Cd tolerance (24).
The biosynthesis of cysteine from sulfate is the major mechanism through which reduced sulfur is incorporated into organic compounds in E. coli (12). During this process, inorganic sulfate is reduced to sulfite and sulfide. To achieve this, sulfate is first activated to 5′-adenylylsulfate (sometimes referred to as 5′-adenosine-phosphosulfate [APS]). This activation is catalyzed by ATP sulfurylase using ATP. A second enzyme, APS kinase, phosphorylates 5′-adenylylsulfate, with the consumption of ATP, to yield phosphoadenosine-phosphosulfate (PAPS). PAPS is reduced by PAPS reductase to yield sulfite. Sulfite is further reduced to sulfide by the action of NADPH-sulfite reductase. Sulfide is incorporated into Cys through the action of O-acetyl-l-serine thiol lyase on OAS and sulfide. SAT catalyzes the production of OAS required for the biosynthesis of Cys. SAT is directly feedback inhibited by Cys, which provides kinetic regulation of this branch in the pathway (12). Additionally, mutant E. coli strains lacking SAT are defective in expression of the enzymes of sulfate assimilation. OAS is the precursor of N-acetyl-l-serine, and N-acetyl-l-serine acts via the transcriptional activator CysB to positively regulate the Cys regulon in E. coli (12).
To understand the mechanism underlying the enhanced Ni and Co resistance observed in E. coli overexpressing sat, we investigated which biochemical steps in the sulfur assimilation pathway are required for Ni resistance. We also performed physiological and biochemical experiments to determine if the chemical form of Ni in the sat-overexpressing strains is altered to affect the observed increase in Ni resistance.
MATERIALS AND METHODS
Bacterial strains.
For general cloning and experiments requiring a “wild type,” TOP10F′ [F′ (Tetr) mcrA Δ(mrr-hsdRMS-mcrBC) phi 80 Δlac ΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara leu)7697 galU galK λ− rpSL (Strr) endA1 nupG] was used, and BM25.8 {[F′ traD36 lacIq lacZ ΔM15 proA+B+] supE thi Δ(lac-proAB) rrnBp1 Cmr hsdR (rK12− mK12+) (λimm434 Kanr)} was used for generation of phagemids. The following strains were kindly provided by the Yale E. coli Genetic Stock Center: CB64 (cysB trp tfr), JM81A (cysC tfr), JM221 (pro lac tsx gal trp his cysD argA rpsL mal xyl ilvA mt1), JM15 (cysE tfr), JM96 (thr leuB fhuA lacY glnV gal LAM trp hisG cysH rpsL malT1 xylA mt1A argH thi), N3002 (cysI), KL267 (LAM cysJ spoT thi deoB), RL165 (thr leuB fhuA lacY glnV gal LAM trp hisG cysK rpsL malT xy1A mt1A argH thi), and strain 821 (thr araC leuB lacY tsx qsr glnV galK rac hisG rfbD mgl gshA rpsL kdgK xylA mtl argE thi).
Expression library screening.
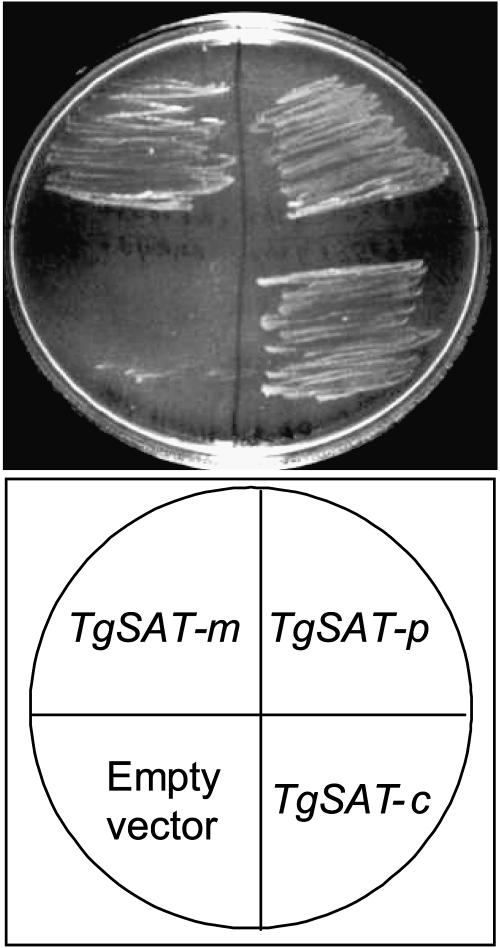
Preparation of the T. goesingense λTriplEx (CLONETECH, Palo Alto, CA) cDNA expression library used in this study was reported by Persans et al. (16). All T. goesingense cDNAs were cloned into the EcoRI and XhoI sites within the λTriplEx multiple cloning site and converted to the phagemid pTriplEx using a Cre-recombinase-expressing E. coli strain (BM25.8). For screening TOP10F′, E. coli was transformed with the T. goesingense pTriplEx cDNA library by electroporation (model EC100 electroporator; EC Apparatus, St. Petersburg, FL) according to the manufacturer's protocol. Transformed cells were selected on Ni selection plates composed of Luria-Bertani (LB) medium containing ampicillin, tetracycline, isopropylthiogalactoside (IPTG), and 2.5 mM Ni-acetate, at 30°C for 40 h. Of the 2 × 106 transformants screened, 2,700 were able to grow on the Ni resistance selection plates. Of the Ni resistance clones observed, 92 were randomly picked and placed into three groups based on fragment banding patterns after restriction digestion with SalI/XbaI, EcoRV/HindIII, and KpnI I/SacI. From these three groups, 10 representative cDNAs were chosen and sequenced. Through sequence homology, these cDNAs were determined to encode T. goesingense SAT, including the predicted mitochondrial (SAT-m), plastid (SAT-p), and cytosolic (SAT-c) isoforms. The identities of these T. goesingense sat cDNAs were confirmed by functional complementation. All three sat cDNAs were expressed in the SAT-minus cysE mutant E. coli plated on yeast nitrogen base synthetic complete (YNB SC) minimal medium minus Cys and Met and containing ampicillin (200 μg/ml) and IPTG (1 mM). Only strains expressing the sat cDNAs were able to grow in the absence of Cys and Met.
Assessment of nickel resistance in E. coli.
For assessment of Ni resistance on solidified medium, the genes encoding all three T. goesingense sat isoforms were expressed in TOP10F′ E. coli from the pTriplEx vector, along with an empty-vector control. Ni resistance was measured in liquid LB medium containing ampicillin (200 μg/ml) and IPTG (1 mM), with or without Ni acetate (2 mM), at 30°C in a shaking incubator for 35 h, and growth was monitored as optical density at 600 nm (OD600). Nickel resistance was also measured semiquantitatively using a modified antimicrobial disk assay. In this assay, a liquid starter culture containing various E. coli strains was initiated in LB medium containing ampicillin (200 μg/ml) and IPTG (1 mM) and subcultured at an OD600 of 0.6 three consecutive times. After a final OD600 of 0.6 was achieved, 200 μl of culture was subcultured into 45 ml of molten (50°C) LB medium containing ampicillin (200 μg/ml), IPTG (1 mM), and 1.5% ultrapure plant tissue grade agar in 50-ml tubes, vortexed, and poured into petri dishes (150 by 50 mm). After solidification, six sterile blank antimicrobial filter paper test disks were placed equidistant in a circle on the surfaces of the plates. All metals were tested separately, using increasing volumes of 400 mM Ni, Co, and Zn and 4 mM Cd acetate stock solutions, which were spotted directly onto the blank disks before the plates were incubated for 24 h at 30°C. After incubation, the diameters of the circular zones of growth inhibition were recorded. Each series was replicated three times. To determine the sulfate requirement for SAT-mediated Ni resistance, sulfate-free YNB SC minimal medium containing ampicillin (200 μg/ml) and IPTG (1 mM) with the addition of various concentrations of sulfate was used.
Modification of the chemical activity of nickel.
To test for release of sulfite into the growth medium by sat-overexpressing strains, YNB SC medium minus Cys and Met and containing ampicillin (200 μg/ml) and IPTG (1 mM) was first inoculated with a cysH mutant E. coli transformed with empty pTriplEx. Plasmids carrying the genes encoding the three sat isoforms and empty pTriplEx vector control were transformed into E. coli TOP10F′, and the strains were streaked onto separate plates previously inoculated with cysH and incubated at 30°C for 24 h. Sulfite was streaked on the plates as a positive control. Growth of cysH was used as an indication that sulfite was present. To test for sulfide release, strains transformed with sat or empty pTriplEx were grown at 30°C in liquid LB medium containing ampicillin (200 μg/ml) and IPTG (1 mM). At an OD600 of 1.0, 25 μl of these cultures was spotted onto solidified LB medium containing ampicillin (200 μg/ml), IPTG (1 mM), and 1% (wt/vol) bismuth ammonium citrate (Sigma catalog no. 09678). Sodium sulfide at a concentration of 10 μM was spotted as a positive control, and the plates were incubated at 30°C for 24 h. Formation of a dark-brown precipitate indicated the presence of sulfide. To test for cysteine release from sat-overexpressing strains, YNB SC medium minus Cys and Met and containing ampicillin (200 μg/ml) and IPTG (1 mM) was first inoculated with cysE transformed with empty pTriplEx. The different sat cDNAs were transformed into TOP10F′, streaked onto plates previously inoculated with cysE, and incubated at 30°C for 24 h. Cysteine was streaked as a positive control. Growth of cysE was used to indicate the presence of cysteine.
To test for the general chemical inactivation of Ni in the growth media by sat-overexpressing strains, a 500-ml culture of TOP10F′ transformed with sat in LB medium containing ampicillin (200 μg/ml), IPTG (1 mM), and Ni acetate (2 mM) was grown to an OD600 of 1.5 at 30°C with shaking for 24 h. The cells were removed by centrifugation at 14,000 × g for 10 min, and the medium was resterilized by filtration through a 0.2-μm filter. This sterilized, previously used medium was reinoculated with an actively growing culture of TOP10F′ transformed with empty pTriplEx or sat and incubated with shaking at 30°C for 24 h. Cell growth was monitored by OD600.
Chemical speciation of Ni in cells using near-edge X-ray absorption spectroscopy.
E. coli TOP10F′ transformed with sat-m was grown in LB medium containing ampicillin (200 μg/ml), IPTG (1 mM), and Ni acetate (2 mM) with shaking for 24 h at 30°C before being cooled rapidly on ice, and the cells were collected by centrifugation at 12,000 × g for 5 min at 4°C. The cells were washed in NaCl (150 mM) and recentrifuged at 12,000 × g for 5 min at 4°C, after which the cells were flash frozen in liquid N2 and shipped on dry ice to the Stanford Synchrotron Radiation Laboratory for analysis by Ni K-edge X-ray absorption spectroscopy (XAS), following our previously described methods (17). Briefly, samples were compacted into Lucite sample holders with Mylar windows and cooled in liquid nitrogen. During data collection, samples were held at 15 K using a liquid helium cryostat. Ni K-edge spectra were measured on beam line 7-3 using an Si(220) double-crystal monochromator, a 1-mm upstream vertical aperture, and no focusing optics, and the absorption spectrum was collected in fluorescence using a 13-element germanium detector. XAS data reduction was performed using the EXAFSPAK suite of programs, and the EXAFS phase and amplitudes were calculated using the program feff7 (20).
RESULTS
Metal resistance conferred by expression of T. goesingense sat in E. coli.
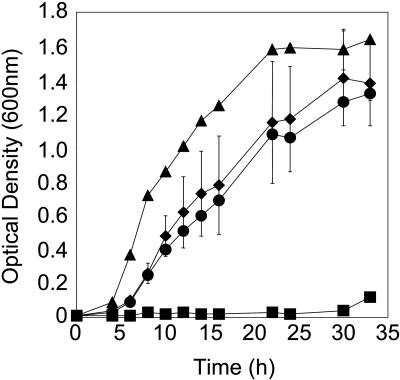
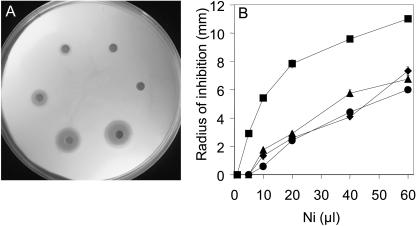
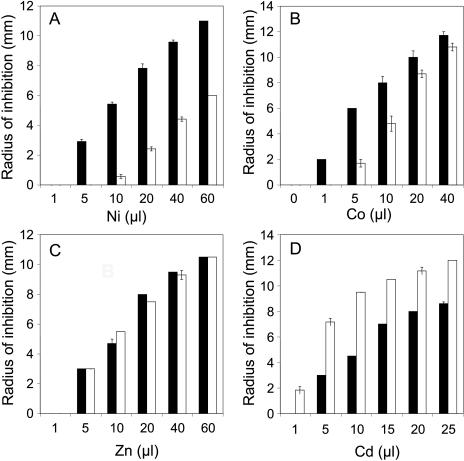
To identify cDNAs involved in Ni resistance, we screened a T. goesingense cDNA expression library for cDNAs that confer enhanced Ni resistance in E. coli. Such a screen identified three full-length sat cDNAs (GenBank accession numbers AY618468, AY618469, and AY618470). All cDNAs were confirmed to encode functional SAT proteins by suppression of the Cys and Met requirements of the cysE auxotroph (Fig. 1). sat cDNAs were confirmed to confer resistance to Ni when overexpressed in E. coli in liquid medium (Fig. 2). All three SAT isoforms conferred similar levels of Ni resistance when measured using the semiquantitative disk assay (Fig. 3). Using this assay system, sat overexpression was also confirmed to confer enhanced resistance to Co (Fig. 4), though it has no significant effect on Zn resistance (Fig. 4). Furthermore, sat overexpression was found to increase the sensitivity of E. coli to Cd (Fig. 4).
FIG. 1.
Thlaspi goesingense sat cDNAs complement the Cys or Met requirement of the cysH E. coli strain. E. coli transformed with pTriplEx containing sat-m, sat-p, or sat-c (TgSAT-m, -p, and -c, respectively) or empty vector was incubated at 30°C on solidified minimal medium containing ampicillin and IPTG and lacking Cys and Met.
FIG. 2.
Thlaspi goesingense sat-m cDNA confers enhanced Ni resistance when expressed in E. coli growing in liquid medium. E. coli transformed with empty pTriplEx (triangles and squares) or pTriplEx containing sat-m (diamonds and circles) was cultured in liquid LB medium containing ampicillin and IPTG, with (squares and circles) or without (triangles and diamonds) 2 mM Ni acetate, at 30°C. The data represent averages of independent cultures (n = 3) ± standard deviations.
FIG. 3.
Thlaspi goesingense sat cDNAs confer enhanced Ni resistance when expressed in E. coli. E. coli transformed with pTriplEx containing sat-m, sat-p, or sat-c or empty vector was incorporated into LB medium containing ampicillin and IPTG. Filter paper disks placed on the solidified medium were soaked with various volumes of Ni acetate (400 mM stock solution), and the zone of growth inhibition was measured after incubation at 30°C for 24 h (A). The radius of inhibition was plotted against the volume of Ni applied to each of the filter paper disks for E. coli transformed with empty pTriplEx (squares) and pTriplEx containing sat-m (circles), sat-p (diamonds), or sat-c (triangles) (B). The data presented represent the averages (n = 3).
FIG. 4.
Effects of expression of T. goesingense sat-m on resistance to various metal ions in E. coli. E. coli was transformed with either empty pTriplEx (black bars) or pTriplEx containing sat-m (white bars). These strains were incorporated into LB medium containing ampicillin and IPTG. Filter paper disks placed on the solidified medium were soaked with various volumes of Ni acetate (A), Co acetate (B), Zn acetate (C) from 400 mM stock solutions, or Cd acetate from a 4 mM stock solution (D), and the zone of growth inhibition was measured after incubation at 30°C for 24 h. The data represent averages of independent cultures (n = 3) ± standard deviations.
Role of chemical speciation in SAT-mediated Ni resistance.
SAT is involved in sulfate assimilation and cysteine biosynthesis, and overexpression of this enzyme would be expected to lead to increases in sulfate assimilation intermediates, including sulfite, sulfide, and cysteine (7). Accumulation of these intermediates in the medium or inside cells could possibly reduce the solution activity of Ni2+, reducing its potential toxicity. Such a mechanism has been observed in yeasts that release sulfite (1). To identify release of sulfite into the medium, we used a simple bioassay. The auxotrophic cysH mutant has an absolute growth requirement for Cys or Met, and this can be suppressed by the addition of sulfite into the growth medium. E. coli TOP10F′ overexpressing sat was streaked on minimal medium (minus Cys and Met) that had previously been inoculated with cysH E. coli prior to being poured. Sodium sulfite (10 μM) solution was also streaked onto these plates as a positive control that could clearly suppress the cysH growth defect, resulting in a strong growth halo. However, E. coli TOP10F′ overexpressing sat or the empty-vector control produced no clear cysH growth. This confirmed that sulfite was not being released in significant amounts by the sat-overexpressing E. coli and thus could not account for the observed Ni resistance.
In order to test for sulfide release into the medium, we used a chemical indicator of sulfide production, bismuth ammonium citrate (10). When bismuth ammonium citrate is complexed with sulfide, it forms a dark-brown precipitate. In order to test for the production of sulfide, we grew E. coli strains overexpressing sat and empty vector in YNB SC medium to an OD600 of 1 before spotting cultures onto filter disks placed on sulfide indicator plates containing bismuth ammonium citrate. A slight brown precipitate was observed; however, no differences between the empty-vector control and the sat-overexpressing strains was apparent. As a positive control, sodium sulfide (10 μM) solution was spotted onto the plates, resulting in a large dark-brown zone. We conclude that sulfide is not released into the medium by the sat-overexpressing E. coli in significant enough amounts to be responsible for the observed SAT-mediated Ni resistance.
To detect if Cys release into the medium plays a role in sat-mediated Ni resistance, we utilized the inability of the cysE E. coli mutant to grow in the absence of Cys. We streaked E. coli TOP10F′ overexpressing sat and empty-vector control onto plates lacking Cys and Met, which had previously been inoculated with cysE. Growth of TOP10F′ cells overexpressing sat did not suppress the Cys or Met requirement of cysE, whereas application of a Cys (10 μM) solution to the plates produced a clear cysE growth halo within 24 h. From this, we conclude that Cys is not released by sat-overexpressing strains in significant enough amounts to account for sat-mediated Ni resistance.
Our experiments suggest that Ni in the sat-overexpressing strains is not being precipitated or coordinated by sulfite, sulfide, or cysteine in the medium. However, the possibility exists that the solution activity of Ni2+ is modified by an unknown ligand produced by the sat-overexpressing cells. To test this, we grew a 500-ml culture of sat-overexpressing E. coli in LB medium containing Ni acetate (2 mM) and IPTG (1 mM) for 24 h. The cells were removed, and the clarified medium was resterilized. This medium was reinoculated with cultures of E. coli overexpressing sat or empty vector, and growth was monitored for 24 h. Both cultures grew in a way similar to that observed with fresh medium (data not shown), with the sat-overexpressing strains showing strong growth in the presence of Ni compared to the empty-vector control. From this, we conclude that sat-mediated resistance is not achieved by detoxification of Ni2+ in the growth medium but rather is an intracellular resistance phenomenon.
Using Ni K-edge XAS, we investigated the cellular speciation of Ni, using methods previously applied to Ni speciation in the Ni hyperaccumulator plant T. goesingense (11). By fitting the near-edge X-ray absorption spectra of various Ni complexes, we estimate that Ni in sat-overexpressing E. coli is coordinated mainly by histidine-like ligands (70% of total Ni), with the remaining Ni being modeled by equal parts Ni(S)4 and hydrated Ni2+. This evidence supports our conclusion that Ni in sat-overexpressing cells is not detoxified by coordination or precipitation with S.
Role of sulfur assimilation in SAT-mediated Ni resistance.
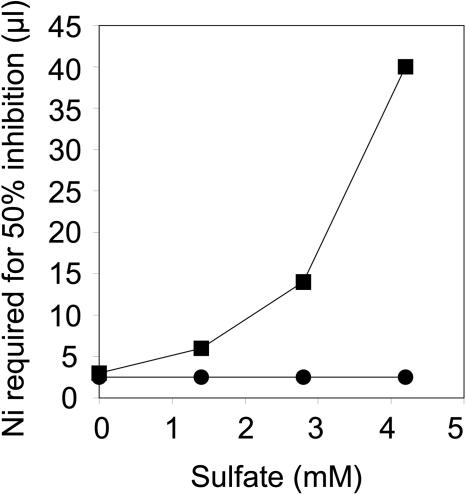
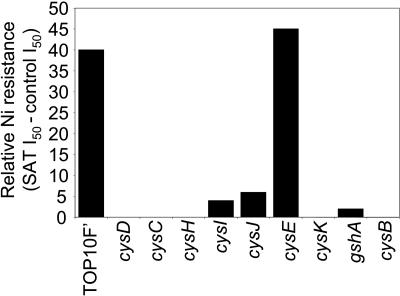
Though Ni is not directly coordinated with S, sat clearly plays a major role in S assimilation. Therefore, to investigate the involvement of reductive S assimilation in the mechanism of sat-mediated Ni resistance, we cultured E. coli strains transformed with either sat or the empty-vector control on YNB SC media with various concentrations of sulfate. Such experiments revealed that sat-mediated Ni resistance requires sulfate, with sat-mediated Ni resistance increasing with increasing sulfate concentrations (Fig. 5). To further understand the role of S assimilation in sat-mediated Ni resistance, the ability of sat to confer Ni resistance in strains lacking enzymes in the S assimilation pathway was investigated. The E. coli CysB transcription factor is required to positively regulate S assimilation and Cys biosynthesis in response to elevated OAS concentrations (12). sat overexpression in the cysB mutant was found not to confer enhanced Ni resistance when it was quantified using the disk assay system (Fig. 6), suggesting that SAT confers Ni resistance in E. coli via elevating OAS concentrations, which in turn enhance the rate of S assimilation and Cys biosynthesis. To confirm this, sat was overexpressed in E. coli lacking either ATP sulfurylase (cysD), APS kinase (cysC), PAPS reductase (cysH), or sulfite reductase (cysI and cysJ), enzymes required for sulfate reduction, and OAS thiol lyase (cysK), required for Cys biosynthesis from sulfide and OAS. Overexpression of sat in all of these mutant strains was found not to confer Ni resistance (Fig. 6). This genetic evidence confirms that sat overexpression acts to enhance Ni resistance via constitutive activation of S assimilation and Cys biosynthetic pathways. Furthermore, overexpression of sat in an E. coli gshA mutant lacking γ-glutamylcysteine synthetase and unable to synthesize GSH did not confer enhanced Ni resistance (Fig. 6). Such evidence supports the conclusion that sat-mediated Ni resistance is dependent on the biosynthesis of GSH.
FIG. 5.
Effect of sulfate on sat-mediated Ni resistance in E. coli. E. coli was transformed with either empty pTriplEx (circles) or pTriplEx containing sat-m (squares). The transformed strains were incorporated into solidified minimal medium containing ampicillin and IPTG with increasing concentrations of sulfate. Six filter paper disks placed on the solidified medium were soaked with different volumes of Ni acetate (1, 5, 10, 20, 40, and 60 μl from a 400 mM stock solution), and the zone of growth inhibition for each of the strains was measured after incubation at 30°C for 24 h. Replicate plates for each sulfate concentration (n = 3) were cultured for each strain, and the average volume of Ni acetate required to cause 50% of the maximal inhibition of the controls was calculated and plotted for each sulfate treatment.
FIG. 6.
Requirement for S assimilation and Cys and GSH biosynthesis of sat-mediated Ni resistance in E. coli. E. coli strains with defects in various genes involved in S assimilation and Cys and GSH biosynthesis were transformed with either empty pTriplEx or pTriplEx containing sat-m. The transformed strains were incorporated into solidified LB medium containing ampicillin and IPTG. Six filter paper disks placed on the solidified medium were soaked with different volumes of Ni acetate (1, 5, 10, 20, 40, and 60 μl from a 400 mM stock solution), and the zone of growth inhibition for each of the strains was measured after incubation at 30°C for 24 h. Replicate plates for each strain (n = 3) were cultured, and the average volume of Ni acetate required to cause 50% of the maximal inhibition of the empty-vector control in the same E. coli background was calculated (I50). The difference in I50 between the strains transformed with either empty vector or sat-m represents the level of sat-m-mediated Ni resistance in a given E. coli background.
Taken together, these results confirm that enhanced S assimilation and Cys and GSH biosynthesis, driven by overexpression of sat, are required for the enhanced Ni resistance observed in sat-overexpressing E. coli. The E. coli strains with mutations in the metE, metH, and metB genes, which represent mutations in the early enzymes required for methionine biosynthesis, the only other branch point in S assimilation, were not required for sat-mediated Ni resistance (data not shown). This supports the conclusion that to be Ni resistant, sat-overexpressing E. coli needs to make OAS, Cys, and GSH, but not methionine.
DISCUSSION
It is important to consider why the increases in S assimilation, caused by SAT overexpression, lead to both Ni and Co resistance, no resistance to Zn, and sensitivity to Cd (Fig. 4). Genetic evidence has established that SAT-mediated Ni resistance requires constitutive activation of S assimilation into Cys, via CysB activation and up regulation of the Cys regulon, and GSH biosynthesis (Fig. 6). The involvement of S assimilation in SAT-mediated Ni resistance is also supported by evidence demonstrating that Ni resistance requires sulfate (Fig. 5).
Reduced S has the ability to form metal ion-ligand complexes in the order Cd > Co > Ni > Zn, based on the classification of Nieboer and Richardson (14), and Cd, Co, Ni, and Zn sulfides and sulfites are all insoluble or partially insoluble in water. However, sat overexpression was found not to confer resistance to either Zn or Cd, suggesting that the mechanism of Ni resistance does not involve direct binding or precipitation with S-containing compounds. This is supported by both the XAS evidence establishing that intracellular Ni is mainly coordinated with N and O ligands and data showing no enhanced release of sulfite, sulfide, or Cys into the growth medium. Experiments also confirmed that Ni2+ in the growth medium is not being detoxified by release of an unknown Ni chelate, after regrowth experiments demonstrated that growth of sat-overexpressing strains does not detoxify Ni2+ in the growth medium.
Taken together, our data support the conclusion that in E. coli, overexpression of sat drives the unregulated overaccumulation of OAS. Such accumulation of this positive regulator activates S assimilation, increasing Cys and GSH production, as observed previously in both plants (2, 5, 8, 24) and E. coli (24). Ni and Co are thought to deplete cellular GSH levels by forming a complex with GSH that promotes its oxidation to oxidized glutathione (22), thus causing increased accumulation of reactive oxygen species. The limited Ni-S coordination observed, using K near-edge X-ray absorption spectroscopy, in sat-overexpressing E. coli is consistent with such a model. Elevated GSH levels, driven by sat overexpression, would then confer enhanced resistance to this Ni- and Co-induced oxidative stress. Furthermore, overexpression of the cytoplasmic, mitochondrial, or chloroplastic sat from T. goesingense (Fig. 3) or the chloroplastic sat from Arabidopsis thaliana (data not shown) in E. coli all conferred equivalent Ni resistance. From this, we conclude that it is the enhanced SAT activity driving increased OAS biosynthesis that is important for metal resistance in E. coli, which in our experiments was achieved by driving gene expression from a strong promoter system. Unique amino acid sequences in the different sat isoforms appear not to be required to achieve enhanced Ni resistance in E. coli.
Interestingly, the proposed mechanism of SAT-mediated Ni and Co resistance in E. coli does not increase either Cd or Zn resistance (Fig. 4). In fact, paradoxically, sat overexpression increases the sensitivity of E. coli to Cd. A possible explanation for this enhanced Cd sensitivity could be that the Cd-GSH complex formed in the sat-overexpressing strain is more toxic than Cd2+, perhaps because intracellular Cd-GSH complexes are not easily effluxed out of cells.
In higher plants, eukaryotic algae, fungi, and nematodes, phytochelatins are produced on exposure to various elements, including Cd and As (18). Phytochelatins are thiol-rich peptides that are enzymatically synthesized from GSH by the action of phytochelatin synthase (18). In plants, it is known that phytochelatin overproduction paradoxically also leads to Cd sensitivity (13). Lee and coauthors concluded that this increased Cd sensitivity could be due to increased oxidative stress resulting from the loss of GSH through its incorporation into phytochelatins. In contrast, neither phytochelatins nor phytochelatin synthase genes have been found in prokaryotes, except in the cyanobacterium Nostoc sp. strain PCC 7120 (23). Recently, E. coli overexpressing the A. thaliana phytochelatin synthase gene showed a marked accumulation of phytochelatins, together with a decrease in cellular GSH levels, and this was correlated with 20- and 50-fold increases in Cd and As accumulation relative to control strains (21). Surprisingly, expression of the A. thaliana phytochelatin synthase gene in E. coli had no significant effect on Cd or As resistance (21). At present, the interaction between GSH, increased Cd-thiol coordination capacity, and Cd resistance is not clear. However, we can conclude that simply increasing the thiol binding capacity in E. coli does not lead to significant increases in Cd resistance.
Acknowledgments
We extend our appreciation to Ingrid Pickering for X-ray absorption spectroscopy data collection and analysis. D.E.S. conceived the experiment with contributions from M.W.P. and J.L.F. J.L.F. was primarily responsible for carrying it out, with M.W.P. and K.N. contributing to the cloning of SAT from T. goesingense. D.E.S. and J.L.F. cowrote the paper.
This work was supported by grants to D.E.S. from the U.S. National Science Foundation (0196310-IBN and 0129747-IBN).
REFERENCES
- 1.Babich, H., and G. Stotzky. 1983. Further studies on environmental factors that modify the toxicity of nickel to microbes. Regul. Toxicol. Pharmacol. 3:82-99. [DOI] [PubMed] [Google Scholar]
- 2.Błaszczyk, A., R. Brodzik, and A. Sirko. 1999. Increased resistance to oxidative stress in transgenic tobacco plants overexpressing bacterial serine acetyltransferase. Plant J. 20:237-243. [DOI] [PubMed] [Google Scholar]
- 3.Brooks, R. R., J. Lee, R. D. Reeves, and T. Jaffré. 1977. Detection of nickeliferous rocks by analysis of herbarium specimens of indicator plants. J. Geochem. Explor. 7:49-77. [Google Scholar]
- 4.Clemens, S. 2001. Molecular mechanisms of plant metal tolerance and homeostasis. Planta 212:475-486. [DOI] [PubMed] [Google Scholar]
- 5.Freeman, J. L., M. W. Persans, K. Nieman, C. Albrecht, W. Peer, I. J. Pickering, and D. E. Salt. 2004. Increased glutathione biosynthesis plays a role in nickel tolerance in Thlaspi nickel hyperaccumulators. Plant Cell. 16:2176-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geslin, C., J. Llanos, D. Prieur, and C. Jeanthon. 2001. The manganese and iron superoxide dismutases protect Escherichia coli from heavy metal toxicity. Res. Microbiol. 152:901-905. [DOI] [PubMed] [Google Scholar]
- 7.Hell, R. 1997. Molecular physiology of plant sulfur metabolism. Planta 202:138-148. [DOI] [PubMed] [Google Scholar]
- 8.Hesse, H., K. Harms, P. Von Ballmoos, C. Brunold, L. Willmitzer, and R. Höfgen. 2000. Serine acetyltransferase: a bottleneck for cysteine and glutathione synthesis, p. 321-323. In C. Brunold, H. Rennenberg, L. J. De Kok, I. Stulen, and J. C. Davidian (ed.), Sulfur nutrition and sulfur assimilation in higher plants. Paul Haupt, Bern, Switzerland.
- 9.Hultberg, B., A. Andersson, and A. Isaksson. 1997. Copper ions differ from other thiol reactive metal ions in their effects on the concentration and redox status of thiols in HeLa cell cultures. Toxicology 117:89-97. [DOI] [PubMed] [Google Scholar]
- 10.Jiravek, V., P. Langridge, and P. Henschke. 1995. Validation of bismuth containing indicator media for predicting H2S-producing potential of Saccharomyces cerevisiae wine yeasts under ecological conditions. Am. J. Enol. Vitic. 46:269-273. [Google Scholar]
- 11.Kramer, U., I. J. Pickering, R. C. Prince, I. Raskin, and D. E. Salt. 2000. Subcellular localization and speciation of nickel in hyperaccumulator and non-accumulator Thlaspi species. Plant Physiol. 122:1343-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kredich, N. 1996. Biosynthesis of cysteine, p. 514-527. In F. C. Neidhardt, et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 13.Lee, S., J. S. Moon, T. S. Ko, D. Pet, P. B. Goldsbrough, and S. S. Korban. 2003. Overexpression of Arabidopsis phytochelatin synthase paradoxically leads to hypersensitivity to cadmium stress. Plant Physiol. 131:656-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nieboer, E., and D. H. S. Richardson. 1980. The replacement of the nondescriptive term heavy metals by a biologically and chemically significant classification of metal ions. Environ. Pollut. Ser. B. 1:3-26. [Google Scholar]
- 15.Nies, D. H. 1999. Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 51:730-750. [DOI] [PubMed] [Google Scholar]
- 16.Persans, M. W., X. Yan, J. M. Patnoe, U. Kramer, and D. E. Salt. 1999. Molecular dissection of the role of histidine in nickel hyperaccumulation in Thlaspi goesingense (Halacsy). Plant Physiol. 121:1117-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pickering, I. J., R. C. Prince, G. N. George, W. E. Rauser, W. A. Wickramasinghe, A. A. Watson, C. T. Dameron, I. G. Dance, D. P. Fairlie, and D. E. Salt. 1999. X-ray absorption spectroscopy of cadmium phytochelatin and model systems. Biochim. Biophys. Acta 1429:351-364. [DOI] [PubMed] [Google Scholar]
- 18.Rea, P. A., O. K. Vatamaniuk, and D. J. Rigden. 2004. Weeds, worms, and more. apain's long-lost cousin, phytochelatin synthase. Plant Physiol. 136:2463-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reeves, R. D., and A. J. M. Baker. 2000. Phytoremediation of toxic metals, p. 193-229. In I. Raskin and B. D. Ensley (ed.), Using plants to clean up the environment. John Wiley and Sons, Inc., New York, N.Y.
- 20.Rehr, J. J., J. Mustre de Leon, S. I. Zabinsky, and R. C. Albers. 1991. Theoretical X-ray absorption fine structure standards. J. Am. Chem. Soc. 113:5135-5140. [Google Scholar]
- 21.Sauge-Merle, S., S. Cuine, P. Carrier, C. Lecomte-Pradines, D. T. Luu, and G. Peltier. 2003. Enhanced toxic metal accumulation in engineered bacterial cells expressing Arabidopsis thaliana phytochelatin synthase. Appl. Environ Microbiol. 69:490-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stohs, S., J., and D. Bagchi. 1995. Oxidative mechanisms in the toxicity of metal ions. Free Radic. Biol. Med. 18:321-336. [DOI] [PubMed] [Google Scholar]
- 23.Tsuji, N., S. Nishikori, O. Iwabe, K. Shiraki, H. Miyasaka, M. Takagi, K. Hirata, and K. Miyamoto. 2004. Characterization of phytochelatin synthase-like protein encoded by alr0975 from a prokaryote, Nostoc sp. PCC 7120. Biochem. Biophys. Res. Commun. 315:751-755. [DOI] [PubMed] [Google Scholar]
- 24.Wawrzynska, A., D. Gaganidze, E. Kopera, K. Piatek, W. Bal, and A. Sirko. 2005. Overexpression of genes involved in phytochelatin biosynthesis in Escherichia coli: effects on growth, cadmium accumulation and thiol level. Acta Biochim. Pol. 52:109-116. [PubMed] [Google Scholar]
- 25.Wirtz, M., and R. Hell. 2002. Production of cysteine for bacterial and plant biotechnology. Application of cysteine feedback-insensitive isoforms of serine acetyltransferase. Amino Acids 24:195-203. [DOI] [PubMed] [Google Scholar]