Abstract
During the fermentation of sake, cells of Saccharomyces cerevisiae are exposed to high concentrations of ethanol, thereby damaging the cell membrane and functional proteins. l-Proline protects yeast cells from damage caused by freezing or oxidative stress. In this study, we evaluated the role of intracellular l-proline in cells of S. cerevisiae grown under ethanol stress. An l-proline-accumulating laboratory strain carries a mutant allele of PRO1, pro1D154N, which encodes the Asp154Asn mutant γ-glutamyl kinase. This mutation increases the activity of γ-glutamyl kinase and γ-glutamyl phosphate reductase, which catalyze the first two steps of l-proline synthesis and which together may form a complex in vivo. When cultured in liquid medium in the presence of 9% and 18% ethanol under static conditions, the cell viability of the l-proline-accumulating laboratory strain is greater than the cell viability of the parent strain. This result suggests that intracellular accumulation of l-proline may confer tolerance to ethanol stress. We constructed a novel sake yeast strain by disrupting the PUT1 gene, which is required for l-proline utilization, and replacing the wild-type PRO1 allele with the pro1D154N allele. The resultant strain accumulated l-proline and was more tolerant to ethanol stress than was the control strain. We used the strain that could accumulate l-proline to brew sake containing five times more l-proline than what is found in sake brewed with the control strain, without affecting the fermentation profiles.
Sake is a traditional Japanese alcoholic beverage made from steamed rice by multiple parallel fermentations of the fungus Aspergillus oryzae and the yeast Saccharomyces cerevisiae, which produce saccharification enzymes and ethanol from glucose, respectively. During sake fermentation, yeast cells are exposed to various stresses under anaerobic conditions, including high concentrations of ethanol (∼20% [vol/vol]) and low temperature (∼15°C). Ethanol is toxic. It damages the cell membrane and functional proteins (22), gradually reducing cell viability and leading to cell death during fermentation. Therefore, the use of ethanol-resistant yeast strains should make it possible to reduce the fermentation time.
Amino acids and organic acids produced by yeast during fermentation influence the taste of sake. The effects of organic acids such as malic, fumaric, and succinic acids have been evaluated by disrupting the genes encoding the appropriate metabolic enzymes in sake yeast strains (2, 21, 23). Little attention has been paid to the effects of amino acids during sake fermentation. By breeding yeasts with various amino acid composition profiles, it may be possible to expand the diversity of sake tastes.
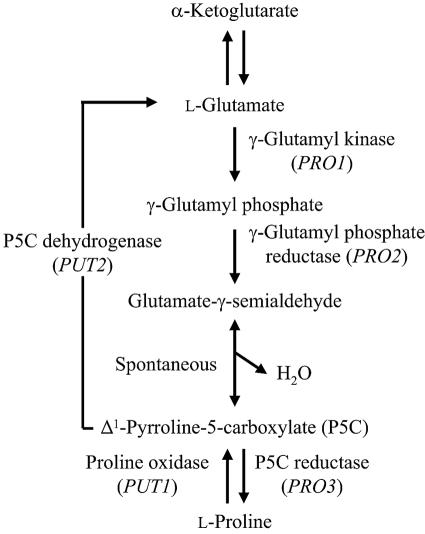
l-Proline is an osmoprotectant (10, 12) and a sweet amino acid that helps protect yeast cells from damage by freezing, desiccation, or oxidative stress (26, 27, 36-38). l-Proline enhances the stability of proteins and membranes in environments with low water activity or high temperature (30) and inhibits aggregation during protein refolding (32). These observations suggest that intracellular l-proline could play a crucial role in reducing ethanol stress by preventing protein denaturation and membrane disorder during sake fermentation. S. cerevisiae synthesizes l-proline from l-glutamate via a pathway consisting of three enzymes: γ-glutamyl kinase (γ-GK) (the PRO1 gene product), γ-glutamyl phosphate reductase (the PRO2 gene product), and Δ1-pyrroline-5-carboxylate (P5C) reductase (the PRO3 gene product) (Fig. 1) (6, 39). l-Proline is converted to l-glutamate within mitochondria in two steps by the enzymes proline oxidase (the PUT1 gene product) and P5C dehydrogenase (the PUT2 gene product) (Fig. 1) (5, 40). We previously isolated a mutant of S. cerevisiae that was resistant to the l-proline analog l-azetidine-2-carboxylic acid (AZC), accumulated l-proline, and tolerated freezing (36). This mutant has a single mutation in PRO1 that results in an Asp154Asn amino acid substitution in γ-GK and increased activity of both γ-GK and γ-glutamyl phosphate reductase, which may be part of a single protein complex in vivo (27, 38).
FIG. 1.
Biosynthesis and metabolism of l-proline in Saccharomyces cerevisiae. Genes encoding enzymes are shown in parentheses.
Our objectives in this study were (i) to determine if l-proline reduces ethanol stress in yeast cells and (ii) to determine if cells that accumulate l-proline alter the sake fermentation process or product. We report here the protective effect of l-proline on ethanol stress in yeast cells. In addition, to test the hypothesis that the improved ethanol tolerance is due to l-proline accumulation, we constructed the l-proline-accumulating sake yeast and analyzed its fermentation profiles during sake brewing.
MATERIALS AND METHODS
Strains and plasmids.
The S. cerevisiae strains used in this study are listed in Table 1. Escherichia coli strain DH5α [F− λ− φ80lacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) supE44 thi-1 gyrA96 relA1] and the plasmid vector pBluescript II SK(+) (Toyobo Biochemicals, Osaka, Japan) were used to subclone the PUT1 gene.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Background and/or description |
|---|---|---|
| MB329-17C | α trp1 ura3-52 put1-54 | S288C and Σ1278b; put1 mutant |
| FH515 | α trp1 ura3-52 put1-54 pro1D154N | MB329-17C; pro1 mutant; the l-proline-accumulating laboratory strain |
| XUW-14 | α ura3 trp1 | Sake yeast strain Kyokai no. 14 (K-14) |
| XUW-TRP | α ura3 TRP1 | XUW-14; TRP1 revertant; the control sake yeast strain |
| XUDput1 | α ura3 trp1 put1::TRP1 | XUW-14; put1 disruptant |
| XUDput1-MT | α ura3 trp1 put1::TRP1 pro1D154N | XUDput1; pro1 mutant, the l-proline-accumulating sake yeast strain |
Plasmid pRS414 (Stratagene, La Jolla, Calif.), which carries the TRP1 gene, was used to disrupt the PUT1 gene. Plasmid pRS404 (Stratagene), which contains the TRP1 gene, was used for the integration of the TRP1 gene into strain XUW-14. Two yeast plasmids, pRS-D154NPRO1, carrying the pro1D154N allele of PRO1 (27), and pRS406 (Stratagene), carrying the URA3 gene, were used to integrate the pro1D154N allele into strain XUDput1. Yeast episomal plasmids pUV2, carrying the URA3 gene, and pTV3, carrying the TRP1 gene, were used to complement the auxotrophic marker (28).
Culture media.
The media used for growth of S. cerevisiae were SD medium (20 g/liter glucose, 6.7 g/liter Bacto yeast nitrogen base without amino acids [Difco Laboratories, Detroit, Mich.]) and YPD medium (20 g/liter glucose, 10 g/liter Bacto yeast extract, 20 g/liter Bacto peptone). The SD medium contains 1 g/liter ammonium sulfate as the nitrogen source. When the put1 disruptant was grown, 1 g/liter monosodium l-glutamate or l-proline was used instead of ammonium sulfate as the sole source of nitrogen. Required supplements were added to the media for auxotrophic strains as necessary. Yeast strains were also cultured on SD agar plates with 100 μg/ml of the l-proline analogue AZC (Sigma Chemical, St. Louis, Mo.) or with 1 mg/ml of 5-fluoroorotic acid (35). E. coli strains were grown in Luria-Bertani medium (31) supplemented with 50 μg/ml ampicillin, as necessary. Media were solidified as necessary with 20 g/liter of agar.
Disruption of PUT1.
The enzymes used for DNA manipulation were obtained from Takara Bio (Ohtsu, Japan) and were used under the conditions recommended by the supplier. Conventional techniques were used for DNA manipulation and transformation as described previously (29). The DNA fragment of PUT1 was prepared by PCR with genomic DNA from S. cerevisiae MB329-17C and oligonucleotide primers based on the available nucleotide sequences. The forward primer was 5′-GAG GAT CCG AAC ACA AAC TCC A-3′, and the reverse primer was 5′-GCG GTA CCC CAA AAT CCT TAC A-3′ (the underlined sequences indicate the positions of the BamHI and KpnI restriction sites, respectively). A unique amplified band of 1.9 kb was digested with BamHI and KpnI and then ligated into the BamHI and KpnI sites of pBluescript II SK(+) to construct pBlue-PUT1. Plasmid pBlueDput1-TRP1 was constructed by deleting the 0.9-kb BalI-AatI fragment in PUT1 from pBlue-PUT1 and inserting the 2.6-kb ScaI-NaeI fragment containing TRP1 of plasmid pRS414 by blunt-end ligation. The 3.6-kb BamHI-KpnI fragment containing put1::TRP1 of pBlueDput1-TRP1 was integrated into the PUT1 locus in strain XUW-14 to construct strain XUDput1 by transformation. The Trp+ phenotype was selected, and the correct disruption was verified by chromosomal PCR analysis. To remove the influence of tryptophan auxotrophy, pRS404 was cut with MfeI in TRP1, and the linearized plasmid was introduced to integrate TRP1 into the control strain (XUW-TRP).
Assay of proline oxidase activity.
Proline oxidase (EC 1.4.3.2) activity was assayed by monitoring P5C-o-aminobenzaldehyde as previously described (7, 26). The enzyme is very unstable, so whole-cell extracts were prepared as previously described (7). Yeast cells were grown in 50 ml of SD medium or SD medium plus l-proline at 30°C for 48 h with shaking, collected on a 0.8-μm nitrocellulose filter (Whatman, Clifton, N.J.), and immediately immersed in liquid N2 for 10 s. Each filter was then placed in a small test tube containing 0.5 ml of 0.1 M HEPES buffer (pH 7.5) with 3 mM MgCl2 and kept on ice. Each tube was vortexed vigorously to transfer the cells from the filter to the buffer. A 0.4-ml portion of 10% l-proline was added to each tube and incubated without shaking at 30°C for 15 min. One hundred microliters of o-aminobenzaldehyde (6 mg/ml in 20% ethanol; Sigma Chemical) was added, followed by 0.5 ml of 10% trichloroacetic acid to stop the reaction. The tube was mixed, and the color was allowed to develop for 30 min. The A443 was recorded against a blank identical to the one described above but lacking l-proline. The millimolar extinction coefficient of the P5C-o-aminobenzaldehyde complex was 2.71. One unit of activity was defined as the amount of enzyme required to produce 1 nmol of P5C per min. Protein concentrations were determined by using a Bio-Rad protein assay kit (Hercules, Calif.) with bovine serum albumin as the standard (4).
Replacement of the wild-type PRO1 gene with the pro1D154N gene.
A two-step method was used to replace the wild-type PRO1 sequence with pro1D154N at the native chromosomal location. The 1.8-kb HindIII-SacI fragment from pRS-D154NPRO1 was ligated into the large fragment of pRS406 digested with HindIII and SacI to yield pRS406-D154NPRO1. Plasmid pRS406-D154NPRO1 was cut with XbaI in the pro1D154N allele, and the linearized DNA was integrated into the PRO1 locus of strain XUDput1 by transformation. The Ura+ phenotype was selected as a single-crossover transformant that duplicates the PRO1 locus (one copy is the wild type and the other is the mutant) with plasmid sequences in between. The transformant was cultured in 1 ml of YPD medium at 30°C for 24 h with shaking to obtain 5-fluoroorotic acid-resistant strains that have excised the plasmid and lost one of the two copies of the duplicated region by homologous crossover. Depending on the location of the crossover, the PRO1 allele that remains may be either the mutant or the wild type. The pro1D154N sake yeast strain XUDput1-MT was selected as an AZC-resistant phenotype due to the overproduction of l-proline (27, 36, 38). The construct was checked by direct sequencing of PCR products amplified from the chromosomal DNA.
Intracellular contents of l-proline.
For the determination of intracellular l-proline, yeast cells were grown to the stationary phase in 10 ml of SD medium, SD medium plus l-proline, or YPD medium at 30°C for 48 h under either shaking (120 rpm) or static conditions. Five milliliters of cell suspension was removed, and the cells were washed twice with 0.9% NaCl and suspended in 0.5 ml of distilled water. The 1.5-ml microcentrifuge tube containing cells was transferred to a boiling-water bath, and intracellular amino acids were extracted by boiling for 10 min. After centrifugation (15,000 × g, 5 min, 4°C), each supernatant was analyzed quantitatively with an amino acid analyzer (model L-8500A; Hitachi, Tokyo, Japan). l-Proline content was expressed as a percentage of dry weight.
Ethanol tolerance test.
Yeast cells were pregrown in 10 ml of YPD medium at 30°C for 2 days with shaking (120 rpm). The cells were washed twice with 0.9% NaCl, suspended in 100 ml of SD medium or SD medium plus ethanol (final concentration, 9% or 18%), and cultured at 15°C under static conditions. After 0, 2, 5, and 8 days, a 0.1-ml aliquot of the culture was removed, diluted in distilled water, and plated onto YPD plates. The number of viable cells was determined after incubation at 30°C for 2 days. The relative cell number was expressed as a percentage, which was calculated as follows: [(number of colonies after addition of ethanol)/(number of colonies before addition ethanol)] × 100.
For cultures growing in SD medium or SD medium plus 9% ethanol, after 0, 2, 5, and 8 days, intracellular amino acids were measured as described above. The amount of trehalose was measured by the anthrone method (9). Intracellular amino acids and trehalose contents were expressed as a percentage of dry weight.
Sake brewing.
Laboratory-scale sake was brewed (20) with a sake mash consisting of 160 g of steamed rice (α-rice), 40 g of koji rice (a culture of A. oryzae on steamed rice), and 260 ml of water added in three steps (at 4, 6, and 7 days). One-third of the amount specified was added each time. Each strain was grown in YPD medium at 30°C for 2 days under static conditions and inoculated into the mash to yield 1 × 107 cells per g of mash. Fermentation profiles were monitored by weight loss in conjunction with CO2 evolution. When about 65 g of mass had been lost (after ∼24 days of fermentation at 15°C), the sake mash was centrifuged (175 × g, 10 min, 4°C), and the supernatant was obtained as sake. Yeast cells in sake mash were isolated by centrifugation (175 × g, 10 min, 4°C) (25), and the number of viable cells and their intracellular l-proline content were determined.
General components of the sake, such as ethanol, glucose, amino acids, organic acids, and aroma compounds, were analyzed by the standard method established by the Japanese National Tax Administration Agency (8). The ethanol concentration in the sake was measured with a gas chromatograph (model GC-15A; Shimadzu, Kyoto, Japan). The amino acid and organic acid compositions were analyzed with an amino acid analyzer and a high-performance liquid chromatograph (model LC-6A; Shimadzu) equipped with a conductivity detector and an SCR-102H column (Shimadzu), respectively. The glucose concentration was determined using a Glucose CII-Test (Wako Pure Chemical Industries, Osaka, Japan). In general, the sake meter indicates the apparent specific gravity of sake and is basically a Baumé meter, which works on the principle that alcohol is lighter than water while glucose is heavier. The lower the sake meter value, the heavier the gravity (20). Water is given a value of 0 at 15°C and a Baumé value of 1 corresponding to a sake meter value of −10.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the PRO1 and PUT1 genes are M85293 and M18107, respectively.
RESULTS
l-Proline accumulation and tolerance to ethanol stress.
We compared the number of viable cells (Fig. 2) and the intracellular l-proline content (Fig. 3) of laboratory strains in the presence of 9% or 18% ethanol. When pregrown in liquid YPD medium with shaking (120 rpm), the mutant strain FH515 accumulated higher levels of l-proline (3.0% of the dry weight) than did the parent strain, MB329-17C (1.3%). A significant amount of l-proline was detected intracellularly even in strain MB329-17C, possibly due to the uptake of l-proline derived from YPD medium. The growth pattern in the absence of ethanol was virtually the same in both strains. However, the number of viable cells at 2, 5, and 8 days after cultivation in SD medium containing ethanol gradually decreased. The ethanol tolerance of strain FH515 was much higher than that of strain MB329-17C (Fig. 2), even though a significant decrease in l-proline content occurred during growth (Fig. 3).
FIG. 2.
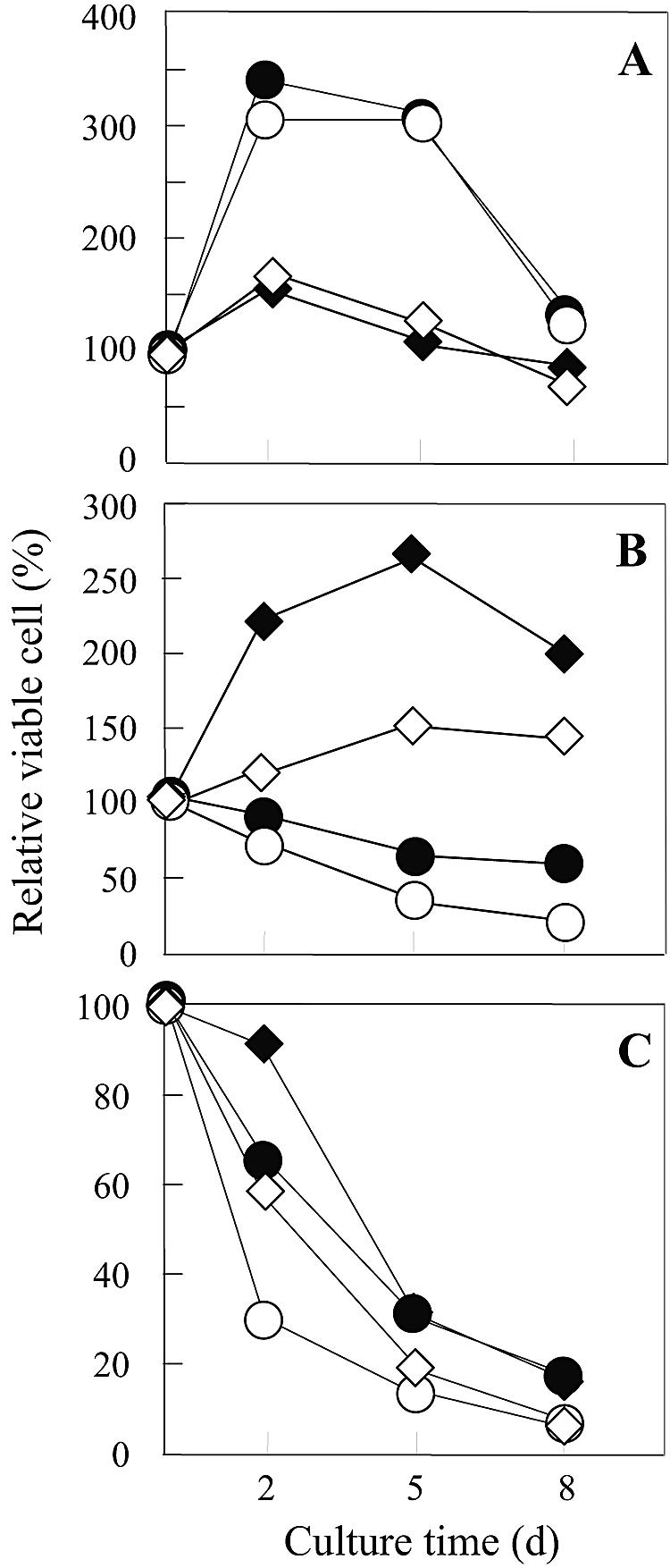
Relative number of viable cells of laboratory and sake strains grown in SD medium without (A) or with 9% (B) and 18% (C) ethanol and incubated under static conditions. The S. cerevisiae strains used were the parent strain MB329-17C (○) and l-proline-accumulating mutant strain FH515 (•) as laboratory strains and the control strain XUW-TRP (◊) and l-proline-accumulating strain XUDput1-MT (⧫) as sake strains. Yeast cells were pregrown in YPD medium at 30°C for 2 days (d) with shaking (120 rpm). The number of viable cells was expressed as a percentage of the initial number of viable cells (day 0). Values are means of results from three independent experiments. The standard deviations for these values were <10% of the value of the point.
FIG. 3.
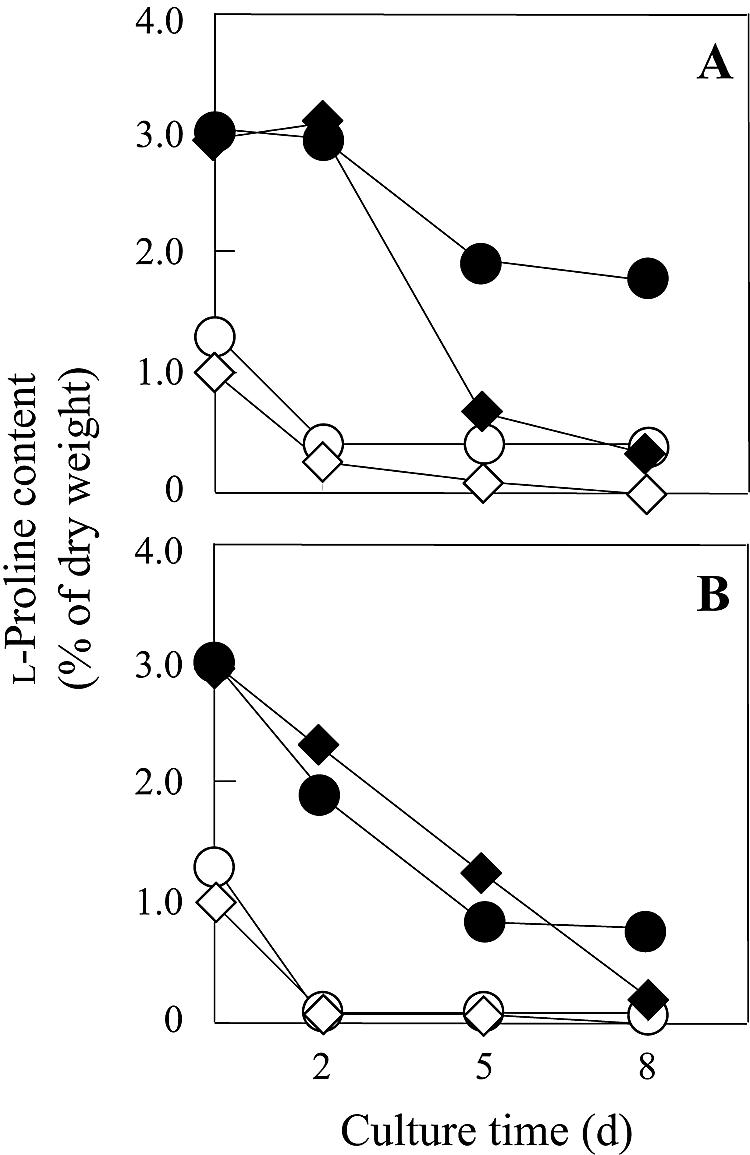
Intracellular l-proline content of laboratory and sake strains grown in SD medium without (A) or with (B) 9% ethanol and incubated under static conditions. The S. cerevisiae strains used were the parent strain MB329-17C (○) and l-proline-accumulating mutant strain FH515 (•) as laboratory strains and the control strain XUW-TRP (◊) and l-proline-accumulating strain XUDput1-MT (⧫) as sake strains. Yeast cells were pregrown in YPD medium at 30°C for 2 days (d) with shaking (120 rpm). Values are means of results from three independent experiments. The standard deviations for these values were <10% of the value of the point.
We also evaluated the intracellular content of trehalose (Fig. 4) and total amino acid content (Table 2) of both strains during cultivation. Trehalose content increased transiently in response to ethanol stress, as previously reported (1). Total amino acid content, except for l-proline, decreased in the presence of ethanol. However, no difference in the contents of the two laboratory strains was detected. These results are consistent with the hypothesis that the accumulation of high levels of l-proline helps protect yeast cells against ethanol stress.
FIG. 4.
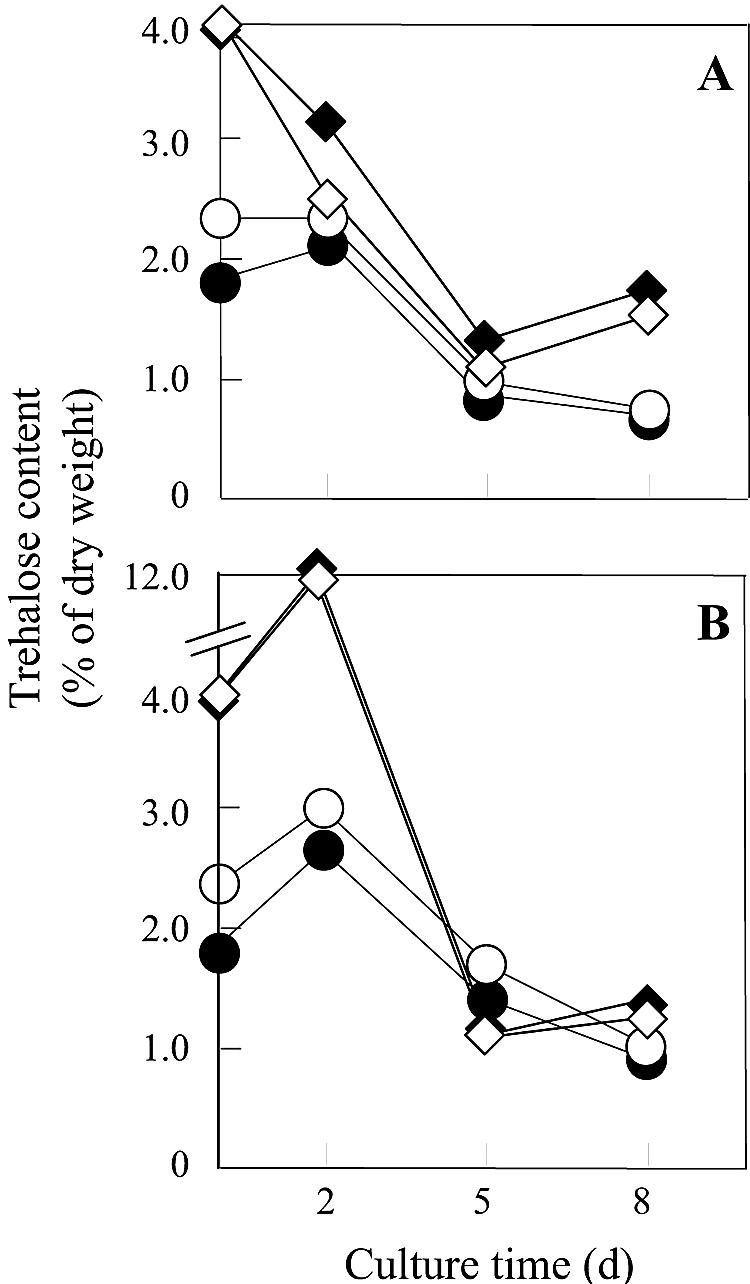
Intracellular trehalose content of laboratory and sake strains grown in SD medium without (A) or with (B) 9% ethanol and incubated under static conditions. The S. cerevisiae strains used were the parent strain MB329-17C (○) and l-proline-accumulating mutant strain FH515 (•) as laboratory strains and the control strain XUW-TRP (◊) and l-proline-accumulating strain XUDput1-MT (⧫) as sake strains. Yeast cells were pregrown in YPD medium at 30°C for 2 days with shaking (120 rpm). Values are means of results from three independent experiments. The standard deviations for these values were <10% of the value of the point.
TABLE 2.
Intracellular total amino acid content of laboratory and sake strains grown in SD medium without or with 9% ethanol and incubated under static conditions
| Strain | Culture time (days) | Total amino acid content (% dry weight)a
|
|
|---|---|---|---|
| Without ethanol | With 9% ethanol | ||
| Laboratory strains | |||
| MB329-17C | 0 | 11 | 11 |
| 2 | 19 | 14 | |
| 5 | 11 | 7.2 | |
| 8 | 10 | 5.5 | |
| FH515 | 0 | 10 | 10 |
| 2 | 18 | 14 | |
| 5 | 11 | 7.7 | |
| 8 | 13 | 6.4 | |
| Sake strains | |||
| XUW-TRP | 0 | 7.3 | 7.3 |
| 2 | 13 | 17 | |
| 5 | 1.8 | 8.5 | |
| 8 | 1.0 | 1.6 | |
| XUDput1-MT | 0 | 9.4 | 9.4 |
| 2 | 15 | 14 | |
| 5 | 3.8 | 7.0 | |
| 8 | 1.4 | 0.9 | |
l-Proline content is not included in total amino acids. Yeast cells were pregrown in YPD medium at 30°C for 2 days with shaking (120 rpm). Values are means of results from three independent experiments. The standard deviations for these values were <10% of the value of the point.
Construction of an l-proline-accumulating sake yeast.
We constructed a novel sake strain that accumulates l-proline and is disrupted in put1. The put1-disrupted strain XUDput1 grew on SD medium plus l-glutamate but not on SD medium plus l-proline. The put1 disruptant also had no detectable proline oxidase activity in cell extracts, although such activity was readily detected when wild-type strain XUW-TRP was grown in a similar manner (0.82 ± 0.10 U/mg of protein). Both the wild-type strain and the put1 disruptant contain trace amounts of intracellular l-proline after cultivation in liquid SD medium with shaking. When strain XUDput1 was grown in SD medium plus l-proline, l-proline accumulated (0.72% ± 0.10% of dry weight), as expected from previous studies (37).
If the wild-type PRO1 allele was replaced with the pro1D154N allele, AZC resistance resulted. Overproduction of l-proline dilutes AZC, which competes with l-proline for incorporation into proteins (13, 27, 36). The PRO1 allele in strain XUDput1-MT has a single base change from G to A at position 460 that results in the replacement of aspartate with asparagine at position 154 in the γ-GK enzyme for pro1D154N.
Ethanol resistance of the l-proline-accumulating sake yeast.
When strain XUDput1-MT was cultured in liquid SD medium under shaking conditions (120 rpm), it accumulated l-proline (0.59% ± 0.12% of dry weight), but strain XUW-TRP did not. When grown in liquid YPD medium under shaking conditions, there was three times as much intracellular l-proline in cells of strain XUDput1-MT (3.38% ± 0.57%) as there was in cells of strain XUW-TRP (1.03% ± 0.15%).
Strain XUDput1-MT was also more tolerant to ethanol (Fig. 2) than was strain XUW-TRP. The growth of neither strain was inhibited for the first 5 days in the presence of 9% ethanol. We also determined the intracellular contents of each strain during cultivation. There was no significant difference between the two sake strains in the intracellular content of trehalose (Fig. 4) or total amino acids (Table 2). These results are consistent with the hypothesis that the improved ethanol tolerance is due to l-proline accumulation by the sake strains.
Sake brewing with the l-proline-accumulating strain.
Under laboratory-scale sake brewing conditions, strain XUDput1-MT accumulated more l-proline (0.31% ± 0.03%) and had higher cell viability (69% ± 9.5%) than did strain XUW-TRP (0.20% ± 0.01% l-proline content and 46% ± 3.8% cell viability). This result is consistent with the conclusion that intracellular l-proline accumulation confers ethanol tolerance under sake brewing conditions. Throughout the fermentation, strain XUDput1-MT evolved CO2 (66 ± 1.6 g), produced ethanol (20% ± 1.2%), and consumed glucose (0.6% ± 0.1%) at the same rate as strain XUW-TRP (69 ± 3.9 g, 20%, and 0.4% ± 0.2%, respectively), suggesting that intracellular l-proline does not inhibit the normal sake brewing process.
Sake brewed with strain XUDput1-MT contained ∼30% more total amino acids (3,700 ± 62 mg/liter) and five times more l-proline (880 ± 12 mg/liter) than did the sake brewed with strain XUW-TRP (2,700 ± 78 mg/liter and 170 ± 10 mg/liter, respectively). The sake brewed with the l-proline-accumulating strain had a sake meter value (+4.1) that was three times lower than that from the sake brewed with the control strain (+13.2). The lower meter value of the sake brewed with strain XUDput1-MT is probably due to significantly higher levels of total amino acids.
The concentrations of organic acids and aromatic compounds produced during sake brewing were also measured (Table 3). Strain XUDput1-MT produced more of the organic acids citrate, malate, and succinate than did strain XUW-TRP. There were no significant differences in the amounts of aromatic compounds produced except that strain XUW-TRP made more isobutyl alcohol.
TABLE 3.
Concentrations of organic acids and aroma compounds in the brewed sake with sake yeast strains
| Organic acid or aroma compound | Concn (mg/liter) (±SD)a
|
|
|---|---|---|
| XUW-TRP | XUDput1-MT | |
| Organic acids | ||
| Citrate | 75 ± 4 | 110 ± 8 |
| Malate | 170 ± 15 | 200 ± 7 |
| Succinate | 800 ± 32 | 1,000 ± 40 |
| Lactate | 370 ± 25 | 400 ± 27 |
| Acetate | 600 ± 41 | 410 ± 27 |
| Aroma compounds | ||
| Acetoaldehyde | 43 ± 3 | 48 ± 2 |
| Ethyl acetate | 28 ± 7 | 33 ± 3 |
| Isobutyl alcohol | 140 ± 6 | 120 ± 3 |
| Isoamyl acetate | 12 ± 4 | 13 ± 1 |
| Isoamyl alcohol | 280 ± 13 | 290 ± 10 |
Values are means ± standard deviations from five independent experiments.
DISCUSSION
Ethanol toxicity affects the growth and fermentation rates of yeast cells as well as their viability (16, 22). The major effect of ethanol seems to increase the permeability of the yeast cell membrane, which is equated with an increase in membrane fluidity (3, 17, 18). The incorporation of certain sterols and long-chain fatty acyl residues, such as ergosterol and palmitoyl-coenzyme A, into cell membranes is important for overcoming changes in fluidity (16, 28). Intracellular ethanol levels can be high enough to denature enzymes and functional proteins. Heat shock treatment increases ethanol tolerance, which induces stress responses such as the expression of heat shock proteins and the accumulation of trehalose (1). The trehalose content increased transiently in response to ethanol stress (Fig. 4). However, there was no significant difference in the content between the l-proline-accumulating and control strains. With respect to the intracellular total amino acid content, there was no significant difference between two sake strains (Table 2). Despite these findings, the mechanism(s) of ethanol toxicity and tolerance remains unclear. Sake yeasts produce more than 20% ethanol in sake brewing, and this high tolerance for ethanol, as inferred from the increased viability of yeast cells in the presence of ethanol, is one of the most important parameters in brewing sake. Increasing ethanol tolerance should improve fermentation performance and product quality while simultaneously simplifying the brewing process.
Intracellular l-proline protects yeast cells from damage by freezing, desiccation, and oxidative stress (26, 27, 36-38). In many plants, osmotic stress induces the rapid accumulation of l-proline resulting from the simultaneous activation of its biosynthesis and the inactivation of its degradation (11). Overproduction of l-proline may lead to increased osmotolerance in transgenic plants (19). In these cases, the l-proline concentration required to increase stress resistance is 10 to 200 μmol per g fresh or dry weight (10, 18, 25, 38). At higher concentrations (>1.5 M), l-proline effectively prevents aggregation during protein refolding (32) and can preserve membrane structure and function during freezing (30). Somewhat lower levels of l-proline (>1.0 M) can increase the stability and the solubility of hydrophobic macromolecules and sparingly soluble proteins (33, 34). In vivo, l-proline in the cell could reduce or prevent ethanol-induced membrane disorder and protein denaturation. We confirmed that l-proline increases cell viability in the presence of ethanol during static incubations of both laboratory and sake yeast strains. The unusually high l-proline content does not appear to stress the cells such that the levels of trehalose or other amino acids increase. Further research to identify the mechanism(s) by which l-proline interacts with membrane phospholipids and proteins is needed to understand how l-proline increases ethanol tolerance. Thus, l-proline might serve as a protective agent for industrial microorganisms and enzymes.
The present study is the first to report the construction of an l-proline-accumulating sake yeast strain. We expected the l-proline-accumulating strain XUDput1-MT to have a shorter fermentation time than the wild-type control strain. However, the sake mash of the two strains showed no significant differences in sake fermentation ability, such as CO2 evolution or ethanol productivity. There are at least two explanations for these observations. First, the degree of ethanol tolerance might depend on culture conditions and the stress responses, and cell metabolism under experimental static and sake brewing conditions could be quite different. Alternatively, ethanol tolerance could vary by strain. The sake yeast strains have a different genetic background than do the laboratory strains. Our results could also reflect the influence of factors other than l-proline on ethanol tolerance. A combination of ethanol protectants, e.g., trehalose (24), ergosterol (16), inositol (14), and l-proline, might result in even higher resistance to ethanol stress.
The large amount of l-proline in sake made by fermenting the l-proline-accumulating strain is of particular interest. l-Proline is probably excreted actively into the sake by proline permeases rather than passively by cell lysis. However, the intracellular metabolite content decreased with culture time, particularly in the presence of ethanol. We calculated dry cell weight by using both the viable and the dead cells. Due to the severe cellular damage caused by ethanol, intracellular l-proline could be leaking from the dead cells into the sake. Thus, at the end of sake brewing, the difference in intracellular l-proline contents between parental and modified strains is less than it was before the sake brewing began. Although we do not know why, total amino acids, including l-aspartate, l-threonine, and l-leucine, were ∼30% higher (1 g/liter) in the sake brewed with strain XUDput1-MT than in the sake brewed with the control strain. A similar tendency was observed for the total organic acids tested, which increased by ∼100 mg/liter in the sake brewed with strain XUDput1-MT. Organic acids are part of the essential flavor components produced by yeast (Table 3). Although the mechanism underlying the significant differences in the amounts of amino acids and organic acids remains unclear, it is possible that enhancement of the l-proline biosynthetic pathway would affect the whole metabolic profile of S. cerevisiae. In addition, the process of sake brewing may affect cellular transport systems, e.g., amino acid permeases and glucose uptake.
l-Proline is the major free amino acid in juice and fermented alcoholic beverages such as wine and beer (11, 15). l-Proline may impart some sensory qualities to sake. In particular, high levels of l-proline may contribute to its perceived sweetness. The taste of sake is determined by a combination of many compounds such as amino acids, organic acids, carbohydrates, nucleotides, and inorganic salts. Amino acids are the main taste components produced by S. cerevisiae. However, a lack of knowledge concerning the mechanism of amino acid production during sake fermentation has made it difficult to develop yeast strains with different amino acid profiles. The development of strains that can produce specific or various amino acids could enable the production of sake with distinctive tastes.
Acknowledgments
We thank M. C. Brandriss (University of Medicine and Dentistry of New Jersey, Newark, N.J.) and J. Nikawa (Kyushu Institute of Technology, Fukuoka, Japan) for providing strain MB329-17C and plasmids pUV2 and pTV3, respectively, and K. Matsuura and S. Nakamori for helpful discussions.
This work was supported by grants to H.T. from PROBRAIN (Program for Promotion of Basic Research Activities for Innovative Biosciences) and the Fukui Prefectural Scientific Research Foundation.
REFERENCES
- 1.Alexandre, H., V. Ansanay-Galeote, S. Dequin, and B. Blondin. 2001. Global gene expression during short-term ethanol stress in Saccharomyces cerevisiae. FEBS Lett. 498:98-103. [DOI] [PubMed] [Google Scholar]
- 2.Arikawa, Y., M. Kobayashi, R. Kodaira, M. Shimosaka, H. Muratsubaki, K. Enomoto, and M. Okazaki. 1999. Isolation of sake yeast strains possessing various levels of succinate- and/or malate-producing abilities by gene disruption or mutation. J. Biosci. Bioeng. 87:333-339. [DOI] [PubMed] [Google Scholar]
- 3.Beavan, M. J., C. Charpentier, and A. H. Rose. 1982. Production and tolerance of ethanol in relation to phospholipid fatty acyl composition in Saccharomyces cerevisiae. J. Gen. Microbiol. 128:1447-1455. [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Brandriss, M. C. 1983. Proline utilization in Saccharomyces cerevisiae: analysis of the cloned PUT2 gene. Mol. Cell. Biol. 3:1846-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandriss, M. C., and D. A. Falvey. 1992. Proline biosynthesis in Saccharomyces cerevisiae: analysis of the PRO3 gene, which encodes Δ1-pyrroline-5-carboxylate reductase. J. Bacteriol. 174:3782-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandriss, M. C., and B. Magasanik. 1979. Genetics and physiology of proline utilization in Saccharomyces cerevisiae: enzyme induction by proline. J. Bacteriol. 140:498-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brewing Society of Japan. 1993. Annotation of the official analytical methods of the National Tax Administration Agency of Japan, 4th ed., p. 16-18. Brewing Society of Japan, Tokyo, Japan. (In Japanese.)
- 9.Brim, M. 1966. Transketolase: clinical aspects. Methods Enzymol. 9:506-514. [Google Scholar]
- 10.Csonka, L. N. 1989. Physical and genetic responses of bacteria to osmotic stress. Microbiol. Rev. 53:121-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dale, C. J., and T. W. Young. 1992. Low molecular weight nitrogenous components and their influence on the stability of beer foam. J. Inst. Brew. 98:123-127. [Google Scholar]
- 12.Delauney, A. J., and D. P. S. Verma. 1993. Proline biosynthesis and osmoregulation in plants. Plant J. 4:215-223. [Google Scholar]
- 13.Fowden, L., and M. H. Richmond. 1963. Replacement of proline by azetidine-2-carboxylic acid during biosynthesis of protein. Biochim. Biophys. Acta 71:459-461. [Google Scholar]
- 14.Furukawa, K., H. Kitano, H. Mizoguchi, and S. Hara. 2004. Effect of cellular inositol content on ethanol tolerance of Saccharomyces cerevisiae in sake brewing. J. Biosci. Bioeng. 98:107-113. [DOI] [PubMed] [Google Scholar]
- 15.Ingledew, W. M., and R. E. Kunkee. 1985. Factors influencing sluggish fermentations of grape juice. Am. J. Enol. Vitic. 36:65-76. [Google Scholar]
- 16.Inoue, T., H. Iefuji, T. Fuji, H. Soga, and K. Satoh. 2000. Cloning and characterization of a gene complementing the mutation of an ethanol-sensitive mutant of sake yeast. Biosci. Bioechnol. Biochem. 64:229-236. [DOI] [PubMed] [Google Scholar]
- 17.Jones, R. P., and P. F. Greenfield. 1987. Ethanol and fluidity of the yeast plasma membrane. Yeast 3:223-232. [DOI] [PubMed] [Google Scholar]
- 18.Kajiwara, S., T. Aritomi, K. Suga, K. Ohtaguchi, and O. Kobayashi. 2000. Overexpression of the OLE1 gene enhances ethanol fermentation by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 53:568-574. [DOI] [PubMed] [Google Scholar]
- 19.Kishor, P. B. K., Z. Hong, G.-H. Miao, C.-A. A. Hu, and D. P. S. Verma. 1995. Overexpression of Δ1-pyrroline-5-carboxylate synthetase increases proline production and confers osmotolerance in transgenic plants. Plant Physiol. 108:1387-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitamoto, K., K. Oda, K. Gomi, and K. Takahashi. 1991. Genetic engineering of a sake yeast producing no urea by successive disruption of arginase gene. Appl. Environ. Microbiol. 57:301-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kubo, Y., H. Takagi, and S. Nakamori. 2000. Effect of gene disruption of succinate dehydrogenase on succinate production in a sake yeast strain. J. Biosci. Bioeng. 90:619-624. [DOI] [PubMed] [Google Scholar]
- 22.Kunkee, R. E., and L. F. Bisson. 1993. Wine-making yeasts, p. 94-99. In A. H. Rose and J. S. Harrison (ed.), The yeasts, vol. 5, 2nd ed. Academic Press, San Diego, Calif. [Google Scholar]
- 23.Magarifuchi, T., K. Goto, Y. Iimura, M. Tadenuma, and G. Tamura. 1995. Effect of yeast fumarase gene (FUM1) disruption on production of malic, fumaric and succinic acids in sake mash. J. Ferment. Bioeng. 80:355-361. [Google Scholar]
- 24.Mansure, J. J. C., A. D. Panek, L. M. Crowe, and J. H. Crowe. 1994. Trehalose inhibits ethanol effects on intact yeast cells and liposomes. Biochim. Biophys. Acta 1191:309-316. [DOI] [PubMed] [Google Scholar]
- 25.Mizoguchi, H., T. Ikeda, and S. Hara. 1995. Differences in the intracellular lipids of sake yeast in main mash seeded respectively with two kinds of seed mash: kimoto and sokujo-moto. J. Ferment. Bioeng. 80:586-591. [Google Scholar]
- 26.Morita, Y., S. Nakamori, and H. Takagi. 2002. Effect of proline and arginine metabolism on freezing stress of Saccharomyces cerevisiae. J. Biosci. Bioeng. 94:390-394. [DOI] [PubMed] [Google Scholar]
- 27.Morita, Y., S. Nakamori, and H. Takagi. 2003. l-Proline accumulation and freeze tolerance in Saccharomyces cerevisiae are caused by a mutation in the PRO1 gene encoding γ-glutamyl kinase. Appl. Environ. Microbiol. 69:212-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nozawa, M., T. Takahashi, S. Hara, and H. Mizoguchi. 2002. A role of Saccharomyces cerevisiae fatty acid activation protein 4 in palmitoyl-CoA pool for growth in the presence of ethanol. J. Biosci. Bioeng. 93:288-295. [DOI] [PubMed] [Google Scholar]
- 29.Rose, M., and J. R. Broach. 1991. Cloning genes by complementation in yeast. Methods Enzymol. 194:195-230. [DOI] [PubMed] [Google Scholar]
- 30.Rudolph, A. S., and J. H. Crowe. 1985. Membrane stabilization during freezing: the role of two natural cryoprotectants, trehalose and proline. Cryobiology 22:367-377. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Samuel, D., T. K. S. Kumar, G. Ganesh, G. Jayaraman, P.-W. Yang, M.-M. Chang, V. D. Trivedi, S.-L. Wang, K.-C. Hwang, D.-K. Chang, and C. Yu. 2000. Proline inhibits aggregation during protein refolding. Protein Sci. 9:344-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samuel, D., T. K. S. Kumar, G. Jayaraman, P.-W. Yang, M.-M. Chang, and C. Yu. 1997. Proline is a protein solubilizing solute. Biochem. Mol. Biol. Int. 41:235-242. [DOI] [PubMed] [Google Scholar]
- 34.Schobert, B., and H. Tschesche. 1978. Unusual solution properties of proline and its interaction with proteins. Biochim. Biophys. Acta 541:270-277. [DOI] [PubMed] [Google Scholar]
- 35.Sikorski, R. S., and J. D. Boeke. 1991. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 194:302-318. [DOI] [PubMed] [Google Scholar]
- 36.Takagi, H., F. Iwamoto, and S. Nakamori. 1997. Isolation of freeze-tolerant laboratory strain of Saccharomyces cerevisiae from proline-analogue-resistant mutants. Appl. Microbiol. Biotechnol. 47:405-411. [DOI] [PubMed] [Google Scholar]
- 37.Takagi, H., K. Sakai, K. Morida, and S. Nakamori. 2000. Proline accumulation by mutation or disruption of the proline oxidase gene improves resistance to freezing and desiccation stresses in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 184:103-108. [DOI] [PubMed] [Google Scholar]
- 38.Terao, Y., S. Nakamori, and H. Takagi. 2003. Gene dosage effect of l-proline biosynthetic enzymes on l-proline accumulation and freeze tolerance in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 69:6527-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomenchok, D. M., and M. C. Brandriss. 1987. Gene-enzyme relationship in the proline biosynthetic pathway of Saccharomyces cerevisiae. J. Bacteriol. 169:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, S.-S., and M. C. Brandriss. 1986. Proline utilization in Saccharomyces cerevisiae: analysis of the cloned PUT1 gene. Mol. Cell. Biol. 6:2638-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]