Abstract
Processing of the phytoplankton-derived organic sulfur compound dimethylsulfoniopropionate (DMSP) by bacteria was studied in seawater microcosms in the coastal Gulf of Mexico (Alabama). Modest phytoplankton blooms (peak chlorophyll a [Chl a] concentrations of ∼2.5 μg liter−1) were induced in nutrient-enriched microcosms, while phytoplankton biomass remained low in unamended controls (Chl a concentrations of ∼0.34 μg liter−1). Particulate DMSP concentrations reached 96 nM in the enriched microcosms but remained approximately 14 nM in the controls. Bacterial biomass production increased in parallel with the increase in particulate DMSP, and nutrient limitation bioassays in the initial water showed that enrichment with DMSP or glucose caused a similar stimulation of bacterial growth. Concomitantly, increased bacterial consumption rate constants of dissolved DMSP (up to 20 day−1) and dimethylsulfide (DMS) (up to 6.5 day−1) were observed. Nevertheless, higher DMSP S assimilation efficiencies and higher contribution of DMSP to bacterial S demand were found in the controls compared to the enriched microcosms. This indicated that marine bacterioplankton may rely more on DMSP as a source of S under oligotrophic conditions than under the senescence phase of phytoplankton blooms. Phylogenetic analysis of the bacterial assemblages in all microcosms showed that the DMSP-rich algal bloom favored the occurrence of various Roseobacter members, flavobacteria (Bacteroidetes phylum), and oligotrophic marine Gammaproteobacteria. Our observations suggest that the composition of the bacterial assemblage and the relative contribution of DMSP to the overall dissolved organic sulfur/organic matter pool control how efficiently bacteria assimilate DMSP S and thereby potentially divert it from DMS production.
During blooms of phytoplankton, and particularly during their senescence, the activity of bacterioplankton is stimulated by the release of dissolved organic matter (DOM) and nutrients. Since bacterioplankton groups differ in their growth requirements, such release can stimulate the growth of specific components of the bacterioplankton assemblage. Given that bacterial taxa differ in their potential for hydrolytic ectoenzyme activities and their capacity to assimilate and process various substrates (10, 22), shifts in the composition of bacterioplankton assemblages under phytoplankton blooms or oligotrophic conditions are likely to have consequences for the turnover of organic matter and biogeochemically active elements such as N and S.
A component of the DOM pool that has attracted much attention for the last two decades is the algal osmolyte dimethylsulfoniopropionate (DMSP). This sulfur-containing compound is released into the water column, where it plays important roles in C and S cycling, through algal lysis or zooplankton predation. A volatile degradation product of DMSP (dimethylsulfide [DMS]) escapes to the atmosphere, where it influences climate through cloud formation (1). Bacterial utilization of DMSP is also important and, along with simple sugars, amino acids, and carboxylic acids, can satisfy an important fraction of the bacterial carbon and sulfur demands in various marine environments (13, 16, 30, 37). DMSP has been shown to support up to 13% of the bacterial carbon demand in marine surface waters (16, 34) and to provide enough S to satisfy all or most of the requirements of this element for bacterial growth (16, 18, 34, 40).
There are two main bacterial biochemical pathways for the utilization of DMSP (17, 39). First, bacteria transform DMSP to methanethiol by the removal of a methyl group and a three-carbon moiety through the demethylation/demethiolation pathway. Methanethiol is a key reactive intermediate in the assimilation of S into the amino acid methionine and thereafter into cellular protein (18). Furthermore, the methyl group as well as the three-carbon moiety (possibly acrylate) may be utilized as C sources for biosynthesis or energy generation. Thus, the demethylation/demethiolation pathway provides bacteria with both S and C necessary for growth. Second, bacteria cleave DMSP to DMS and acrylate using the enzyme DMSP lyase. Both DMS and acrylate may be utilized by bacteria, although DMS is relatively unreactive and most is lost from the cells without appreciable assimilation of the sulfur (18). Therefore, the cleavage pathway may primarily provide bacteria with some amount of cellular C. It may thus be expected that heterotrophic bacteria switch between the two pathways for DMSP utilization depending on their cellular demands for reduced sulfur and carbon and the availability of substrates (17).
Present inventories of the phylogenetic composition of marine bacterioplankton offer a fairly good picture of the overall diversity among bacteria that exist in the sea (8, 12). However, little progress has been made in explaining the specific functions of particular bacteria in the microbial food web. Bacteria in the Roseobacter clade have been implicated as important actors in the processing of DMSP in both open sea and coastal waters (11, 23, 40). Microautoradiography with [35S]DMSP combined with fluorescence in situ hybridization (FISH) (MicroFISH) recently confirmed the active role of the Roseobacter clade in DMSP assimilation (21, 38). Nevertheless, significant assimilation of DMSP was observed in several other groups of bacteria in these studies, suggesting that many taxa contribute to control the transformation rates of this compound.
Using samples from the microcosm experiments that are reported on in the present study, a MicroFISH protocol with [35S]DMSP was developed (38). Vila et al. (38) showed that nearly half of the bacterioplankton assemblage assimilated [35S]DMSP during the course of an induced phytoplankton bloom and that members of the Roseobacter clade only partially accounted for this activity. Here, we quantify DMSP assimilation and transformation by bacteria during the microcosm experiment and investigate how changes in DMSP processing relate to changes in the composition of the bacterial assemblage during the induced bloom compared to nonbloom conditions.
MATERIALS AND METHODS
Seawater collection.
On 8 June 2001, surface seawater was collected outside the Mobile River plume in the vicinity of Dauphin Island, Alabama (10 miles offshore; salinity, 33.3), located in the northern Gulf of Mexico (30°15′N, 88°05′W). Water was collected with a rinsed 10-liter polyethylene bucket submerged to 0.5 m and transferred to polycarbonate carboys (Nalgene). Water was kept at in situ temperature in cooling boxes until it reached the laboratory, where it was processed within 3 h.
Microcosm experiment.
Water was filtered through a 150-μm mesh to exclude large zooplankton and was partitioned into four 25-liter polycarbonate carboys (Nalgene). The carboys were placed in a climate-controlled room set at 27°C (the in situ temperature at the time of sampling) with an artificial light source (12 h light and 12 h dark; 200 μE m−2 s−1 inside the carboys) and were maintained for 8 days. Gentle mixing was accomplished once daily prior to sampling by turning carboys upside down twice. Two carboys were maintained as unenriched controls (microcosms C1 and C2), and two carboys were enriched with 10 μM N (final concentration; added as NaNO3) and 0.6 μM P (final concentration; added as Na2HPO4) (microcosms NUT1 and NUT2). In control microcosms C1 and C2, the initial concentrations of NO3 plus NO2, NH4, PO4 and Si were less than 0.05, 1.5, 0.1, and 3.0 μM, respectively. Similar concentrations were found in both the control and the enriched microcosms by the end of the experiment. Dissolved organic carbon (DOC) averaged 115 ± 8 μM during the experiment.
For most variables (e.g., chlorophyll a [Chl a], bacterial abundance, and concentrations of DMSP and DMS), the microcosms were sampled daily, immediately before the light was turned on, by withdrawing a 500-ml water sample through a submerged acid-rinsed glass pipette at the end of a silicon tube. Samples for microbial community DNA that required larger volumes (1.5 liters) were sampled every 2 days, but the replicates were sampled out of phase to obtain one sample every day from one replicate (i.e., control microcosm C1 and enriched microcosm NUT1 were sampled on day 1, control microcosm C2 and enriched microcosm NUT2 were sampled on day 2, etc.).
Nutrient limitation bioassay.
The effect of nutrient additions on the growth of heterotrophic bacteria was examined using excess initial sample water (also filtered through 150-μm mesh). Sample water was transferred to acid-rinsed 500-ml polycarbonate bottles (Nalgene) that had been thoroughly rinsed with MilliQ water and sample water. Nutrients were added to final concentrations of 10 μM C (as glucose or DMSP), 2 μM N (NH4Cl), and 0.6 μM P (Na2HPO4), singly and in combination. Each nutrient addition was tested in duplicate. Control bottles received no nutrients. After a 24-h incubation in darkness at 27°C, bacterial biomass production was determined (see below). No significant changes in bacterial abundance were observed during the 24-h bioassay.
Chlorophyll a.
Phytoplankton Chl a was estimated fluorometrically from 50-ml samples filtered through Whatman GF/F filters. Filters were placed in ice-cold 90% acetone, and Chl a was extracted for 24 h at −20°C before fluorescence was measured in the extract.
Bacterial abundance and heterotrophic biomass production.
Samples for bacterial abundance determinations by flow cytometry (1.6 ml) were preserved with 1% paraformaldehyde and 0.05% glutaraldehyde (final concentrations) and thereafter stored frozen. Cell counts were done with a FACSCalibur flow cytometer after staining with Syto13 (6).
Bacterial biomass production was measured using the [3H]leucine incorporation method (19), modified as previously described by Smith and Azam (36). For each sample, triplicate aliquots (1.7 ml) and a trichloroacetic acid (TCA)-killed control were incubated with 20 nM [3H]leucine (final concentration) for 1 to 2 h at 27°C in darkness. The factor for converting leucine incorporation into bacterial biomass production was empirically determined to be 3.05 kg C mol−1 (the average from duplicate seawater cultures, 2.80 and 3.29 kg C mol−1), assuming the following carbon-to-volume relationship as derived previously by Norland (25) from the data of Simon and Azam (35): pg C cell−1 = 0.12 × (μm3 cell−1)0.7. Bacterial volumes were estimated from flow cytometry as described previously by Gasol and del Giorgio (6).
Respiration.
Respiration was measured as the consumption of oxygen over 24 h in 60-ml biological oxygen demand bottles incubated in the dark at the experimental temperature. Oxygen was measured by Winkler titration using a Mettler DL 20 Autotitrator.
Sulfur analyses.
Particulate DMSP (DSMPp) (i.e., DMSP mainly contained in algal cells) was measured by filtering 50 ml of sample onto 25-mm GF/F filters (Whatman) and placing the filters in 14-ml serum vials that were capped with rubber stoppers and sealed. Two milliliters of 5 M NaOH was added to each vial to quantitatively cleave DMSP into DMS. DMS in the headspace was quantified by injecting 100-μl subsamples into a gas chromatograph equipped with a flame photometric detector. DMSP standards were prepared by the same procedure used for the samples and dispensed in the same kind of vials. DMS analysis was made by the headspace sweeping method as described previously (15).
Dissolved DMSP (DSMPd) consumption was determined by incubating 30-ml water samples from the microcosms in the dark with tracer levels of [35S]DMSP. At each of four time points over 3 h, 4-ml subsamples were removed, acidified, and stored for >10 h to preserve [35S]DMSP and to remove volatile 35S species. The [35S]DMSP remaining in each stored subsample was determined by treating the sample with NaOH in a sealed bottle and trapping the volatile [35S]DMS in 3% H2O2-soaked wicks suspended in well cups (16). Rate constants (day−1) were calculated from the exponential loss of [35S]DMSP with time. DMSP flux (micromoles day−1) was calculated as the product of the dissolved DMSP concentration and the loss rate constant (14).
The DMSP sulfur assimilation efficiency of microbial populations was determined by incubating 5-ml water samples with tracer levels of [35S]DMSP for 24 h and then measuring the fraction of added 35S that was incorporated into cellular, TCA-insoluble macromolecules (18). After a 24-h incubation, most if not all of the added DMSP tracer was utilized (consumption rate constants were >10 day−1) (see Results); therefore, the fraction of tracer assimilated should represent the DMSP S assimilation efficiency. The rate of sulfur assimilated from DMSP into biomass was calculated by multiplying the DMSP S assimilation efficiency by the DMSP consumption rate.
Microbial DMS consumption was determined based on the conversion of [35S]DMS into nonvolatile products (16). Duplicate subsamples (5 ml) from each microcosm were amended with tracer levels of [35S]DMS and incubated in the dark for 5 to 8 h. The rate constant (day−1) was taken as the slope of the natural log of the fraction of [35S]DMS converted to nonvolatile products versus time. The DMS consumption rate (nanomoles day−1) was calculated as the product of the DMS concentration and the rate constant.
DMS production (nanomoles day−1) was estimated as the production rate needed to support the concentration change between two consecutive days plus the average consumption rates during the same two days. The assumption was that no DMS losses other than microbial consumption occurred in the UV-opaque closed carboys.
FISH.
Samples for FISH were preserved with formaldehyde (4% final concentration), and 1-ml aliquots were filtered onto 0.2-μm-pore-size polycarbonate filters (Nuclepore). Bacteria on filter sections from the microcosms on days 1 and 7 of the experiment were hybridized with the general bacterial probe EUB338 (5′-GCTGCCTCCCGTAGGAGT) and the group-specific oligonucleotide probes CF319a (5′-TGGTCCGTGTCTCAGTAC) for Bacteroidetes relatives and ROS536 (5′-CAACGCTAACCCCCTCCG) for the Roseobacter clade. The day 7 sample was chosen for FISH analysis so that it coincided with the 16S rRNA gene clone library construction (see below) and differs from the samples analyzed previously by Vila et al. (38). Probes labeled with the cyanine dye CY3 at the 5′ end were purchased from Thermo Hybaid GmbH. Cells were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) (final concentration, 1 μg ml−1) following hybridization and mounted for microscopy (27).
Collection of microbial community DNA.
A 500- to 700-ml volume of the sample water was filtered through a 0.2-μm-pore-size polycarbonate filter at <200 mm Hg to collect bacterial cells. Filters were immediately frozen at −20°C until further processing. DNA was extracted using the Ultra Clean Soil DNA kit (MO BIO Laboratories Inc.), according to instructions from the manufacturer.
16S rRNA gene clone library construction and sequencing.
Selected clone libraries were constructed to allow putative identification of peaks in the more comprehensive terminal restriction fragment length polymorphism (T-RFLP) surveys (see below), but they also provided independent data on the composition of the bacterial assemblages in the microcosms. The 16S rRNA genes from community DNA samples collected on day 7 from microcosms C1 and NUT1 were amplified by means of PCR with the general bacterial primers 27F and 1522R (9), according to the protocol previously described in detail by González et al. (11). Clone libraries were constructed using a TA cloning kit (Invitrogen Corporation). A total of 20 clones from each sample were sequenced using primer 27F to obtain approximately 500 bp of sequence information. Chimeras were detected by generating phylogenetic trees with different regions of the gene. Only 1 of the 40 clones was chimeric, and this sequence was excluded from the analysis. To allow comparisons to sequences obtained from denaturing gradient gel electrophoresis (DGGE), selected Roseobacter clone sequences were also sequenced with the primer 358f (see below).
T-RFLP.
T-RFLP was carried out by restriction enzyme digestion of bacterial 16S rRNA genes amplified by PCR, as previously described (11). Primers were the same as for the clone libraries, except that the concentration of primer 27F was reduced to 0.02 μM and 0.18 μM of primer 8F-FAM (20) (PE Corporation) was added. A total of 30 ng of PCR product was digested for 3 h with 10 U of HhaI (Boehringer Mannheim). The length of the terminal restriction fragment was determined with an ABI PRISM 310 genetic analyzer in Genescan mode. Prediction of T-RFLP fragment sizes for the clone sequences and DGGE bands (see below) were as previously described (11). Chromatogram peaks were identified based on the predicted length of the fragment.
DGGE.
DGGE and gel analysis were performed essentially as previously described (32). Briefly, 16S rRNA gene fragments (around 550 bp in length) were amplified by PCR using the universal primer 907rm, complementary to positions 927 to 907 (5′-CCGTCAATTCA/CTTTGAGTTT), and the bacterium-specific primer 358f, complementary to positions 341 to 358, with a GC clamp (underlined) (5′-CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGGCCTACGGGAGGCAGCAG). PCR products were loaded onto a 6% polyacrylamide gel with a DNA denaturant gradient ranging from 40 to 80%. The gel was run at 100 V for 16 h at 60°C in 1× Tris-acetate-EDTA running buffer. DGGE gel images were analyzed using Diversity Database software (Bio-Rad). Since the relative abundance of bacterial populations is likely to change in a successive manner during microcosm experiments, both the presence/absence and intensity of bands were used in the analysis. DGGE bands were excised, reamplified, and sequenced using primer 358f without the GC clamp with the BigDye Terminator cycle sequencing kit (Perkin-Elmer Corporation) and an ABI PRISM model 377 (v3.3) automated sequencer.
RESULTS
Nutrient limitation bioassay.
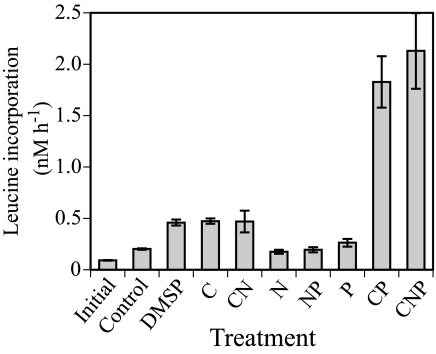
The addition of DMSP or glucose to the initial water used for the microcosm experiment caused a significant increase in bacterial biomass production, whereas mineral nutrients alone did not induce a significant response (Fig. 1). This suggested that C was the primary limiting nutrient for bacterial growth and that DMSP could elicit as strong a response as glucose. The response in the combined glucose and phosphate treatments indicated that P was the second limiting nutrient, once C limitation was alleviated.
FIG. 1.
Comparison of leucine incorporation rates 24 h after the addition of nutrients to surface water from coastal Gulf of Mexico (Alabama). Treatments included the addition of DMSP, glucose (C), ammonia (N), and phosphate (P), singly and in different combinations. Error bars represent standard deviations for the pooled measurements of triplicate subsamples from each duplicate treatment (n = 6).
Chorophyll a, bacterial biomass production, and abundance.
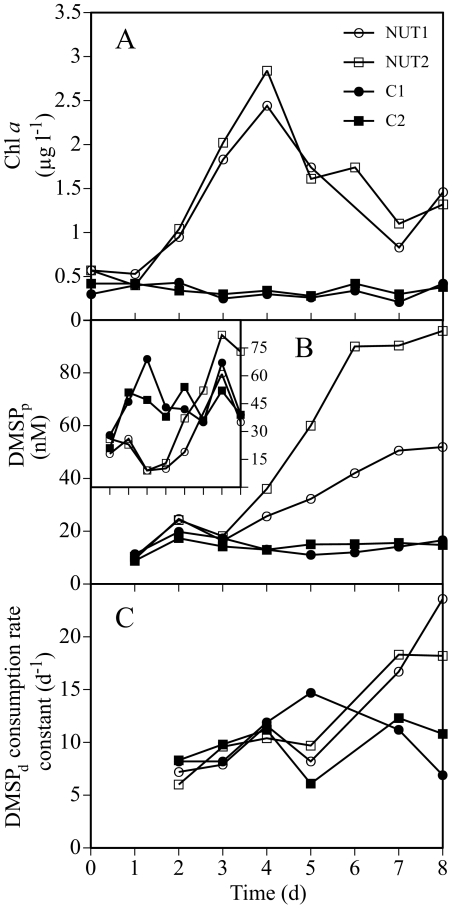
The initial concentration of Chl a in the microcosms was ∼0.5 μg liter−1, consistent with normal offshore concentrations in this region of the Gulf of Mexico. Chl a concentrations in the control microcosms remained low during the experiment. Nutrient enrichment caused a small but significant bloom of phytoplankton; Chl a concentrations peaked on day 4 at about 2.5 μg liter−1 (Fig. 2A), which is within the range of values commonly found in coastal Gulf of Mexico systems (26).
FIG. 2.
Dynamics in Chl a, DMSP concentrations, and DMSP consumption rates during the microcosm experiment. (A) Variability in Chl a concentrations; (B) concentrations of DMSPp; (C) consumption rate constants of DMSPd. Inset figure in panel B shows the daily values of the DMSPp-to-Chl a ratio, expressed as nanomoles per microgram. l, liter; d, days.
Initial bacterial biomass production in the four microcosms averaged 0.68 μM C day−1. Bacterial biomass production in the controls remained low throughout the experiment. Production in the enriched microcosms started to increase on day 3 and reached maximum values of approximately 3.8 μM C day−1 on day 6 (for details on the development of bacterial biomass production, see Fig. 4B in reference 38).
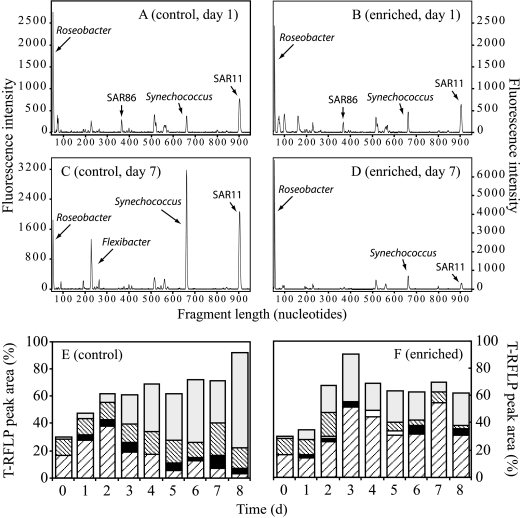
FIG. 4.
Bacterioplankton composition in the microcosms as indicated by T-RFLP analysis using the restriction enzyme HhaI on PCR-amplified 16S rRNA genes. Chromatograms represent samples collected on day 1 (A, B) and day 7 (C, D). Lower panels indicate the relative contribution of major microbial groups to the total integrated area of peaks in the chromatograms in the control (E) or enriched (F) microcosms for each day during the experiment. Coding for T-RFLP peaks in panels E and F is as follows: light gray, Synechococcus; thinly spaced diagonal lines, SAR11; white, SAR86; black, Flexibacter; broadly spaced diagonal lines, Roseobacter. d, days.
Initial bacterial abundance in the microcosms was approximately 1.4 × 106 cells ml−1 and remained relatively stable in the control microcosms throughout the experiment. A higher abundance of bacteria was found in the enriched microcosms towards the end of the experiment, with up to 2.4 × 106 cells ml−1 on day 8 in microcosm NUT2. Flow cytometry analysis showed that the proportion of high-DNA bacteria, i.e., bacteria with a relatively large size and high nucleic acid content (7), averaged 39.9% ± 4.7% (n = 36) in both control and enriched microcosms during the experiment.
Bacterial respiration and growth efficiency.
Integrated bacterial biomass production during the last 24 h of the experiment was approximately 0.27 μM C in the control microcosms, with 10-fold-higher values measured in the enriched microcosms (Table 1). The respiration during the same period differed only threefold between the control and enriched microcosms (averaging 2.6 and 8.3 μM O2, respectively). Based on these values, a bacterial growth efficiency of 10% and 28% was calculated for the bacteria in the control and enriched microcosms, respectively (see Table 1 for calculations and underlying assumptions).
TABLE 1.
Comparison of heterotrophic bacterial biomass production, respiration, and bacterial growth efficiency in the control and enriched microcosms integrated during the last 24 h of the microcosm experimentc
| Characteristic | Control microcosms
|
Enriched microcosms
|
||
|---|---|---|---|---|
| C1 | C2 | NUT1 | NUT2 | |
| BBP (μM C) | 0.24 | 0.30 | 3.05 | 3.04 |
| Respiration (μM O2) | 2.46 | 2.80 | 7.90 | 8.73 |
| BGEa | 0.09 | 0.10 | 0.28 | 0.26 |
| DMSP contribution to C demand (%) | 6 | 5 | 5 | 4 |
| [35S]DMSP sulfur assimilation efficiency into TCA insolubles (%) | 29 | 29 | 13 | 12 |
| DMSP contribution to S demand (%)b | 860 | 690 | 102 | 95 |
Assuming the BGE = BBP/(BBP + respiration), a respiratory quotient of 1, and that nonbacterial respiration was negligible. If nonbacterial respiration was significant, actual bacterial growth efficiencies would be higher than calculated. BBP, bacterial biomass production; BGE, bacterial growth efficiency.
Assuming a C:S ratio of 240 (2).
Also shown is the proportion of the bacterial C and S demands that could be derived from the measured DMSP consumption rates.
DMSP concentrations and consumption rates.
Concentrations of DMSPp in the enriched microcosms started to increase on day 4, coinciding with the peak of the phytoplankton bloom, and eventually reached 52 nM in microcosm NUT1 and 96 nM in microcosm NUT2 as the bloom declined (Fig. 2B). DMSPp concentrations in the control microcosms remained at around 14 nM throughout the experiment. In the control microcosms, DMSPp-to-Chl a ratios remained relatively constant at 40 to 70 nmol μg−1 (Fig. 2B, inset). The enriched microcosms exhibited DMSPp-to-Chl a ratios as low as 9 nmol μg−1 until day 4. Beginning on day 5, the ratio in the enriched microcosms increased and eventually reached levels similar to those of the controls.
Concentrations of DMSPd varied between 3 and 6 nM throughout the experiment, with no significant differences between treatments (data not shown). Microbial DMSPd consumption rate constants in the control microcosms were mostly around 10 day−1, while in the enriched microcosms, they increased to around 20 day−1 during the last 2 days of the experiment (Fig. 2C).
Contribution of DMSP to bacterial C and S demand.
The sum of bacterial biomass production and bacterial respiration over the final 24 h of the experiment (Table 1) provided an estimate of bacterial C demand of approximately 3.0 and 11.0 μM C day−1 in the control and enriched microcosms, respectively. Concurrent microbial DMSPd consumption rates were 0.03 and 0.10 μM day−1 in the control and enriched microcosms. Since one DMSP molecule contains five C atoms, these DMSPd consumption rates represented 0.15 and 0.50 μM C day−1. Hence, DMSP consumption contributed approximately 5% of the bacterial C demand in both the control and the enriched microcosms (Table 1).
The efficiency of assimilation of S from DMSP into bacterial proteins (TCA-insoluble macromolecules) was determined on the final day of the experiment and was found to be 29% in controls and 12 to 13% in enriched microcosms (Table 1). This assimilation efficiency, applied to the DMSPd consumption rates on these same days, resulted in DMSP assimilation rate estimates of 8 and 13 nM S day−1 in control and enriched microcosms, respectively. Assuming that heterotrophic bacterial growth is driven by a C:S stoichiometry of 240 (2), DMSP contributed around 900% and 700% of the bacterial S demand in control microcosms C1 and C2, respectively. In the enriched microcosms, DMSP contributed around 100% of the S demand (Table 1). It should be noted that these numbers would change significantly should the actual C:S stoichiometry be different from the value used in this paper or if we used other values reported in the literature. For example, the assumption of a C:S ratio of 86 as reported by Fagerbakke et al. (5) would result in S demand calculations down to 250% and 34% in the control and enriched microcosms, respectively. In addition, these calculations do not consider the possibility of accumulation of S into storage inclusions rather than incorporation into bacterial biomolecules. Nonetheless, this would not alter the sizeable difference (around eightfold) in the contribution of DMSP to bacterial S requirements observed between the two treatments.
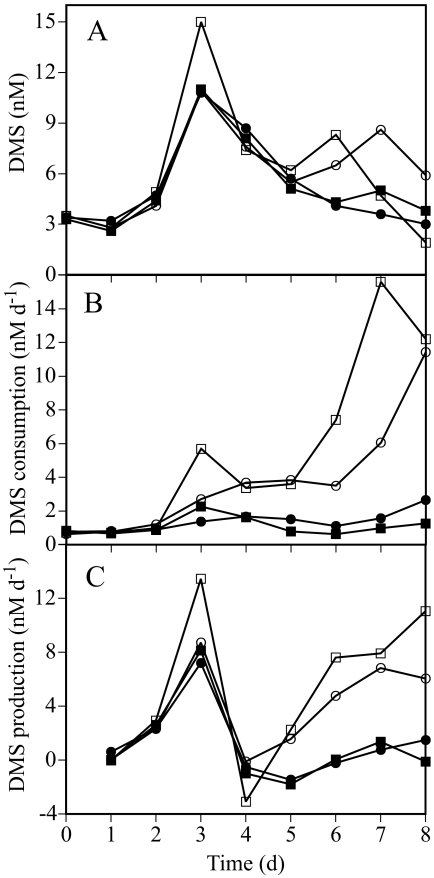
DMS dynamics.
The DMS concentrations increased to around 11 nM on day 3 in all microcosms (up to 15 nM in NUT2) and thereafter decreased slowly to around 3 nM on day 8 (Fig. 3A). DMS consumption rate constants in the enriched microcosms increased from day 3 on, reaching 1.9 to 6.5 day−1 on day 8, while the rate constants in the control microcosms remained around 0.2 day−1 throughout the experiment. These rate constants translated into DMS consumption rates up to 15 nM day−1 in the enriched microcosms and less than 2 nM day−1 in the controls (Fig. 3B).
FIG. 3.
Changes in DMS concentrations and process rates during the experiment. (A) Concentrations of DMS; (B) DMS consumption; (C) DMS production rates estimated from the DMS concentrations and DMS consumption. Symbols are as shown in Fig. 2. d, days.
DMS production rates estimated from DMS concentrations and consumption showed a first peak at about 8 nM day−1 on day 3 (14 nM day−1 in NUT2), coinciding with a maximum in DMS concentrations in all microcosms (Fig. 3C). On day 4, DMS production decreased sharply in all microcosms to values approaching zero and thereafter remained very low in the control microcosms. However, in the enriched microcosms, DMS production steadily increased to approximately 8 nM day−1 on day 8.
FISH.
The general bacterial probe EUB338 hybridized with 51% of the cells (DAPI counts) on day 1 of the experiment (Table 2). This probe still hybridized with half of the cells in the enriched microcosms on day 7, while detection rates in the controls decreased slightly by this time. By day 7, bacteria in the Roseobacter clade accounted for approximately 20% of the DAPI counts in the enriched microcosms but remained at initial values (around 5%) in the control microcosms. Thus, this taxon increased approximately fourfold during the development of the bloom. Bacteria in the Bacteroidetes phylum accounted for a similar proportion of the bacterial assemblage (around 10%) both in the controls and in the enriched microcosms throughout the experiment (but only 3% in control microcosm C2).
TABLE 2.
Relative abundance of bacteria in the Roseobacter and Bacteroidetes groups determined by FISH with group-specific probes compared to the abundance of bacteria during the microcosm experiment
| Sample date (2001) | Day of expt | Microcosm | Abundance (% of DAPI counts)
|
||
|---|---|---|---|---|---|
| Bacteria | Roseobacter | Bacteroidetes | |||
| 9 June | 1 | Control (C1) | 51 | 5 | 10 |
| 15 June | 7 | Control (C1) | 46 | 8 | 12 |
| 15 June | 7 | Control (C2) | 35 | 1 | 3 |
| 15 June | 7 | Enriched (NUT1) | 54 | 23 | 12 |
| 15 June | 7 | Enriched (NUT2) | 50 | 18 | 11 |
Phylogenetic diversity of 16S rRNA gene clones.
The SAR11 clade dominated the 16S rRNA gene clone library made from control microcosm C1 on day 7, accounting for 8 out of 19 clones (Table 3). These clones were distributed into two separate clusters (sequence similarity between clusters of approximately 88 to 91%), indicating that a diverse set of SAR11 clones were present in the microcosms. Other sequences found included Synechococcus (four clones), Roseobacter (three clones), and four other taxa (Table 3). The Roseobacter clade was the single most dominant taxon in the clone library from enriched microcosm NUT1 (6 out of 20 clones), and clone sequences covered a wide diversity of the clade. A total of four clones showed 93 to 98% sequence similarity to the widespread oligotrophic marine Gammaproteobacteria clades SAR92 and OM60, and two clones belonged to the SAR86 clade (Table 3). Most of these clones were found in the enriched microcosms. One Synechococcus clone was the only sequence that was identical between the control (clone BG27-2) and the enriched (clone BG29-2) microcosms.
TABLE 3.
Diversity among 16S rRNA gene clonesa
| Clone | GenBank accession no. | Closest relative in GenBank; accession no. | Similarity (%) | Clade | Group |
|---|---|---|---|---|---|
| Control | |||||
| BG27-10 | AY904490 | Uncult. Alphaproteobacteria clone PI_RT68; AY580445 | 99.4 | Roseobacter | Alpha |
| BG27-14 | AY904494 | Uncult. Alphaproteobacteria clone CD2AC12; AY038571 | 99.8 | Roseobacter | Alpha |
| BG27-16 | AY904495 | Isolate SRF3; AJ002565 | 99.8 | Roseobacter | Alpha |
| BG27-4 | AY904484 | Uncult. Alphaproteobacteria clone MB-C2-128; AY093481 | 98.8 | SAR11 | Alpha |
| BG27-5 | AY904485 | Uncult. Alphaproteobacteria clone MB11B07; AY033299 | 97.1 | SAR11 | Alpha |
| BG27-6 | AY904486 | Uncult. Alphaproteobacteria clone EBAC40E09; AF268220 | 98.9 | SAR11 | Alpha |
| BG27-8 | AY904488 | Uncult. Alphaproteobacteria clone RAI-14; AY499416 | 99.6 | SAR11 | Alpha |
| BG27-12 | AY904492 | Uncult. Alphaproteobacteria clone PI_RT324; AY580474 | 99.8 | SAR11 | Alpha |
| BG27-13 | AY904493 | Uncult. Alphaproteobacteria clone PI_4g12g; AY580483 | 99.3 | SAR11 | Alpha |
| BG27-19 | AY904498 | Uncult. Alphaproteobacteria clone PLY43; U13159 | 99.5 | SAR11 | Alpha |
| BG27-20 | AY904499 | Uncult. Alphaproteobacteria clone F2CB7; AY697876 | 100 | SAR11 | Alpha |
| BG27-1 | AY904481 | Uncult. Deltaproteobacteria clone JL-WNPG-T23; AY664120 | 97.9 | ND | Delta |
| BG27-7 | AY904487 | Uncult. Gammaproteobacteria clone BTM43; AY193157 | 91.8 | ND | Gamma |
| BG27-17 | AY904496 | Uncult. Gammaproteobacteria clone HRV1; Z88579 | 98.5 | OM60 | Gamma |
| BG27-2 | AY904482 | Isolate, Synechococcus sp. strain WH 8109; AY172836 | 99.6 | Synechococcus | Cyanobacteria |
| BG27-9 | AY904489 | Identical to BG27-2, Synechococcus sp. strain WH 8109 | 99.6 | Synechococcus | Cyanobacteria |
| BG27-18 | AY904497 | Identical to BG27-2, Synechococcus sp. strain WH 8109 | 99.6 | Synechococcus | Cyanobacteria |
| BG27-3 | AY904483 | Isolate, Synechococcus sp. strain WH 8109; AY172836 | 99.2 | Synechococcus | Cyanobacteria |
| BG27-11 | AY904491 | Uncult. actinobacteria clone MB11A03; AY033296 | 100 | SAR432 | Actinobacteria |
| Enriched | |||||
| BG29-3 | AY904502 | Isolate K2-53B; AY345413 | 96.8 | Roseobacter | Alpha |
| BG29-4 | AY904503 | Isolate RED15; AY136124 | 98.7 | Roseobacter | Alpha |
| BG29-7 | AY904506 | Uncult. Alphaproteobacteria clone 128-19L; AF473915 | 97.9 | Roseobacter | Alpha |
| BG29-9 | AY904508 | Uncult. Alphaproteobacteria clone CD4C7; AY038531 | 97.1 | Roseobacter | Alpha |
| BG29-10 | AY904509 | Isolate HRV4# HpaAS2; Z88582 | 99.6 | Roseobacter | Alpha |
| BG29-15 | AY904513 | Uncult. Alphaproteobacteria clone PI_RT68; AY580445 | 99.6 | Roseobacter | Alpha |
| BG29-13 | AY904512 | Uncult. Alphaproteobacteria clone MB11B03; AY033298 | 99.1 | SAR116 | Alpha |
| BG29-18 | AY904516 | Uncult. Roseospirillum sp. clone 1B9; AJ627987 | 90.6 | ND | Alpha |
| BG29-5 | AY904504 | Uncult. Gammaproteobacteria clone PI_4r3b; AY580815 | 96.5 | ND | Unknown |
| BG29-16 | AY904514 | Uncult. Gammaproteobacteria clone JH10_C68; AY568820 | 93.7 | OM60 | Gamma |
| BG29-17 | AY904515 | Uncult. Gammaproteobacteria clone CRO-FL8; AF141586 | 96.4 | SAR92 | Gamma |
| BG29-20 | AY904518 | Isolate HTCC2080; AY386339 | 93.4 | OM60 | Gamma |
| BG29-1 | AY904500 | Uncult. Gammaproteobacteria clone HRV33; Z88589 | 99.2 | SAR86 | Gamma |
| BG29-19 | AY904517 | Uncult. Gammaproteobacteria clone HRV33; Z88589 | 99.1 | SAR86 | Gamma |
| BG29-6 | AY904505 | Uncult. Proteobacteria clone CD5F2; AY038415 | 89.3 | ND | Unknown |
| BG29-2 | AY904501 | Uncult. Synechococcus sp. clone A715015; AY125371 | 99.4 | Synechococcus | Cyanobacteria |
| BG29-11 | AY904510 | Isolate, Synechococcus sp. strain WH 8109; AY172836 | 99.5 | Synechococcus | Cyanobacteria |
| BG29-8 | AY904507 | Uncult. Planctomycetes clone K2-30-19; AY344412 | 88.2 | ND | Planctomycetes |
| BG29-12 | AY904511 | Uncult. Planctomycetes clone PI_4e8e; AY580433 | 93.2 | ND | Planctomycetes |
| BG29-14 | AY573531 | Psychroserpens burtonensis ACAM188; U62913 | 94.4 | Flavobacteria | Bacteroidetes |
Clone libraries from the control microcosm C1 and the enriched microcosm NUT1 were constructed from community DNA samples collected on day 7. Alpha, Alphaproteobacteria; Gamma, Gammaproteobacteria; Delta, Deltaproteobacteria; ND, not determined; Uncult., uncultured.
T-RFLP analysis.
The T-RFLP signature from day 1 (and the initial water) (data not shown) demonstrated the presence of eight distinguishable peaks. Based on sequence data from the day 7 clone libraries, the dominant peak could be assigned to bacteria in the Roseobacter clade. Other peaks that could be identified belonged to the SAR86 clade, Synechococcus, and the SAR11 clade (Fig. 4A and B). On day 7, the T-RFLP signature from control microcosm C1 had four distinct peaks of similar height, indicating an abundance of bacteria in the Roseobacter clade, Synechococcus, the SAR11 clade, and a Flexibacter relative (Fig. 4C). In contrast, the Roseobacter peak largely dominated the T-RFLP fingerprint from enriched microcosm NUT1 (Fig. 4D).
The relative abundance of the dominant microbial groups in the T-RFLP chromatograms was tracked each day (Fig. 4E and F). Roseobacter was the dominant group in the control microcosms during the first few days of incubation, but its relative abundance decreased from day 3, concomitant with an increase in the relative abundance of Synechococcus. In contrast, the Roseobacter peak in the enriched microcosms increased substantially from day 2 onwards and remained the dominant peak until day 8. Synechococcus was also present in the enriched microcosms but did not dominate the T-RFLP signatures as much as in the controls. The peaks corresponding to a Flexibacter relative and the SAR11 clade remained evident throughout the experiment, although they were more pronounced in the control microcosms.
DGGE analysis.
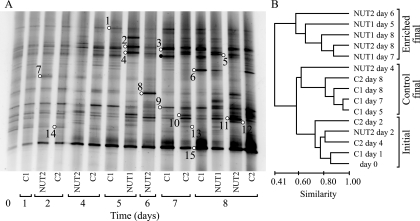
Analysis of the composition of the bacterial assemblages in the microcosms using DGGE of PCR-amplified 16S rRNA gene fragments revealed the presence of 25 to 33 bands in each sample (Fig. 5A). The DGGE fingerprints indicated that the samples formed three clusters (Fig. 5B), essentially separating the early samples in all microcosms (days 0 to 4) from the later samples (days 5 to 8) in the enriched microcosms and later samples in the control microcosms. The sample from enriched microcosm NUT2 at day 4 deviated from this pattern and clustered with the final control samples. Changes were less pronounced in the control microcosms.
FIG. 5.
DGGE analysis of the composition of the bacterial assemblage over 8 days. (A) DGGE profiles in the enriched and control microcosms. (B) Dendrogram comparing the bacterial DGGE fingerprints of samples from the microcosm experiment. Bands marked by open circles denote phylotypes identified by sequencing; adjoined numbers refer to phylotypes with the prefix DI in Table 4. The band marked by a filled black circle could be assigned to Bacteroidetes, but poor sequence quality prevented detailed identification.
The phylogenetic affiliations of major phylotypes in the DGGE gel are shown in Table 4. Two phylotypes (DI-1 and DI-3) that belonged to the SAR11 clade were primarily found in the control microcosms (Fig. 5A). An unidentified Bacteroidetes phylotype was also confined to the control microcosms. Three Bacteroidetes phylotypes (DI-2, DI-4, and DI-5) were affiliated with different members of the genus Tenacibaculum in the family Flavobacteriaceae and were found mainly in the enriched microcosms. This is consistent with recent findings that members of the Flavobacteriaceae are selectively favored during phytoplankton blooms (29). Roseobacter clade phylotypes (i.e., DI-10, DI-11, and DI-12) showed the strongest band intensities in the enriched microcosms, although they were also detected in the controls. These phylotypes were closely related to Roseobacter sequences in our 16S rRNA gene clone libraries (i.e., ≤2 mismatches over approximately 500 bp to clone BG27-14, BG29-15, and BG27-10, respectively). The conspicuous band at the bottom of the gel (DI-15) was identified as Synechococcus. This band increased in intensity from day 5 onwards, more so in the control microcosms.
TABLE 4.
Phylogenetic affiliations of bacterial 16S rRNA gene sequences from bands excized from the DGGE gel, as indicated by BLAST analyses in GenBanka
| Phylotype | GenBank accession no. | Closest relative in GenBank; accession no. | Similarity (%) | Group | Microcosms where mostly found |
|---|---|---|---|---|---|
| DI-1 | AY919593 | Uncult. Alphaproteobacteria clone AEGEAN_233; AF406547 | 93.5 | SAR11 | Control |
| DI-2 | AY573534 | Uncult. Bacteroidetes clone BY-65; AJ298376 | 98.8 | Bacteroidetes | Enriched |
| DI-3 | AY919594 | Uncult. Alphaproteobacteria clone MB11D08; AY033303 | 98.8 | SAR11 | Control |
| DI-4 | AY919595 | Uncult. Bacteroidetes clone BY-65; AJ298376 | 98.9 | Bacteroidetes | Enriched |
| DI-5 | AY573533 | Uncult. Bacteroidetes clone BY-65; AJ298376 | 97.9 | Bacteroidetes | Enriched |
| DI-6 | AY573532 | Uncult. Bacteroidetes clone ARKDMS-428; AF468245 | 90.1 | Bacteroidetes | Control |
| DI-7 | AY919596 | Prasinophycean chloroplast; AF268228 | 97.4 | Algae | Control/enriched |
| DI-8 | AY573535 | Uncult. Bacteroidetes clone KB35; AB074939 | 98.1 | Bacteroidetes | Enriched |
| DI-9 | AY919597 | Uncult. Alphaproteobacteria clone PI_4f4d; AY580536 | 95.2 | SAR116 | Control |
| DI-10 | AY919598 | Uncult. Roseobacter clone NAC11-1; AF245630 | 99.8 | Roseobacter | Enriched |
| DI-11 | AY919599 | Uncult. Alphaproteobacteria clone SW8; AB047143 | 99.4 | Roseobacter | Enriched |
| DI-12 | AY919600 | Uncult. Alphaproteobacteria clone SW8; AB047143 | 99.8 | Roseobacter | Enriched |
| DI-13 | AY919601 | Alteromonas sp. strain UST010723-005; AY241400 | 100 | Gammaproteobacteria | Enriched |
| DI-14 | AY919602 | Uncult. Alphaproteobacteria clone ZA3318c; AF382114 | 97.7 | Rhodobacterales | Control |
| DI-15 | AY919603 | Uncult. Synechococcus clone NAC1-5; AF245618 | 99.0 | Cyanobacteria | Control/enriched |
Also shown are the microcosms where the phylotypes were mostly found. Uncult., uncultured.
DISCUSSION
The addition of inorganic nutrients and the subsequent phytoplankton bloom and its decay profoundly altered the conditions for growth of bacteria in the microcosms. This was evident not only from variables such as bacterial biomass production but also from the changes in consumption rates of dimethylated sulfur compounds and the composition of the bacterial assemblage.
The bacterial growth efficiencies calculated at the end of the experiment, approximately 10% in the control microcosms and 27% in the enriched microcosms, were an indication of different conditions for bacterial growth as a consequence of the phytoplankton bloom. del Giorgio and Cole (3) showed that the majority of bacterial growth efficiency values from open ocean samples fall between 9 and 32%. Growth efficiencies of approximately 10% were recently found for bacteria growing in lake water cultures amended with aged DOC (i.e., DOC that was refractory or of low quality) (4). Using a dilution culture approach similar to that used by Eiler et al. (4), but with freshly collected seawater from an estuarine site in the northern Baltic Sea, bacterioplankton growth efficiencies averaging 25 to 30% were found (41). The significant difference in bacterial growth efficiency found between the control and enriched microcosms in our experiment may have been due to different concentrations and/or quality of the available DOM or possibly to differences in the availability of mineral nutrients.
A large fraction of phytoplankton DMSP is transformed by free-living bacteria upon its release from algal cells. As a consequence, the composition of bacterial assemblages and their physiological demands could be major determinants of the fate of DMSP carbon and sulfur (17, 33, 39). Three separate observations suggest that DMSP was an important source of C for bacterial growth in our experiment. First, approximately 5% of the bacterial C demand could be met by DMSP alone. Second, the nutrient addition bioassay experiment indicated that the addition of DMSP caused an increase in bacterial biomass production similar to that with glucose. Similar results were recently found in the northwest Mediterranean Sea (J. Pinhassi et al., unpublished data). Third, microautoradiography showed that 27 to 51% of the bacteria were actively assimilating [35S]DMSP, which was similar to or slightly lower than the proportion of cells assimilating [3H]leucine (38). We therefore conclude that DMSP was readily assimilated by a major fraction of the microcosm bacterioplankton assemblage.
Bacterial activity responded to the onset of the phytoplankton bloom in the enriched microcosms beginning on day 3 and continued to increase during senescence of the bloom. Microbial consumption of DMSP and DMS followed the increase in bacterial biomass production with a time lag of 2 to 3 days (i.e., Fig. 2 and 3). This implied that the early DMS production pulse that resulted in a transient accumulation of DMS up to day 3 could not be attributed to bacterial activity. Similar DMS accumulation occurred in all microcosms and therefore most likely resulted from enclosure effects on the initial algal assemblage (e.g., stress and mortality), possibly in combination with grazing by tunicates that escaped the prefiltration. Notably, the peak in DMS concentration was similar to the initial concentration of DMSPp (∼10 nM). Following the algal bloom, DMS production increased again in the enriched microcosms in conjunction with increased DMSPp and DMSPd turnover and bacterial biomass production, indicating the central role of microorganisms during the latter phase of the experiment.
During the initial phase of the phytoplankton bloom, the DMSPp-to-Chl a ratio decreased substantially (to 9 nmol μg−1) compared to that in the controls (40 to 70 nmol μg−1) (Fig. 2B, inset). Indeed, DMSPp-to-Chl a ratios in the range of 3 to 30 nmol μg−1 are generally found in productive coastal or estuarine waters, while ratios on the order of 100 nmol μg−1 or higher are typical of open-ocean, oligotrophic waters (17). It is reasonable to assume that sulfur-rich DOM components other than DMSP (e.g., amino acids in proteins) were also released as the bloom decayed and could contribute significantly to bacterial S demand. This could explain the observation that the contribution of DMSP to bacterial S demand in the enriched microcosms was only about one-eighth of that in the controls (Table 1). Since phytoplankton abundance and composition largely determine the rate of primary production and DMSP synthesis in the sea, we infer that the DMSPp-to-Chl a ratio can be used as a proxy for the quantitative role of DMSP as a source of organic S to bacteria over time scales of phytoplankton blooms and longer.
Further insight into the relative importance of DMSP for bacterial S budgets can be gained from an analysis of the efficiency of DMSP S assimilation into macromolecules. During the first days of the experiment, the efficiency at which heterotrophic bacteria incorporated 35S from [35S]DMSP into TCA-insoluble macromolecules (e.g., proteins) was approximately 30 to 40% in all microcosms (data not shown). Starting at day 5 and coinciding with the decay of the bloom and the associated changes in bacterioplankton composition, the assimilation efficiency decreased to 13% in the enriched microcosms. At the same time, the efficiencies remained at 29% in the control microcosms. This indicated that the DMSP S assimilation efficiencies did not depend on the concentration of inorganic nutrients per se but were related to the availability of alga-derived DOM. We did not determine concentrations of S-containing organic molecules other than DMSP and DMS. However, previous experiments have shown that bacterial aminopeptidase activity (responsible for the transformation of polypeptides into easily usable amino acids) typically increases during the decay of phytoplankton blooms (28, 29). Thus, a likely explanation for the lower DMSP S assimilation efficiency in the enriched microcosms would be that bacteria associated with decaying phytoplankton relied less on DMSP as a source of S because their metabolism was geared towards the use of other readily available dissolved organic S compounds.
Kiene et al. (17) hypothesized that assimilation of DMSP S by marine bacterioplankton through the demethylation pathway depends on the bacterial S demand, which is a function of bacterial growth and biomass stoichiometry and the availability of dissolved DMSP. In the original hypothesis, availability of DMSP was represented by its concentration, but Simó (33) redefined availability as the rate of release of DMSP from algal cells. The recognition of what controls DMSP assimilation is important, since the demethylation pathway (which results in S assimilation) generally dominates bacterial DMSP transformations in the surface ocean and diverts DMSP from being cleaved to climatically active DMS. This “bacterial switch” can be regarded as a major biotic control on DMS production. Our observations strongly suggest that it is neither the DMSP concentration per se nor its turnover but its relative contribution to the overall dissolved organic S/organic matter pool that controls how efficiently bacteria assimilate DMSP S and thereby potentially divert it from DMS production.
The lower efficiency of bacterial DMSP S assimilation following the phytoplankton bloom was unexpected. This finding might reflect a shift in active species toward those less specialized in DMSP uptake or a switch to the use of alternative S sources such a sulfur-containing amino acids by the active species. Following massive algal mortality at the end of a phytoplankton bloom and the subsequent release of DOM, substantial changes in the composition of bacterioplankton assemblages have been shown to occur and to coincide with changes in cell-specific growth rates and hydrolytic ectoenzyme activities (28, 31). Whether an analogous argument could be applied to the changes in DMSP utilization was investigated in the present study by tracking the compositional changes of bacterioplankton in the microcosms using a series of molecular biology techniques with different levels of phylogenetic resolution.
Results from both FISH and T-RFLP analyses showed that the abundance of the Roseobacter clade nearly doubled in the enriched microcosms but remained constant or decreased in the control microcosms. This was in agreement with results from the DGGE analysis (changes both in the presence/absence and in the relative abundance of particular phylotypes) and the 16S rRNA gene clone libraries (as a doubling in the number of Roseobacter clones in enriched compared to control microcosms). For Bacteroidetes, FISH results indicated that they constituted approximately 10% of the bacterial assemblage throughout the experiment. Nevertheless, as observed for the Roseobacter clade, the DGGE analysis indicated that there were substantial changes in the relative abundance of phylotypes within the Bacteroidetes phylum during the experiment. Taken together, our analysis of bacterial diversity revealed that there was a shift from a bacterial assemblage dominated by SAR11, Roseobacter, and Bacteroidetes before the phytoplankton bloom to an assemblage largely dominated by Roseobacter, but also with notable contributions of certain Bacteroidetes and Gammaproteobacteria, during and after the bloom. These changes were largely consistent with the observed changes in bacterial activity measurements in general and with the changes in DOM dynamics, as evidenced by DMSP turnover, in particular.
Several studies have indicated the importance of bacteria in the Roseobacter clade for the transformation of DMSP and other S-rich compounds (23). González et al. (10) found that Roseobacter species could mediate several steps in the processing of DMSP. Notably, several members of this clade harbor both the cleavage and the demethylation/demethiolation pathways for DMSP degradation (10). MicroFISH using [35S]DMSP has now linked bacteria in the Roseobacter clade to the active uptake and processing of DMSP (21, 38). In our experiments, MicroFISH counts with a Roseobacter-specific probe previously showed that up to 87% of the Roseobacter cells in the enriched microcosms were actively incorporating [35S]DMSP (38), despite the fact that lower DMSP assimilation efficiencies in these microcosms would make detection of DMSP S uptake less sensitive. Although bacteria in the Roseobacter clade on the whole were an important group in both the control and the enriched microcosms, there appeared to be a shift in the relative abundance of phylotypes in the enriched microcosms towards taxa that utilized DMSP S less efficiently, possibly because of the availability of other sources of reduced sulfur (e.g., amino acids and proteins).
While previous studies using MicroFISH with [35S]DMSP established that bacteria in the Roseobacter clade are involved in the processing of DMSP, they also indicated that DMSP assimilation is a widespread characteristic among marine Bacteroidetes and Gammaproteobacteria (21, 38). However, only a few studies have documented the role of specific bacterial taxa at the genus or species level (24). In our study, nearly half of the bacterial cells incorporated [35S]DMSP, while Roseobacter cells accounted for no more than 20% of the total bacterial abundance. These findings support the position that diverse taxa, in addition to Roseobacter, contribute to the transformation of DMSP. Sequencing of 16S rRNA gene clones and DGGE bands from the enriched microcosms identified specific members of the Roseobacter clade that are candidates for an active role in the processing of DMSP. Still, we are unaware of any studies aimed at discerning the physiological adaptations of Bacteroidetes or Gammaproteobacteria to perform DMSP transformations. Nevertheless, our analyses suggest that flavobacteria (Bacteroidetes phylum) and oligotrophic marine Gammaproteobacteria (e.g., OM60 and SAR92 clade bacteria) may also be relevant taxa to consider, in addition to the Roseobacter clade, when addressing the role of bacterioplankton community dynamics in controlling DMSP cycling and, from the opposite perspective, the role of DMSP in the growth and composition of heterotrophic bacterioplankton assemblages.
Acknowledgments
The skillful efforts of Laura Linn during the experiment are gratefully acknowledged. This study was made possible by the logistic and housing support provided by the Dauphin Island Sea Laboratory. We are grateful for the constructive criticism and comments on the manuscript from three anonymous reviewers.
This project was supported by grants from the U.S.-Spain Science and Technology Program 2000 (grant 20185), the European Union (grant EVK3-CT-2002-00078 BASICS), and the Swedish Research Council (grant 2003-2692) to J.P. and by a grant (MCB-0315200) from the U.S. National Science Foundation to M.A.M. Additional support for R.P.K. was provided by the U.S. National Science Foundation (grants OCE-9907471 and OPP-0221748).
REFERENCES
- 1.Charlson, R. J., J. E. Lovelock, M. O. Andreae, and S. G. Warren. 1987. Oceanic phytoplankton, atmospheric sulphur, cloud albedo and climate. Nature 326:655-661. [Google Scholar]
- 2.Cuhel, R. L., P. B. Ortner, and D. R. S. Lean. 1984. Night synthesis of protein by algae. Limnol. Oceanogr. 29:731-744. [Google Scholar]
- 3.del Giorgio, P. A., and J. J. Cole. 1998. Bacterial growth efficiency in natural aquatic ecosystems. Annu. Rev. Ecol. Syst. 29:503-541. [Google Scholar]
- 4.Eiler, A., S. Langenheder, S. Bertilsson, and L. J. Tranvik. 2003. Heterotrophic bacterial growth efficiency and community structure at different natural organic carbon concentrations. Appl. Environ. Microbiol. 69:3701-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fagerbakke, K. M., M. Heldal, and S. Norland. 1996. Content of carbon, nitrogen, oxygen, sulfur and phosphorous in native and cultured bacteria. Aquat. Microb. Ecol. 10:15-27. [Google Scholar]
- 6.Gasol, J. M., and P. A. del Giorgio. 2000. Using flow cytometry for counting natural planktonic bacteria and understanding the structure of planktonic bacterial communities. Sci. Mar. 64:197-224. [Google Scholar]
- 7.Gasol, J. M., U. L. Zweifel, F. Peters, J. A. Fuhrman, and Å. Hagström. 1999. Significance of size and nucleic acid content heterogeneity as measured by flow cytometry in natural planktonic bacteria. Appl. Environ. Microbiol. 65:4475-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giovannoni, S., and M. Rappé. 2000. Evolution, diversity and molecular ecology of marine prokaryotes, p. 47-84. In D. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss, New York, N.Y.
- 9.Giovannoni, S. J. 1991. The polymerase chain reaction, p. 177-201. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, N.Y.
- 10.González, J. M., R. P. Kiene, and M. A. Moran. 1999. Transformation of sulfur compounds by an abundant lineage of marine bacteria in the α-subclass of the class Proteobacteria. Appl. Environ. Microbiol. 65:3810-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.González, J. M., R. Simó, R. Massana, J. S. Covert, E. O. Casamayor, C. Pedrós-Alió, and M. A. Moran. 2000. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 66:4237-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagström, Å., T. Pommier, F. Rohwer, K. Simu, W. Stolte, D. Svensson, and U. L. Zweifel. 2002. Use of 16S ribosomal DNA for delineation of marine bacterioplankton species. Appl. Environ. Microbiol. 68:3628-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho, T.-Y., M. I. Scranton, G. T. Taylor, R. Varela, R. C. Thunell, and F. Muller-Karger. 2002. Acetate cycling in the water column of the Cariaco Basin: seasonal and vertical variability and implication for carbon cycling. Limnol. Oceanogr. 47:1119-1128. [Google Scholar]
- 14.Kiene, R. P. 1996. Production of methane thiol from dimethylsulfoniopropionate in marine surface waters. Mar. Chem. 54:69-83. [Google Scholar]
- 15.Kiene, R. P., and G. Gerard. 1994. Determination of trace levels of dimethylsulfoxide (DMSO) in seawater and rainwater. Mar. Chem. 47:1-12. [Google Scholar]
- 16.Kiene, R. P., and L. J. Linn. 2000. Distribution and turnover of dissolved DMSP and its relationships with bacterial production and dimethylsulfide in the Gulf of Mexico. Limnol. Oceanogr. 45:849-861. [Google Scholar]
- 17.Kiene, R. P., L. J. Linn, and J. A. Bruton. 2000. New and important roles for DMSP in marine microbial communities. J. Sea Res. 43:209-224. [Google Scholar]
- 18.Kiene, R. P., L. J. Linn, J. González, M. A. Moran, and J. A. Bruton. 1999. Dimethylsulfoniopropionate and methanethiol are important precursors of methionine and protein-sulfur in marine bacterioplankton. Appl. Environ. Microbiol. 65:4549-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirchman, D. L., E. Knees, and R. Hodson. 1985. Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic systems. Appl. Environ. Microbiol. 49:599-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, W.-T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malmstrom, R. R., R. P. Kiene, and D. L. Kirchman. 2004. Identification and enumeration of bacteria assimilating dimethylsulfoniopropionate (DMSP) in the North Atlantic and Gulf of Mexico. Limnol. Oceanogr. 49:597-606. [Google Scholar]
- 22.Martinez, J., D. C. Smith, G. F. Steward, and F. Azam. 1996. Variability in ectohydrolytic enzyme activities of pelagic marine bacteria and its significance for substrate processing in the sea. Aquat. Microb. Ecol. 10:223-230. [Google Scholar]
- 23.Moran, M. A., J. M. González, and R. P. Kiene. 2003. Linking a bacterial taxon to sulfur cycling in the sea: studies of the marine Roseobacter group. Geomicrobiol. J. 20:1-14. [Google Scholar]
- 24.Mou, X., M. A. Moran, R. Stepanauskas, J. M. González, and R. E. Hodson. 2005. Flow-cytometric cell sorting and subsequent molecular analyses for culture-independent identification of bacterioplankton involved in dimethylsulfoniopropionate transformations. Appl. Environ. Microbiol. 71:1405-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Norland, S. 1993. The relationship between biomass and volume of bacteria, p. 303-307. In P. F. Kemp, B. F. Sherr, E. B. Sherr, and J. J. Cole (ed.), Handbook of methods of aquatic microbial ecology. Lewis Publishers, Chelsea, Mich.
- 26.Pennock, J. R. 1999. Nutrient behavior and phytoplankton production in Gulf of Mexico estuaries, p. 109-162. In T. S. Bianchi, J. R. Pennock, and R. R. Twilley (ed.), Biogeochemisty of Gulf of Mexico estuaries. John Wiley & Sons, New York, N.Y.
- 27.Pernthaler, J., F. O. Glöckner, W. Schönhuber, and R. Amann. 2001. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. Methods Microbiol. 30:207-226. [Google Scholar]
- 28.Pinhassi, J., F. Azam, J. Hemphälä, R. A. Long, J. Martinez, U. L. Zweifel, and Å. Hagström. 1999. Coupling between bacterioplankton species composition, population dynamics, and organic matter degradation. Aquat. Microb. Ecol. 17:13-26. [Google Scholar]
- 29.Pinhassi, J., M. M. Sala, H. Havskum, F. Peters, O. Guadayol, A. Malits, and C. Marrasé. 2004. Changes in bacterioplankton composition under different phytoplankton regimens. Appl. Environ. Microbiol. 70:6753-6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rich, J. H., H. W. Ducklow, and D. L. Kirchman. 1996. Concentrations and uptake of neutral monosaccharides along 140′W in the equatorial Pacific: contribution of glucose to heterotrophic bacterial activity and the DOM flux. Limnol. Oceanogr. 41:595-604. [Google Scholar]
- 31.Riemann, L., G. F. Steward, and F. Azam. 2000. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 66:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schauer, M., R. Massana, and C. Pedrós-Alió. 2000. Spatial differences in bacterioplankton composition along the Catalan coast (NW Mediterranean) assessed by molecular fingerprinting. FEMS Microbiol. Ecol. 33:51-59. [DOI] [PubMed] [Google Scholar]
- 33.Simó, R. 2001. Production of atmospheric sulfur by oceanic plankton: biogeochemical, ecological and evolutionary links. Trends Ecol. Evol. 16:287-294. [DOI] [PubMed] [Google Scholar]
- 34.Simó, R., S. D. Archer, C. Pedrós-Alió, L. Gilpin, and C. E. Stelfox-Widdicombe. 2002. Coupled dynamics of dimethylsulfoniopropionate and dimethylsulfide cycling and the microbial food web in surface waters of the North Atlantic. Limnol. Oceanogr. 47:53-61. [Google Scholar]
- 35.Simon, M., and F. Azam. 1989. Protein content and protein synthesis rates of planktonic marine bacteria. Mar. Ecol. Prog. Ser. 51:201-213. [Google Scholar]
- 36.Smith, D. C., and F. Azam. 1992. A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar. Microb. Food Webs 6:107-114. [Google Scholar]
- 37.Suttle, C. A., A. M. Chan, and J. A. Fuhrman. 1991. Dissolved free amino acids in the Sargasso Sea: uptake and respiration rates, turnover times, and concentrations. Mar. Ecol. Prog. Ser. 70:189-199. [Google Scholar]
- 38.Vila, M., R. Simó, R. P. Kiene, J. Pinhassi, J. M. González, M. A. Moran, and C. Pedrós-Alió. 2004. Use of microautoradiography combined with fluorescence in situ hybridization to determine dimethylsulfoniopropionate incorporation by marine bacterioplankton taxa. Appl. Environ. Microbiol. 70:4648-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoch, D. C. 2002. Dimethylsulfoniopropionate: its sources, role in the marine food web, and biological degradation to dimethylsulfide. Appl. Environ. Microbiol. 68:5804-5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zubkov, M. V., B. M. Fuchs, S. D. Archer, R. P. Kiene, R. Amann, and P. H. Burkill. 2001. Linking the composition of bacterioplankton to rapid turnover of dissolved dimethylsulphoniopropionate in an algal bloom in the North Sea. Environ. Microbiol. 3:304-311. [DOI] [PubMed] [Google Scholar]
- 41.Zweifel, U. L., B. Norrman, and Å. Hagström. 1993. Consumption of dissolved organic carbon by marine bacteria and demand for inorganic nutrients. Mar. Ecol. Prog. Ser. 101:23-32. [Google Scholar]