Abstract
Genomic analysis has revealed heterogeneity among bacterial 16S rRNA gene sequences within a single species; yet the cause(s) remains uncertain. Generalized transducing bacteriophages have recently gained recognition for their abundance as well as their ability to affect lateral gene transfer and to harbor bacterial 16S rRNA gene sequences. Here, we demonstrate the ability of broad-host-range, generalized transducing phages to acquire 16S rRNA genes and gene sequences. Using PCR and primers specific to conserved regions of the 16S rRNA gene, we have found that generalized transducing phages (D3112, UT1, and SN-T), but not specialized transducing phages (D3), acquired entire bacterial 16S rRNA genes. Furthermore, we show that the broad-host-range, generalized transducing phage SN-T is capable of acquiring the 16S rRNA gene from two different genera: Sphaerotilus natans, the host from which SN-T was originally isolated, and Pseudomonas aeruginosa. In sequential infections, SN-T harbored only 16S rRNA gene sequences of the final host as determined by restriction fragment length polymorphism analysis. The frequency of 16S rRNA gene sequences in SN-T populations was determined to be 1 × 10−9 transductants/PFU. Our findings further implicate transduction in the horizontal transfer of 16S rRNA genes between different species or genera of bacteria.
16S rRNA gene sequences are used extensively to construct phylogenetic trees as well as in the study of microbial communities (6). These uses of the 16S rRNA gene have been based upon three assumptions. First, most bacteria contain either a single 16S rRNA gene or multiple genes that are highly similar. Second, there has been no horizontal gene transfer (HGT) of these genes. Third, 16S rRNA genes are unique to prokaryotes and mitochondria. Comparative nucleotide sequence analysis has shown the first assumption to be untrue. This was first reported as intraspecific variation in small-subunit rRNA (3). It is now recognized that many bacteria possess more than one and up to 15 copies of the 16S rRNA gene (1, 9). Comparisons of 16S rRNA gene operons from strains belonging to the same species have shown nucleotide differences ranging from a single nucleotide difference in Pseudomonas aeruginosa to 188 nucleotide differences in Thermoanaerobacter tengcongensis (1). The causes of 16S rRNA gene heterogeneity are suggested to be twofold: (i) misincorporation of nucleotides into DNA during replication and (ii) horizontal gene transfer (15). HGT has been implicated in the transfer of an entire rRNA gene in Thermomonospora chromogena (18). Although HGT of 16S rRNA gene sequences is accepted as a probable cause of 16S rRNA gene heterogeneity, it is believed to occur at a low frequency. According to the complexity hypothesis, because 16S rRNA genes interact with several other gene products, they have a low probability of successfully undergoing a transfer event (7, 11). It has also been suggested that even if 16S rRNA genes had a high frequency of HGT, processes that normalize genes could mask this (1). Concerning the third assumption, bacteria are not the only organisms in the environment harboring 16S rRNA gene sequences; phages in soil have also been shown to contain 16S rRNA gene sequences that are assumed to be of bacterial origin (12). The presence of 16S rRNA gene sequences in phages would likely add to the overestimation of bacterial cells in microbial communities.
Broad-host-range, generalized transducing phages are a possible vector for the transfer of genes between strains, species, and even genera. In the past, it was thought that phages rarely, if at all, crossed species boundaries. However, it has recently been shown that cyanophages are capable of infecting across species (13). Because of their prevalence, it has been suggested that phages may play an important role in gene transfer in the environment. For example, in one study, 9 of 10 phages isolated from sewage were found to display a broad host range (8). One such broad-host-range phage is SN-T, a phage with a hexagonal head and a long flexible tail. SN-T was isolated from sewage and enriched on Sphaerotilus natans (16). In addition to S. natans, SN-T is capable of infecting Rhodospirillum rubrum, Shigella flexneri, Proteus vulgaris, P. aeruginosa, and Escherichia coli (8). While all the bacterial hosts of SN-T belong to the phylum Proteobacteria, they differ in classes, which include Gammaproteobacteria (S. flexneri, P. vulgaris, E. coli, and P. aeruginosa), Alphaproteobacteria (R. rubrum), and Betaproteobacteria (S. natans). Here, we report that the broad-host-range, generalized transducing phage SN-T can acquire 16S rRNA gene sequences from P. aeruginosa and S. natans. We have also determined the frequency of 16S rRNA gene acquisition by this phage.
MATERIALS AND METHODS
Bacterial strains and bacteriophages.
To detect the presence of 16S rRNA gene sequences in phages, Pseudomonas aeruginosa PAO1 and phages known to infect this bacterium (D3112, UT1, SN-T, and D3) were used (Table 1). P. aeruginosa was grown in LB medium or on LB 1.5% agar plates. The broad-host-range, generalized transducing phage SN-T was chosen and used in all sequential infection experiments because it is capable of infecting both P. aeruginosa and Sphaerotilus natans ATCC 133388 (Table 1). S. natans was cultivated on tryptone-yeast (TY) 1.5% agar plates.
TABLE 1.
Bacterial strains and phages
| Organism | Bacterial characteristic
|
Phage characteristic
|
|||
|---|---|---|---|---|---|
| Strain | Class | Type of transduction | Host(s) | Source | |
| P. aeruginosa | PAO1 | Gammaproteobacteria | |||
| S. natans | ATCC 133388a | Betaproteobacteria | |||
| D3112 | Generalized | P. aeruginosa | Al Darzins | ||
| UT1 | Generalized | P. aeruginosa | Cory Pfeiffer | ||
| D3 | Specialized | P. aeruginosa | Cory Pfeiffer | ||
| SN-T | Generalized | P. aeruginosa, S. natans, E. coli, S. flexneri, and P. vulgaris | Ken Nickerson | ||
ATCC, American Type Culture Collection.
Phage preparation.
Phages were prepared using the top agar technique: 100 μl of a known dilution of phage was mixed with an equal volume of a culture of P. aeruginosa grown in broth overnight or resuspended, plate-grown cells of S. natans. The mixture of bacterial host and phage was immediately added to 4 ml of 0.7% top agar (LB for P. aeruginosa or TY for S. natans). The top agars were poured onto LB or TY 1.5% agar plates, and the plates were incubated for 24 h at 37°C in the case of P. aeruginosa or for 2 days at room temperature for S. natans. Control plates which contained bacterial cells that had not been infected with phage were also prepared; these will henceforth be referred to as “phage-free, cell-only” controls. These phage-free, cell-only controls were processed in parallel through the phage isolation protocol with cells that had received phage. Phages were harvested by resuspending the soft agar in 4 ml of LB or TY broth and vortexing to release phage, followed by incubation on ice for 30 min. Bacterial cells were then removed by room temperature centrifugation at 10,000 × g for 10 min. The supernatant was then filtered through a 0.2-μm filter and stored at 4°C. Sterility of phage preparations was determined by plating 100 μl onto an LB or TY plate and incubating at the appropriate temperature and time. Phage preparations that were shown to be free of bacterial host cells were used in subsequent infections and DNA preparations.
Sequential infection.
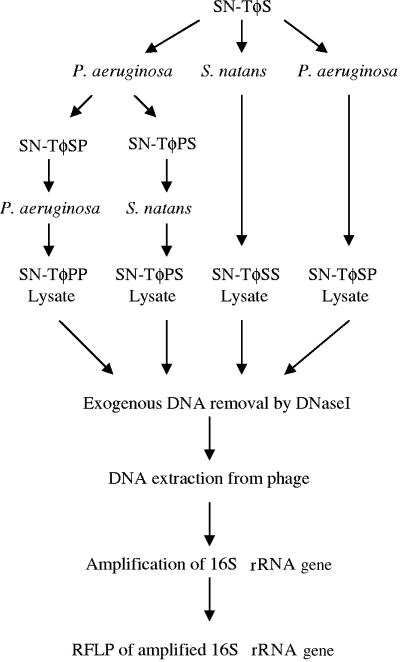
In order to determine whether a phage could acquire 16S rRNA gene sequences from two different genera, we used the broad-host-range, generalized transducing phage SN-T. Several different series of sequential infections were performed (Fig. 1). The phages harvested from these sequential infections were given designations based on the order of the bacterial host(s) used in the infection: SN-ΤφPP (P. aeruginosa infection twice in sequence), SN-ΤφSS (S. natans infection twice in sequence), SN-ΤφSP (S. natans infection followed by P. aeruginosa infection), and SN-TφPS (P. aeruginosa infection followed by S. natans infection). DNA was then extracted from all SN-T phage preparations as well as from the uninfected, cell-only controls.
FIG. 1.
Protocol for phage infection (where SN-ΤφPP denotes sequential infection of P. aeruginosa by SN-T, SN-ΤφSP denotes sequential infection of S. natans followed by P. aeruginosa, etc.) and subsequent DNA preparation, amplification, and analysis by RFLP.
DNA preparation and amplification by PCR.
Genomic DNA was isolated from E. coli, P. aeruginosa, and S. natans cells according to the procedure described previously by Woo et al. (17).
To ensure that only phage DNA was obtained, SN-T lysates were treated with DNase I prior to lysis of phage particles and DNA extraction (2). Phage-free, cell-only controls were run in parallel. To test for the presence the entire 16S rRNA gene, the universal sequence primers 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1525R (5′-AAGGAGGTGATCCAGCC-3′) were used (4; N. Pace, personal communication). This set of primers amplifies a 1,517-bp sequence. Taq DNA polymerase was used for the PCR amplifications to determine the presence of 16S rRNA gene sequences in phages; however, Accuprime Pfx DNA polymerase was used for amplification from SN-T for restriction fragment length polymorphism (RFLP) analysis because of its high fidelity. Thermocycler conditions were as follows: an initial denaturation step (94°C for 12 min) followed by 33 cycles of denaturation (94°C for 1 min), annealing (54°C for 45 s), and extension (72°C for 2 min). Finally, the tubes were incubated at 72°C for 12 min to ensure complete synthesis of the entire sequence.
P. aeruginosa or E. coli purified genomic DNA was used as a positive control in all PCRs. A negative control consisting of PCR mix with no DNA added was run to ensure that there were no contaminating 16S rRNA gene sequences in the reagents. Also, phage-free, cell-only controls were run in parallel with the phage-containing samples to again make certain that the DNase step was effective and that there were no contaminating bacterial 16S rRNA gene sequences in the phage preparations.
RFLP analysis of 16S rRNA gene sequences.
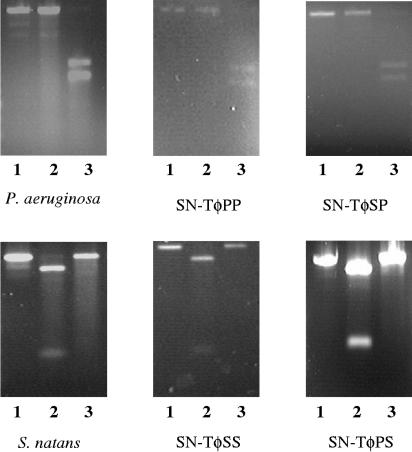
To determine the source of the bacterial donor 16S rRNA genes acquired by SN-T, the 1,517-bp PCR products were recovered from agarose gels and purified using a QIAEX II gel extraction kit. RFLP analysis was performed on the purified DNA fragments. The known 16S rRNA gene sequences from P. aeruginosa PAO1 (GenBank accession number NC_002516) and S. natans (accession number Z18534) were analyzed for restriction cut sites using NEB Cutter version 2.0 (http://tools.neb.com/NEBcutter2/index.php). A single unique enzyme that would result in two unique bands upon digestion of either P. aeruginosa or S. natans 16S rRNA gene sequences was identified (see Fig. 5). BamHI was predicted to cut the P. aeruginosa sequence once, producing two bands of 600 and 900 bp. AgeI was predicted to cut the S. natans sequence once, producing two bands of 1,300 and 200 bp. 16S rRNA gene sequences amplified from the genomic DNA of each bacterial donor were digested and analyzed by agarose gel electrophoresis. In both cases, the predicted bands were present. This allowed us to then analyze PCR-amplified 16S rRNA gene sequences acquired by SN-T to determine their bacterial origin.
FIG. 5.
Restriction fragments of the PCR-amplified region (8 to 1,525 bp) of the 16S rRNA gene from bacterial and phage genomic DNA. Lanes: 1, uncut; 2, cut with AgeI; 3, cut with BamHI.
Detection of frequency of 16S rRNA gene sequences in phage populations using PCR.
Viable phages of both SN-ΤφPP and SN-TφSS were enumerated using standard PFU plate counts. Serial dilutions were then prepared, and the DNA was extracted from 1 ml of lysates of each dilution as described above. The highest dilution of phage to contain 16S rRNA gene sequences was determined by PCR.
RESULTS AND DISCUSSION
Generalized transducing phages acquire 16S rRNA gene sequences.
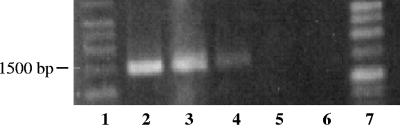
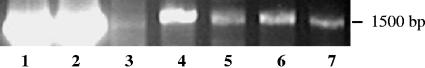
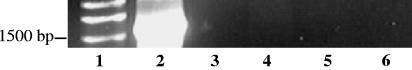
16S rRNA gene PCR products of 1,517 bp, representing >99% of the 16S rRNA gene, were amplified from the bacteriophage genomic DNA of all generalized transducing phages: D3112, UT1, and SN-T. In contrast, no PCR product was amplified from the DNA of the specialized transducing phage D3 (Fig. 2 and 3). That generalized transducing phage and not specialized transducing phage carried 16S rRNA gene sequences was to be expected; this further establishes the absence of contaminating 16S rRNA gene sequences. Additionally, 16S rRNA gene PCR products were absent in all phage-free, cell-only controls (Fig. 4) and in control 16S rRNA gene PCRs containing no DNA. The latter was important, as we had found some commercial polymerases to be contaminated with 16S sequences resulting in PCR products in the negative PCR controls. This is not unusual, as Tanner et al. (14) amplified several different bacterial 16S rRNA genes from PCRs lacking template DNA. Furthermore, Sander and Schmeiger (12), who previously showed that generalized transducing phages in activated sludge contain 16S rRNA genes, found contaminating 16S rRNA gene sequences to be a problem. We too detected 16S rRNA gene sequences in phages isolated from wetland soil samples (data not shown) and experienced problems with contaminating 16S rRNA gene sequences. We did not detect 16S rRNA gene PCR products in our phage-free, cell-only DNase-treated controls. Thus, we concluded that the DNase treatment was effective at degrading exogenous DNA, which would include any 16S rRNA gene sequences.
FIG. 2.
Agarose gel electrophoresis of 16S rRNA gene PCR-amplified region (8 to 1,525 bp). Lanes: 1, 1-kb ladder; 2, E. coli; 3, D3112; 4, UT1; 5, D3; 6, no DNA; 7, 1-kb ladder.
FIG. 3.
Agarose gel electrophoresis of 16S rRNA gene PCR-amplified region (8 to 1,525 bp). Lanes: 1, P. aeruginosa; 2, S. natans; 3, SN-ΤφPP; 4, SN-ΤφSS; 5, SN-ΤφSP; 6, SN-ΤφPS; 7, 1-kb ladder.
FIG. 4.
Agarose gel electrophoresis of SN-T uninfected cell-only controls. Lanes: 1, 1-kb ladder; 2, P. aeruginosa genomic positive control; 3, SN-ΤφPP; 4, SN-ΤφSS; 5, SN-ΤφSP; 6, SN-ΤφPS.
Source and frequency of 16S rRNA genes in SN-T phage populations.
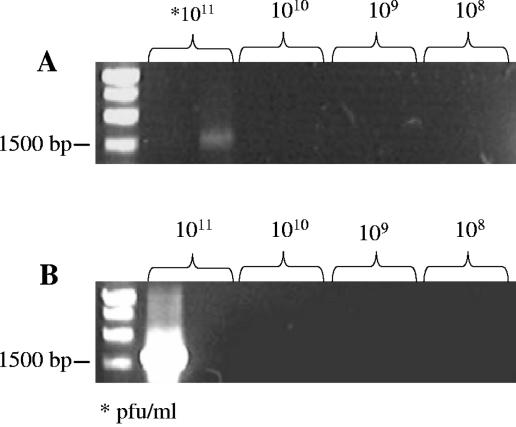
Using RFLP analysis, we were able to determine the source of the 16S rRNA gene sequence. We found that SN-T was able to acquire either P. aeruginosa or S. natans 16S rRNA genes (Fig. 3). Therefore, it is possible for a broad-host-range, generalized transducing phage to acquire and carry 16S rRNA gene sequences from different bacterial genera. Interestingly, in all sequential infections, SN-T was found to contain the 16S rRNA gene sequence from the bacterial host it had last infected and not more than one detectable 16S rRNA gene sequence (Fig. 5). However, the 16S rRNA gene (8 to 1,525 bp) was detected only in SN-T populations containing greater than or equal to 1011 PFU/ml (Fig. 6). This was true regardless of which host was used. Assuming that one copy of the 16S rRNA gene sequence led to amplification and because only 1/100 of the total DNA extracted from 1 ml of phage lysate was used, the frequency of SN-T acquiring the 16S rRNA gene is no less than 1 in 109 phages. This is comparable to the transduction rate of 3 × 10−9 transductants/PFU for SN-T as determined through auxotrophic conversion (16). Since the PCR is not sensitive enough to detect a single copy of a sequence, it is likely that the actual frequency is actually higher and thus comparable to transduction frequencies of 10−8 to 10−10 transductants/PFU that have previously been reported for UT1 transduction of chromosomal alleles in freshwater environments in P. aeruginosa (10). Thus, our finding that SN-T contained 16S rRNA gene sequences only from the final bacterial host was not surprising, given the low frequency of 16S rRNA gene sequence acquisition by SN-T. These results indicate that the mechanism of acquisition of 16S rRNA genes by SN-T is likely mistaken packaging of the bacterial 16S rRNA gene instead of phage genomic DNA.
FIG. 6.
Relative frequencies of each bacterial 16S rRNA gene sequence in SN-T. Frequencies were determined by dilution series of SN-T followed by DNA extraction and PCR amplification. Replicates were run for each dilution.
Transduction frequency is also known to change with environmental parameters and multiplicity of infection. To further understand the frequency of broad-host-range phages acquiring 16S rRNA gene sequences, various natural environmental conditions and multiplicities of infection need to be studied.
Evolutionary and ecological implications.
We have demonstrated that a broad-host-range, generalized transducing phage can acquire and carry 16S rRNA gene sequences from bacteria belonging to different genera. We believe that SN-T is a good model phage and that P. aeruginosa and S. natans are good model hosts for the study of HGT for the following reasons. First, SN-T was isolated from sewage, and S. natans is also found in sewage and grows well in fully aerated and low-oxygen environments (5). Second, P. aeruginosa is ubiquitous in the environment, including sewage. Therefore, all of these organisms could be expected to cohabitate. Finally, P. aeruginosa and S. natans are from different classes of the phylum Proteobacteria; P. aeruginosa is classified as Gammaproteobacteria, while S. natans is classified as Betaproteobacteria. Thus, our model could be extended to study transduction under a variety of environmental conditions as well as to probe the ecological significance of 16S rRNA gene horizontal transfer via phages.
It has been suggested that HGT may contribute to the presence of heterogeneous 16S rRNA gene sequences in bacteria. Our results support this possibility, although the frequency of such events appears to be low. Furthermore, the presence of broad-host-range, generalized transducing phage carrying 16S rRNA gene sequences from more than one genera of bacteria could lead to a misrepresentation of microbial diversity in the environment. Sander and Schmeiger (12) found that phages isolated from sewage carry 16S rRNA gene sequences, and we have seen the same results in samples from wetland soils. Furthermore, our experiments show that phages can acquire 16S rRNA gene sequences from more than one genera of bacteria. Thus, assessment of a microbial community based on 16S rRNA gene sequences must take into account the presence of generalized transducing phages, especially those that are broad host range, in that community.
REFERENCES
- 1.Acinas, S. G., L. A. Marcelino, V. Klepac-Ceraj, and M. F. Polz. 2004. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 186:2629-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashelford, K. E., M. J. Day, M. J. Bailey, A. K. Lilley, and J. C. Fry. 1999. In situ population dynamics of bacterial viruses in a terrestrial environment. Appl. Environ. Microbiol. 65:169-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clayton, R. A., G. Sutton, P. S. Hinkle, Jr., C. Bult, and C. Field. 1995. Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa. Int. J. Syst. Bacteriol. 45:595-599. [DOI] [PubMed] [Google Scholar]
- 4.de la Torre, J. R., B. M. Goebel, E. I. Friedman, and N. R. Pace. 2003. Microbial diversity of cryptoendolythic communities from McMurdo Dry Valleys, Antarctica. Appl. Environ. Microbiol. 69:3858-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dias, F. F., N. C. Dondero, and M. S. Finstein. 1968. Attached growth of Sphaerotilus and mixed population in continuous-flow apparatus. Appl. Microbiol. 16:1191-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Head, I. M., J. R. Saunders, and R. W. Pickup. 1998. Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb. Ecol. 35:1-21. [DOI] [PubMed] [Google Scholar]
- 7.Jain, R., M. C. Rivera, and J. A. Lake. 1999. Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl. Acad. Sci. USA 96:3801-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen, E. C., H. S. Schrader, B. Rieland, T. L. Thompson, K. W. Lee, K. W. Nickerson, and T. A. Kokjohn. 1998. Prevalence of broad-host-range lytic bacteriophages of Sphaerotilus natans, Escherichia coli, and Pseudomonas aeruginosa. Appl. Environ. Microbiol. 64:575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rainey, F. A., N. L. Ward-Rainey, P. H. Janssen, H. Hippe, and E. Stackebrandt. 1996. Clostridium paradoxum DSM 7308T contains multiple 16S rRNA genes with heterogeneous intervening sequences. Microbiology 142:2087-2095. [DOI] [PubMed] [Google Scholar]
- 10.Ripp, S., O. A. Ogunseitan, and R. V. Miller. 1994. Transduction of a freshwater microbial community by a new Pseudomonas aeruginosa generalized transducing phage, UT1. Mol. Ecol. 3(2):121-126. [DOI] [PubMed] [Google Scholar]
- 11.Rivera, M. C., R. Jain, J. E. Moore, and J. A. Lake. 1998. Genomic evidence for two functionally distinct gene classes. Proc. Natl. Acad. Sci. USA 95:6239-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sander, M., and H. Schmeiger. 2001. Method for host-independent detection of generalized transducing bacteriophages in natural habitats. Appl. Environ. Microbiol. 67:1490-1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sullivan, M. B., J. B. Waterbury, and S. W. Chisholm. 2003. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature 424:1047-1050. [DOI] [PubMed] [Google Scholar]
- 14.Tanner, M. A., B. M. Goebel, M. A. Dojka, and N. R. Pace. 1998. Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl. Environ. Microbiol. 64:3110-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ueda, K., T. Seki, T. Kudo, T. Yoshida, and M. Kataoka. 1999. Two distinct mechanisms cause heterogeneity of 16S rRNA. J. Bacteriol. 181:78-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winston V., and T. L. Thompson. 1979. Isolation and characterization of a bacteriophage specific for Sphaerotilus natans which contains an unusual base in its deoxyribonucleic acid. Appl. Environ. Microbiol. 37:1025-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woo, T. H., A. F. Cheng, and J. M. Ling. 1992. An application of a simple method for the preparation of bacterial DNA. BioTechniques 13:696-698. [PubMed] [Google Scholar]
- 18.Yap, W. H., Z. Zhang, and Y. Wueng. 1999. Distinct types of rRNA operons exist in the genome of the actinomycete Thermomonospora chromogena and evidence for horizontal transfer of an entire rRNA operon. J. Bacteriol. 181:5201-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]