Abstract
We isolated a novel gram-positive bacterium, Brevibacillus texasporus, that produces an antibiotic, BT. BT is a group of related peptides that are produced by B. texasporus cells in response to nutrient limitation. We report here purification and determination of the structure of the most abundant BT isomer, BT1583. Amino acid composition and tandem mass spectrometry experiments yielded a partial BT1583 structure. The presence of ornithine and d-form residues in the partial BT1583 structure indicated that the peptide is synthesized by a nonribosomal peptide synthetase (NRPS). The BT NRPS operon was rapidly and accurately identified by using a novel in silico NRPS operon hunting strategy that involved direct shotgun genomic sequencing rather than the unreliable cosmid library hybridization scheme. Sequence analysis of the BT NRPS operon indicated that it encodes a colinear modular NRPS with a strict correlation between the NRPS modules and the amino acid residues in the peptide. The colinear nature of the BT NRPS enabled us to utilize the genomic information to refine the BT1583 peptide sequence to Me2-4-methyl-4-[(E)-2-butenyl]-4,N-methyl-threonine-L-dO-I-V-V-dK-V-dL-K-dY-L-V-CH2OH. In addition, we report the discovery of novel NRPS codons (sets of the substrate specificity-conferring residues in NRPS modules) for valine, lysine, ornithine, and tyrosine.
Novel antibiotics are needed to combat infections caused by bacteria that are resistant to conventional antibiotics. It is well known that microbes produce a huge variety of antibiotics to wage chemical warfare against competing microbes. We screened soil microorganisms for strains that produce novel antibiotics. Bacillus sp. strain E58 (= ATCC PTA-5854) was isolated for its ability to produce the antibiotic BT against Staphylococcus aureus strains that cause life-threatening infections (Jiang and Munoz-Romero, unpublished results). This strain was named Brevibacillus texasporus based on its relatedness to Brevibacillus laterosporus.
Many peptide antibiotics of microbial origin (mostly from actinomycetes, bacilli, and fungi) are synthesized by nonribosomal peptide synthetases (NRPS), and they contain unusual amino acids. NRPS usually have a colinear modular architecture (15). The N-terminal to C-terminal order and the specificities of the individual modules correspond to the sequential order and identities of the amino acid residues in the peptide product. Each NRPS module recognizes a specific amino acid and catalyzes stepwise condensation to form a growing peptide chain. The identity of the amino acid recognized by a particular module can be predicted by comparisons to other modules having known specificities (3). Such strict correlation made it possible to identify genes encoding the NRPS enzymes for a number of microbial nonribosomal peptides with known structures, as demonstrated by identification of the mycobactin biosynthesis operon in the genome of Mycobacterium tuberculosis (18). Conversely, an NRPS operon may be a source of information that allows researchers to determine certain structural details of the peptide product. In this study, we show that identification of the BT NRPS operon resulted in critical refinements of the BT1583 peptide structure.
The modules of an NRPS are composed of smaller units or “domains,” each of which has a specific role in the recognition, activation, modification, or joining of amino acid precursors to form the peptide product. One type of domain, the adenylation domain (A-domain), is responsible for selectively recognizing and activating the amino acid that is to be incorporated by a particular module of the NRPS. Through analysis of the substrate-binding pocket of the A-domain of the PheA subunit of the gramicidin S NRPS in combination with sequence comparison with other A-domains, it was possible to define 10 residues that are the main determinants of the substrate specificity for an A-domain (4, 21). The 10 residues are considered an NRPS “codon.” The NRPS codon collection is still growing as new NRPS codons continue to be discovered. In this paper, we describe novel NRPS codons for valine, lysine, ornithine, and tyrosine. The amino acid activation step is ATP dependent and involves the transient formation of an aminoacyl adenylate. The reaction consumes one ATP and generates one PPi. An ATP-PPi exchange assay has been developed to measure the substrate specificity for an A-domain (10, 14).
The activated amino acid is covalently attached to the peptide synthetase through another type of domain, the thiolation domain (T-domain), which is generally located adjacent to the A-domain. The T-domain is posttranslationally modified by covalent attachment of a phosphopantetheinyl prosthetic arm to a conserved serine residue. The activated amino acid substrates are tethered to the NRPS via a thioester bond to the phosphopantetheinyl prosthetic arm of the corresponding T-domains. Amino acids joined to successive units of the NRPS are subsequently covalently linked together by the formation of amide bonds catalyzed by another type of domain, the condensation domain (C-domain). NRPS modules can also occasionally contain additional functional domains that carry out auxiliary reactions, and the most common of these is epimerization of an amino acid substrate from the l form to the d form. This reaction is catalyzed by a domain referred to as an epimerization domain (E-domain), which is generally located adjacent to the T-domain of a given NRPS module. Thus, a typical NRPS module has the following domain organization: C-A-T-(E).
Product assembly by NRPS involves three distinct phases, namely, chain initiation, chain elongation, and chain termination (8). Peptide chain initiation is carried out by specialized modules termed “starter modules” that comprises an A-domain and a T-domain. Elongation modules have, in addition, a C-domain that is located upstream of the A-domain. It has been experimentally demonstrated that such elongation domains cannot initiate peptide bond formation due to interference by the C-domain (12). All the growing peptide intermediates are covalently tethered to the NRPS during translocations as an elongating series of acyl-S-enzyme intermediates. To release the mature peptide product from the NRPS, the terminal acyl-S-enzyme bond must be broken. This process is the chain termination step and is usually catalyzed by a C-terminal thioesterase domain (TE-domain). Thioesterase-mediated release of the mature peptide from the NRPS enzyme involves transient formation of an acyl-O-TE intermediate that is then hydrolyzed or hydrolyzed and concomitantly cyclized to release the mature peptide (7). An alternative termination scheme involves reduction of the tethered C-terminal residue by a reductase domain (R-domain) that resides in the last NRPS module, resulting in release of a peptide with an alcoholic C-terminal residue (5, 9). Such a reductase-mediated termination/C-terminal modification occurs in BT biosynthesis and contributes to superprotease resistance of the BT peptides.
MATERIALS AND METHODS
Partial purification of BT.
B. texasporus E58 cells were grown in 1 liter of LB in an air shaker at 37°C for 3 days. The culture was spun in a clinical centrifuge at 3,000 rpm for 15 min. The supernatant was collected, and 500 g of ammonium sulfate was added and dissolved. The sample was spun in the clinical centrifuge at 3,000 × g for 15 min. The pellet was dissolved in 200 ml of distilled water. The solution was then boiled for 15 min and then cooled on ice. The sample was filtered with a 0.2-μm filter (Nalgene). The filtrate was mixed with 0.2 liter of chloroform at room temperature for 20 min with a stir bar. The mixture was separated into two phases by centrifugation in the clinical centrifuge at 3,000 rpm for 15 min. The organic phase was collected and dried in a vacuum evaporator.
C18 reverse-phase HPLC.
The dried chloroform extract was dissolved in 2 ml of sterile distilled water. The solution was fractionated on a C18 reverse-phase high-performance liquid chromatography (HPLC) column by using a gradient from 30% solution B to 55% solution B (solution B was 0.075% trifluoroacetic acid in acetonitrile, and solution A was 0.1% trifluoroacetic acid in water). The resulting fractions were dried, dissolved in sterile distilled water, and analyzed for anti-S. aureus activity with a Kirby-Bauer assay. The peak fraction (fraction 33) was subjected to amino acid composition, mass spectrometry (MS), tandem mass spectrometry, and chirality analyses.
Detection of d-form amino acid residues.
The chiral analysis of amino acid residues in BT was performed by Commonwealth Biotechnologies, Inc., Richmond, VA. BT was subjected to hydrolysis in 6 N HCl in vacuo for 18 h at 110°C. The amino acids were derivatized to 9-fluorenylmethoxy carbonyl amino acids and separated by HPLC. The elution profile of each amino acid was then determined on a chiral column. For both types of chromatography columns, peaks were identified by comparison with appropriate standards.
Genomic DNA preparation.
Log-phase E58 cells were harvested from an LB culture and lysed with lysis buffer (10 mM Tris [pH 8.0], 100 mM EDTA, 0.5% sodium dodecyl sulfate). RNase A was added to digest contaminating RNA. Genomic DNA was extracted with phenol-chloroform and then precipitated with ethanol. Dried DNA was resuspended in Tris-EDTA, and an aliquot was electrophoresed in a 0.5% agarose gel for quality control.
Library construction and genome sequencing.
E58 genomic library construction, shotgun sequencing, and assembly were performed by Agencourt Biosciences Corporation (Beverly, MA). Briefly, the whole genome library was constructed with an average insert length of around 5 kb. A total of 10,000 clones were subjected to automated DNA sequencing from both ends of the insert; 16,901 successfully sequenced reads were obtained.
Nucleotide sequences and data analysis.
All BLAST analyses with the E58 genome were performed by using the WU BLAST software package (version 2.0) installed on a local computer (W. Gish, 1996 to 2003; http://BLAST.wustl.edu). Amino acid sequence homology searches were performed by using the BLAST server at the National Center for Biotechnology Information (Bethesda, Md.) and a nonredundant protein sequence database with default parameter values (1). Amino acid sequence alignment was performed by using the CLUSTALW program (2) running at the NPS@ web server at the Institute of Biology and Chemistry of Proteins (Lyon, France).
Cloning, overexpression, and purification of His10-tagged BT A-domain proteins.
DNA fragments encoding the A-domains of BT NRPS modules 8, 5, 7, 4, and 2 (Bt8A, Bt5A, Bt7A, Bt4A, and Bt2A) were PCR amplified, and the PCR products were inserted into His10-tagged recombinant protein expression vector pET16b (Novagen). The A-domain borders were determined as described by Konz et al. (10). The expression constructs were transformed into the Escherichia coli BL21-AI strain (Invitrogen). Transformants were grown in L broth at 37°C to an A600 of 0.6 and then induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) plus 0.2% l-arabinose. The cells were allowed to grow for two additional hours at 30°C before they were harvested. The His10-tagged recombinant proteins were purified by using TALON metal affinity resins (BD Biosciences) under conditions recommended in the manual.
ATP-PPi exchange assay.
ATP-PPi exchange assays were performed to determine the substrate specificity of an A-domain. ATP-PPi exchanges were assayed as previously described (20).
MIC determination assays.
S. aureus was grown to the mid-log phase in LB at 37°C, diluted 500-fold with fresh LB, and dispensed into 96-well microtiter plates. Different concentrations of peptides were added, and the microtiter plates were incubated at 37°C with shaking. The MIC was the lowest peptide concentration that produced a clear well. All experiments were performed in triplicate, and highly consistent MICs were obtained.
Nucleotide sequence accession number.
The DNA sequence of the BT NRPS operon has been deposited in the GenBank database under accession no. AY953371.
RESULTS
Identification of the BT peptide.
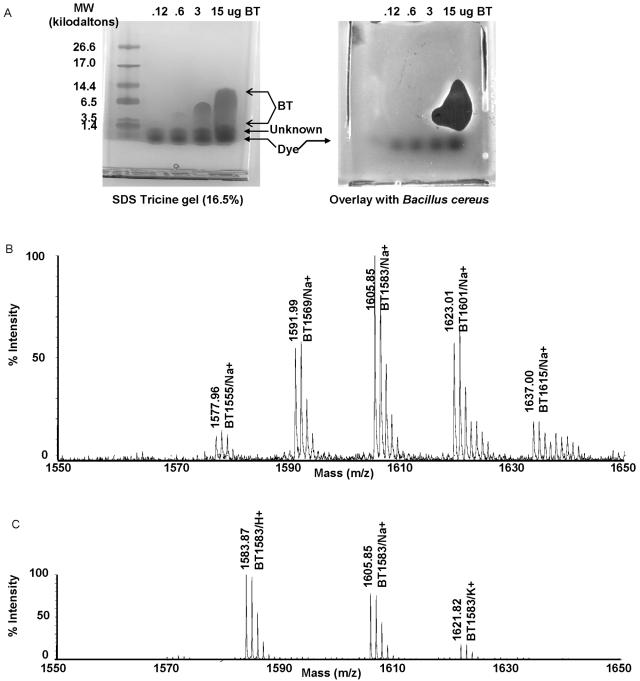
Bacterial strain E58 was isolated from soil in an effort to identify soil microorganisms that produce novel antibiotics for use against S. aureus. E58 was found to be closely related to B. laterosporus based on 16S rRNA gene sequence homology (98.5% identity) (Jiang and Ballard, unpublished results). E58 was named B. texasporus and deposited in the ATCC (catalog number ATCC PTA-5854). The antibiotic produced by E58 was named BT, and its activity could be detected in the supernatant of a liquid LB E58 culture. The antibiotic activity was precipitated by ammonium sulfate, which suggested that the antibiotic is a protein or peptide (data not shown). The activity was further extracted with chloroform, and the chloroform extract was analyzed by Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The two halves of a gel with identical lanes in each half were either stained for proteins and peptides or overlaid with agar containing the BT-sensitive bacterium Bacillus cereus to test for antibiotic activity (Fig. 1A). The following three species were visible after staining: the bromophenol blue dye that originated from the gel loading buffer, an unknown peptide with a molecular mass of <1.4 kDa, and a third species with antibiotic activity. The third species produced a ∼1.5-kDa band at low concentrations (clearly visible on the original gel) and was later shown to be made up of a group of related peptides (see below). The apparent mass increased with concentration, suggesting that the peptides aggregate at higher concentrations. An antibiotic activity was associated with the peptides at higher concentrations, and we therefore concluded that the peptides likely conferred the BT antibiotic activity. The peptides were referred as the BT peptides. The BT peptides apparently were not toxic to B. cereus at lower concentrations in this assay. Since the smallest detectable BT band was at ∼1.5 kDa, we concluded that the BT peptides contained approximately 13 residues.
FIG. 1.
Purification of peptide BT. (A) Tricine gel separation of BT and the associated antibiotic activity. Chloroform-treated extracted peptides were separated on a precast 16.5% Tricine gel (purchase from Bio-Rad). One gel was stained with Coomassie blue to show peptide bands. Another gel was overlaid with agar containing B. cereus. Clear zones in the bacterial lawn correspond to the species that collapses into a single band at a molecular weight of ∼1,500. The following molecular mass markers were used: triose phosphate isomerase (26.6 kDa), myoglobin (17.0 kDa), alpha-lactalbumin (14.4 kDa), aprotinin (6.5 kDa), insulin b chain (oxidized) (3.5 kDa), and bacitracin (1.4 kDa). SDS, sodium dodecyl sulfate. (B) Mass spectrometry of chloroform-extracted BT. Chloroform-extracted BT was treated with sodium by addition of sodium chloride and then subjected to mass spectrometry analysis. Five sodium-containing BT isomers (BT1555, BT1571, BT1583, BT1599, and BT1613) were detected and labeled. (C) Mass spectrometry of purified BT1583. Fraction 33 from C18 reverse-phase HPLC was subjected to mass spectrometry analysis. Only protonated, sodium-containing, and potassium-containing BT1583 were detected.
The chloroform-extracted BT was subject to a mass spectrometry assay. A group of peptides were detected in the range between 1,550 and 1,650 Da (Fig. 1B). The main species had a molecular weight of 1,583, and it was designated BT1583. The other peptides were later shown to be isomers of BT1583.
Determination of partial BT sequence.
The chloroform-extracted BT was purified further by C18 reverse-phase HPLC (see Materials and Methods for details). BT1583 was purified to homogeneity in fraction 33 by C18 HPLC (Fig. 1C). An amino acid composition analysis of BT1583 (fraction 33) showed that BT1583 contained Tyr, Lys, Leu, Ile, Val, and Orn residues (Table 1). BT1583 was refractory to N-terminal sequencing and resistant to degradation by aminopeptidase M, suggesting that there was a nonstandard N-terminal residue. BT1583 was also resistant to cleavage by carboxypeptidase Y, suggesting that there was a nonstandard C-terminal amino acid. Carboxyl-terminal sequencing was, therefore, not attempted.
TABLE 1.
Amino acid composition of purified BT1583 peptide
| Amino acid | Amt (nmol) | Molar ratios normalized to:
|
No. of residues per peptide | |
|---|---|---|---|---|
| Tyr | Ile | |||
| Tyrosine | 1.75 | 1.00 | 1.16 | 1 |
| Valine | 4.58 | 2.62 | 3.05 | 3 |
| Isoleucine | 1.50 | 0.86 | 1.00 | 1 |
| Leucine | 5.32 | 3.04 | 3.54 | 3 |
| Lysine | 3.57 | 2.04 | 2.38 | 2 |
| Ornithine | 1.2 | 0.69 | 0.80 | 1 |
| Total for derivatizable residues | 10.25 | 11.93 | 11 | |
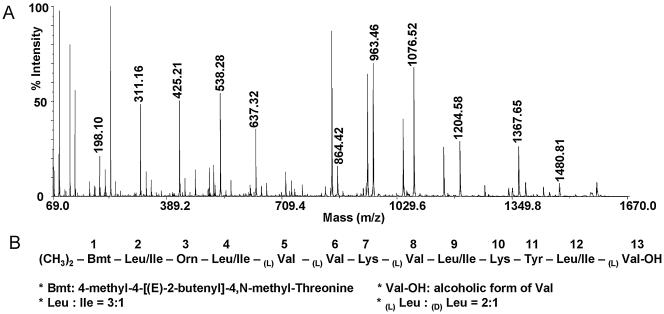
Tandem mass spectrometry (MS-MS) was then used to sequence the BT1583 peptide. MS-MS data were obtained for BT1583, and they are shown in Fig. 2A and Table 2. The MS-MS data indicated that BT1583 contained 13 amino acid residues that correlated well with the amino acid composition. As expected, the molecular masses of residues 1 and 13 did not correspond to the molecular masses of any standard amino acids. The last residue had a molecular mass of 103 Da, which appeared to be compatible with valinol (a valine whose C terminus is reduced from a carboxylic acid to an alcohol). The presence of a C-terminal valinol was further confirmed by the presence of a reductase domain in the 13th valine-specific module of the BT NRPS (see below). The identity of the N-terminal residue was more difficult to determine. Nonetheless, an N-terminal residue with a molecular mass of 198 seemed to be compatible with the N,N-methylated form of 4-methyl-4-[(E)-2-butenyl]-4,N-methyl-threonine (Bmt) (16, 17).
FIG. 2.
(A) BT1583 tandem mass spectrometry data. (B) Partial BT1583 sequence structure deduced from amino acid composition and MS-MS experiments. See Tables 1 and 2.
TABLE 2.
Tandem mass spectrometry of BT1583
| M/H+ b series | ΔM | Possible amino acid residue | M/H+ y series | ΔM | Possible amino acid residue | Compiled (N to C) |
|---|---|---|---|---|---|---|
| 198.10 | (CH3)2-Bmt (?) | (CH3)2-Bmt (?) | ||||
| 311.16 | 113.06 | L/I | 1386.73 | 113.12 | L/I | L/I |
| 425.21 | 114.05 | O | 1273.61 | 114.04 | O | O |
| 538.28 | 113.07 | L/I | 1159.57 | 113.05 | L/I | L/I |
| 637.32 | 99.04 | V | 1046.52 | 198.08 | V+V | V |
| V | ||||||
| 864.42 | 227.10 | V+K | 848.44 | 128.07 | K | K |
| 963.46 | 99.04 | V | 720.37 | 99.04 | V | V |
| 1076.52 | 113.06 | L/I | 621.33 | L/I | ||
| 1204.58 | 128.06 | K | K | |||
| 1367.65 | 163.07 | Y | Y | |||
| 1480.81 | 113.16 | L/I | L/I | |||
| 1583.87 | 103.06 | Val-CH2OH | Val-CH2OH |
The presence of ornithine and possibly Bmt in BT1583 indicated that BT1583 could not be synthesized by ribosomes. The presence of d amino acids would strengthen this idea. We chose to assess the chiral properties of two of the most abundant residues in BT1583, Val and Leu. Chiral analyses revealed uniform l-Val residues, but there were both l- and d-Leu residues at a ratio of 2:1.
The biochemical and structural analyses described above provided a partial BT1583 peptide sequence (Table 2 and Fig. 2B). The structures of the N- and C-terminal residues were not fully determined. Isoleucine and leucine could not be distinguished. The position of the d form of Leu was not specified. Chiral properties of other residues in the peptide were not determined.
Shotgun sequencing of the E58 genome.
To better understand the structure and biosynthesis of the BT1583 peptide, we decided to identify the gene or operon that is responsible for BT biosynthesis. As mentioned above, BT1583 is likely to be synthesized by the NRPS in vivo (13). Most of the NRPS genes are colinear, reflecting a strict correlation between NRPS modules and the amino acid residues in the peptide product. If the BT NRPS operon is colinear, it should encode 13 modules corresponding to the 13 amino acid residues in the BT1583 peptide. Assuming that each module is encoded by an average 3.5-kb DNA fragment, a DNA fragment that is 46 kb long would be needed to accommodate the BT NRPS operon. The traditional method used to identify an NRPS operon involves probing a cosmid library with a generic probe. Since an imperfect generic probe may miss the target gene and there are usually multiple NRPS operons in a bacterial genome, researchers often waste time chasing the wrong NRPS operons. To avoid this pitfall, we adopted a genomic approach that provided an unbiased in silico assessment of all NRPS operons in a genome to allow direct comparisons of the NRPS operons and therefore rational candidate operon selection. This novel approach resulted in rapid and accurate identification of the BT NRPS operon.
The E58 genome was estimated to be 5 Mb long. An E58 genomic library was constructed with an average insert size of 5 kb. The whole genome was sequenced for twofold coverage. After sequence assembly, the E58 genome was represented by 1,919 contigs whose sizes ranged from 700 bp to 22.6 kb and 932 singlets. Such coverage allowed 99.995% of the genome to be represented by clones. Also, the average length of the gap between two neighboring contigs was as short as 250 bp, so that supercontigs could be constructed (see below). Moreover, supercontigs at this resolution should contain sufficient information to allow accurate in silico NRPS operon identification.
In silico identification of the BT NRPS operon.
A three-step procedure was used to select the candidate BT NRPS operon. First, all contigs and singlets were searched for sequences encoding NRPS modules. Since E58 is related to Bacillus subtilis, the putative peptide synthetase PPS1 sequence from B. subtilis was chosen as the query sequence for BLAST analysis with a database containing all assembled E58 contigs. A total of 128 contigs showed translated amino acid sequence similarity to PPS1, with P values ranging from 0 to 1.
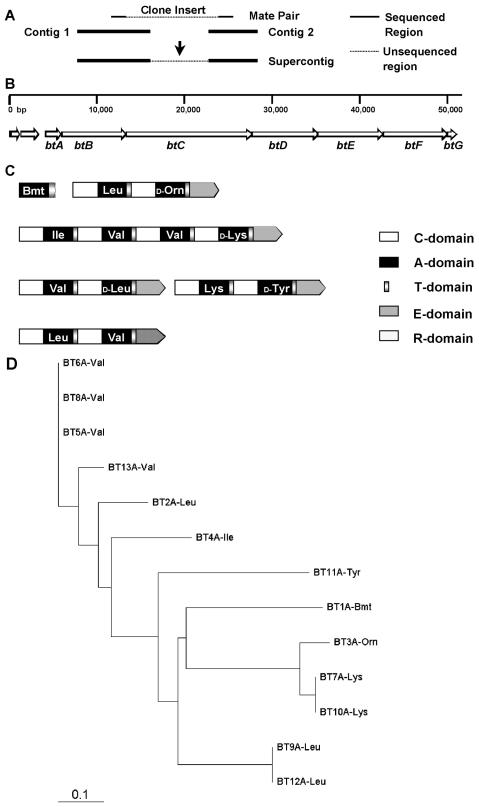
Second, supercontigs were constructed from the 128 contigs. Two sequencing reads from the ends of the same insert formed a mate pair. A supercontig is a collection of contigs joined by mate pairs that reside in different contigs. Identification of mate pairs allowed neighboring contigs to be ordered and orientated to form a supercontig (Fig. 3A). Thirty-one supercontigs were successfully constructed to represent the whole E58 NRPS operon portfolio.
FIG. 3.
BT NRPS operon. (A) Construction of a supercontig from two contigs linked by a mate pair. Contig 1 and contig 2 share a mate pair from a clone. The contigs are ordered and arranged to form a supercontig, which contains the sequences of contig 1 and contig 2, which are separated by an unsequenced gap region. (B) Region sequenced in this work and location of 10 ORFs found in the region. Six ORFs (btA through btF) encode the BT NRPS subunits. (C) Domain organization of the BT NRPS subunits. The predicted amino acid substrate specificity of each module is indicated in each A-domain. (D) Phylogenetic tree for a multiple-sequence alignment of all 13 binding pocket constituents described in Table 3. The putative specificity was assigned using the partial BT1583 sequence. The binding pockets of A-domains that supposedly activate the same or similar substrate cluster together.
Third, the candidate BT NRPS operon was selected from the E58 NRPS operon portfolio. The 31 supercontigs were examined for the possibility that they harbored the BT NRPS operon, and supercontig 3 (whose genetic features based on finished sequence are shown in Fig. 3B and C) was chosen as the candidate based on the following findings.
(i) Supercontig 3 was big enough to potentially contain a DNA sequence encoding 13 NRPS modules.
(ii) The available information regarding the A-domian substrate specificities of supercontig 3 was compatible with the partial BT1583 sequence. Complete sets of substrate specificity-conferring amino acid residues could be identified for 11 modules (all modules except modules 2 and 13 due to an incomplete DNA sequence). Although not all specificity predictions could be made, good correlations between predicted NRPS amino acid substrates and the partial BT1583 sequence were established. Specifically, module 4 was predicted to incorporate Ile, and modules 9 and 12 were predicted to incorporate Leu (Table 3; see below for details). The partial BT1583 sequence had Leu or Ile at positions 4, 9, and 12. Phylogenetic analysis of the substrate-conferring amino acids of the 11 modules showed that modules expected to incorporate the same or very similar amino acids did group together (Fig. 3D). For example, modules 5, 6, and 8, which were all predicted to incorporate Val, formed a cluster, and modules 7, 10, and 3, which were predicted to incorporate similar cationic amino acids (Lys and Orn), formed another cluster.
TABLE 3.
Predicted BT NRPS module substrate specificities and refinement of the BT1583 peptide structure
| Module | Amino acid at PheA residuea:
|
Predicted substrate specificity | Partial BT1583 sequence | Refined BT1583 sequence | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 235 | 236 | 239 | 278 | 299 | 301 | 322 | 330 | 331 | 517 | ||||
| 1 | D | F | W | N | I | G | M | V | H | K | Thr/Dht | (CH3)2-Bmt?b | (CH3)2-Bmt? |
| 2 | D | G | F | L | L | G | G | V | F | K | Ile/Leu | Leu/Ile | Leuc |
| 3 | D | S | G | P | S | G | A | V | D | K | Ornb | Orn | |
| 4 | D | G | F | F | L | G | V | V | Y | K | Ileb | Leu/Ile | Ile |
| 5 | D | G | F | F | V | G | G | V | F | K | Ile/Leu/Val | Valb | Val |
| 6 | D | G | F | F | V | G | G | V | F | K | Ile/Leu/Val | Valb | Val |
| 7 | D | A | G | P | S | G | A | V | D | K | Lysb | Lys | |
| 8 | D | G | F | F | V | G | G | V | F | K | Ile/Leu/Val | Valb | Val |
| 9 | D | A | W | F | L | G | N | V | V | K | Leub | Leu/Ile | Leu |
| 10 | D | A | G | P | S | G | A | V | G | K | Lysb | Lys | |
| 11 | D | A | A | A | V | V | G | V | A | K | Phe/Trp/Tyr | Tyrb | Tyr |
| 12 | D | A | W | F | L | G | N | V | W | K | Leub | Leu/Ile | Leu |
| 13 | D | G | F | F | A | G | G | V | F | K | Ile/Leu/Val | Val-CH2OHb | Val-CH2OH |
The residues were numbered according to the corresponding residues of PheA (4).
The information was used for BT1583 peptide sequence refinement.
The Leu at this position was deduced from the fact that the only Ile had been assigned to position 4.
(iii) The E-domain positions in the NRPS encoded by supercontig 3 showed compatibility with the partial BT1583 peptide structure. Four E-domains were found in modules 3, 7, 9, and 11 (Fig. 3C). Their positions were consistent with the BT1583 chiral properties of all l-Val residues mentioned above and a ratio of l-Leu to d-Leu of 2:1.
Supercontig 3 was therefore identified as the candidate locus for the BT NRPS operon. Primer extension and genome walking were performed to obtain a high-quality sequence of the locus. These efforts resulted in a 50,674-bp contig covering the putative BT NRPS operon (GenBank accession no. AY953371).
Putative BT NRPS subunits.
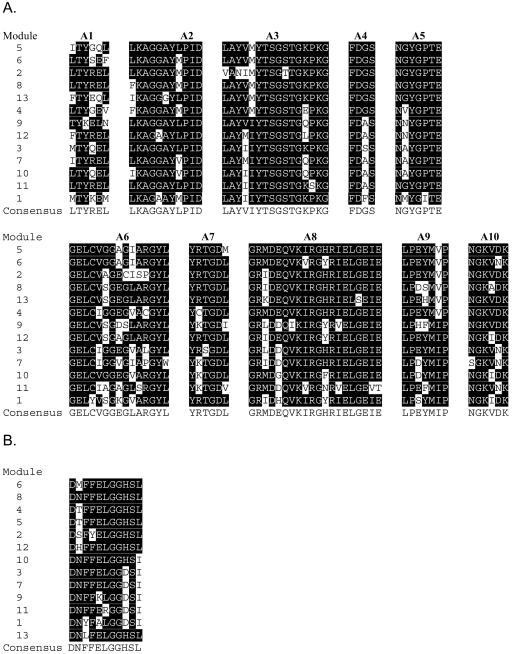
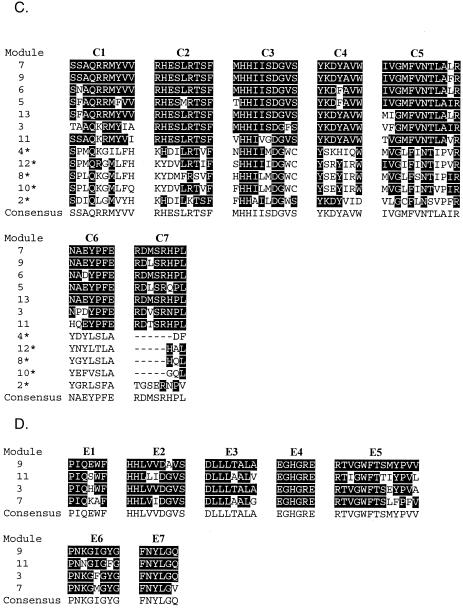
Ten open reading frames (ORFs) were identified in the sequenced region through translation analysis and BLAST searches (2) (Fig. 3B). The middle six ORFs (designated btA through btF) were predicted to encode six subunits of the BT NRPS (BtA through BtF), and their coordinates are shown in Table 4. Sequence analysis of the putative subunits confirmed the modular structure of a typical colinear NRPS (Fig. 3C). The modules, each containing an A-domain and a T-domain, are linked by a C-domain. The loading module BtA has an A-domain followed by a T-domain. There are two noticeable overall features of the putative BT NRPS subunits. First, four of the six subunits exhibit a two-module structure. Second, all auxiliary E-domains are present at the end rather than in the middle of the putative NRPS subunits. Sequence alignments of conserved domains are shown in Fig. 4.
TABLE 4.
ORFs of the BT NRPS operon
| ORF | Start (nucleotide) | End (nucleotide) | Length (bp) | Gene product
|
||
|---|---|---|---|---|---|---|
| Length (amino acids) | Mol wt (103) | Homology | ||||
| btA | 2861 | 4786 | 1,926 | 641 | 72.87 | NRPS |
| btB | 4789 | 12381 | 7,593 | 2,530 | 288.99 | NRPS |
| btC | 12410 | 26263 | 13,854 | 4,617 | 526.68 | NRPS |
| btD | 26293 | 33918 | 7,626 | 2,541 | 289.31 | NRPS |
| btE | 33948 | 41528 | 7,581 | 2,526 | 288.45 | NRPS |
| btF | 41556 | 49031 | 7,476 | 2,491 | 284.46 | NRPS |
| btG | 49092 | 49814 | 723 | 240 | 26.95 | ABC transporter |
FIG.4.
Conserved motifs and alignments of the adenylation, condensation, thiolation, epimerization, and reductase domains from the BT NRPS modules. Conserved motifs were identified as described by Marahiel (13). Consensus sequences are indicated under each alignment. Residues that are the same as consensus residues are indicated by a black background. All 12 C-domains were aligned, and asterisks indicate the start C-domains that are known to be less conserved.
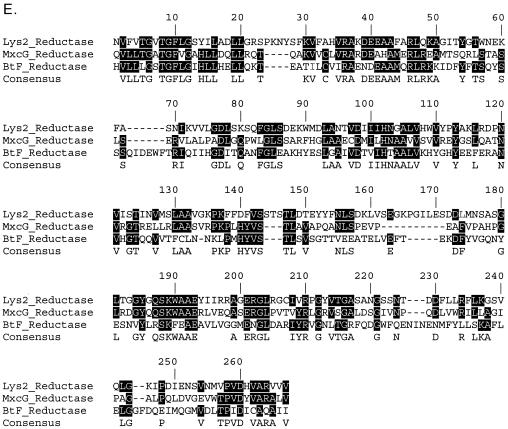
Reductase domain in module 13.
A domain consisting of about 500 amino acids was identified at the C terminus of BtF or module 13. BLAST analysis showed that this domain has high levels of similarity with several NADPH-dependent reductases from other NRPSs and polyketide synthetases. An alignment of this domain with the reductase domains of MxcG of Stigmatella aurantiaca and Lys2 of Saccharomyces cerevisiae is shown in Fig. 4E. A similar reductase domain has also been identified in the gramicidin A NRPS (9). All three reductases have been experimentally demonstrated to reduce their substrates to corresponding aldehydes in an NADPH-dependent reaction (5, 9, 19). For myxochelin A and gramicidin A, the aldehydes are further reduced to alcohols. The exact mechanism for the second reduction step has not been identified. Either the reductases themselves or other proteins carry out the second reduction step or the second reduction step is spontaneous. The MS-MS experiment suggested that the C-terminal residue of BT1583 might be valinol (Fig. 2B). The A-domain specificity prediction for the last putative BT NRPS module and the presence of a reductase domain in the module confirmed this suggestion.
btG encodes an ABC transporter.
btG is an ORF that is immediately downstream of btF, and it is transcribed in the same direction as other BT ORFs. The initiation codon, ATG, is located 61 bp downstream of the btF stop codon. The translated amino acid sequence exhibited high levels of similarity to members of the ATP-binding cassette (ABC) transporter superfamily (data not shown). ABC transporter ORFs are found in typical NRPS operons. They have been proposed to provide the host with resistance to the peptide antibiotic product by pumping the peptide out of the cells. The exact role of the putative BtG ABC transporter needs to be established.
BT1583 peptide sequence refinement.
The substrate specificity-conferring residues (21) were extracted from all 13 A-domains and were compared to the collection of amino acid-binding pocket constituents in the public NRPS codon database (raynam.chm.jhu.edu/∼nrps/index.html) (3). Substrate specificity predictions were made based on the sequence alignments, and they are shown in Table 3. The amino acid-binding pocket constituents of the first module showed a perfect match with an NRPS codon for threonine/dehydrothreonine, and it was predicted that module 1 incorporates a threonine derivative. N,N-methylated Bmt was proposed to be the N-terminal amino acid residue according to the MS-MS data (Fig. 2B and Table 2). Although the two proposals do not completely agree with each other, both indicate that a threonine derivative is the N-terminal amino acid residue.
As mentioned above, three unambiguous specificity assignments could be made for module 4 (Ile), module 9 (Leu), and module 12 (Leu) according to the NRPS codon database. These assignments were compatible with the partial BT1583 sequence; accordingly, positions 4, 9, and 12 of the BT1583 peptide were refined to Ile, Leu, and Leu, respectively. Since the only Ile of the BT1583 peptide had been assigned to position 4, the remaining Leu was assigned to position 2. The A-domain specificity of module 2 was therefore deduced to be Leu. These assignments, in conjunction with the E-domain position information, allowed us to refine the BT1583 peptide sequence to (CH3)2-Bmt-Leu-dOrn-Ile-Val-Val-dLys-Val-dLeu- Lys-dTyr-Leu-Val-CH2OH.
Novel NRPS codons in BT biosynthesis.
As mentioned above, the amino acid-binding pocket constituents of modules 5, 6, and 8 are identical. They differ from those of module 13 by only one residue. No good matches were found for these sets of amino acid-binding pocket constituents in the NRPS codon database. However, they showed similarities to certain Ile, Leu, or Val NRPS codons in the database. Since the partial BT1583 peptide sequence had Val residues at positions 5, 6, 8, and 13, modules 5, 6, 8, and 13 were deduced to incorporate Val. The amino acid-binding pocket constituents of modules 5, 6, 8, and 13 represent potential novel NRPS codons for Val.
The amino acid-binding pocket constituents of modules 7 and 10 are identical, and they differ from those of module 3 by only one residue. No match was found for these sets of amino acid-binding pocket constituents in the NRPS codon database. Since the partial BT1583 peptide sequence had Lys residues at positions 7 and 10, the specificities of these modules were deduced to be Lys. Likewise, the partial BT1583 peptide sequence had an Orn residue (whose structure is very similar to that of Lys) at position 3, and the specificity of module 3 was therefore deduced to be Orn. The amino acid-binding pocket constituents of modules 7 and 10 represent potentially the first NRPS codon for Lys, while those of module 3 represent a potential novel NRPS codon for Orn.
The specificity prediction for module 11 was quite ambiguous according the NRPS codon database. No good match was found for this set of amino acid-binding pocket constituents in the NRPS codon database. However, module 11 showed similarities to certain Phe, Trp, or Tyr NRPS codons in the database (data not shown). Since the partial BT1583 peptide sequence had Tyr residues at position 11, the A-domain specificity of module 11 was therefore deduced to be Tyr. The amino acid-binding pocket constituents of module 11 represent a potential novel NRPS codon for Tyr.
Experimental testing of the novel NRPS codons.
Since BT biosynthesis involves novel NRPS codons, experimental establishment of the novel codons (especially the novel valine and lysine codons) is critical for verifying the identity of the BT NRPS operon. In addition, since the placement of Ile at position 4 in BT1583 affects the placement of three Leu residues, the module 4 codon also needs to be tested. The A-domain specificity of module 2 was deduced to be Leu, and it also needs to be tested.
Since a purified recombinant A-domain of an NRPS module can selectively and efficiently activate the cognate amino acid substrate of the NRPS module in an ATP-PPi exchange assay (10, 14), ATP-PPi exchange assays were used to experimentally establish NRPS module specificities and novel NRPS codons. Recombinant A-domains of modules 8, 5, 7, 4, and 2 of the BT NRPS were produced and purified as described in Methods and Materials. Almost completely soluble recombinant A-domain proteins were obtained. A-domain specificities were determined in ATP-PPi exchange and amino acid Km assays (see Methods and Materials). All 20 proteinogenic amino acids and l-Orn were tested for each A-domain protein, and the background noise in the experiments was usually less than 1%.
The module 8 A-domain protein was shown to activate l-Val (100%), and there was minor activation of l-Lys (10%) and l-Ile (4%). The apparent Km was determined to be 2.75 mM for l-Val. These results confirmed the novel valine NRPS codon. Similarly, the module 5 A-domain protein was found to activate l-Val (100%), l-Ile (23%), and l-Leu (17%). The apparent Km values were determined to be 1.11 mM for l-Val and 2.78 mM for l-Ile, clearly showing that l-Val is the preferred substrate for module 5.
l-Lys was the only amino acid that activated by the module 7 A-domain protein. The apparent Km was determined to be 1.12 mM. These results established the first Lys NRPS codon.
The module 4 A-domain protein was shown to selectively activate l-Ile (100%), and there was minor activation of l-Val (9%) and l-Leu (7%). The apparent Km for l-Ile was 0.5 mM.
The module 2 A-domain protein was found to be quite ambiguous. It activated l-Leu (98%) and l-Met (100%) with nearly equal efficiency, and there was significant activation of l-Val (67%) and minor activation of l-Ile (19%) and l-Phe (3.5%).
In general, all purified A-domain proteins were found to selectively activate predicted amino acid substrates in the ATP-PPi exchange assays. These results experimentally confirmed the identity of the BT NRPS operon.
Verification of the identity of the BT NRPS operon.
The results described above allowed us to propose a degenerate formula for the isomers of BT1583 (Table 5). Based on the relative substrate selectivity of each module, the BT isomers likely to be produced by B. texasporus in significant amounts were predicted, and most of the predicted BT isomers have been detected by MS-MS (such as BT1555, BT1571, BT1583, BT1599, and BT1613 in Fig. 1B).
TABLE 5.
Degenerate formula for the BT isomers
| Amino acid at positiona:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
| Me2-Bmt | L | dO | I | V | V | dK | V | dL | K | dY | L | V-CH2OH |
| M | V | I | ||||||||||
| V | L | |||||||||||
| I | ||||||||||||
| F | ||||||||||||
The numbers indicate the amino acid residues from the N terminus to the C terminus. Entries indicate possible amino acid residues at each position.
Synthetic peptides.
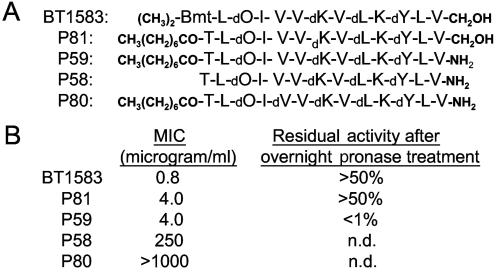
To further verify the BT peptide sequence as well as the identity of the BT NRPS operon, synthetic peptide P81 (Fig. 5) was made by Biomer Technology (Concord, CA) and tested to determine its properties. Since Bmt is not commercially available, we were not able to synthesize a peptide according to the refined BT1583 sequence, and we used octanic acid-modified threonine to synthesize the lipopeptide P81 to mimic BT1583. P81 showed antibiotic activities and pronase resistance similar to that of BT1583. These results support the refined BT1583 peptide sequence and the identity of the BT NRPS operon.
FIG. 5.
Synthetic BT variants. (A) Structures of synthetic BT variants. (B) Properties of the BT variants. The antibiotic activities were measured by determining the MICs for S. aureus as described in Materials and Methods. For pronase resistance assays, the peptides were incubated with pronase overnight at 37°C in phosphate-buffered saline, and the residual antibiotic activities against S. aureus were determined.
To investigate the significance of the C-terminal alcoholic modification, an amide form of P81 (P59) was synthesized. P59 displayed antibiotic activity but no pronase resistance. These results indicated that the C-terminal alcoholic modification plays a key role in conferring protease resistance to P81 and likely to BT1583 as well.
Since the codon for the first BT NRPS module matches known Thr NRPS codons perfectly, the possibility of an active BT isomer with an unmodified Thr at position 1 needed to be investigated. An N-terminally unmodified form of P59 (P58) was therefore synthesized, and P58 displayed poor antibiotic activity. This result confirmed that a derivative of (rather than unmodified) Thr had to be at position 1 to confer full BT antibiotic activity.
We noticed that the l- and d-form residues alternate in the middle of BT1583 with the exception of position 5 (Val). Since alternating chirality is a key structural feature for the peptide antibiotic gramicidin A, we investigated whether we missed the coding sequence of an E-domain for module 5. A d/l alternating version of P59 (P80) was synthesized. P80 displayed no antibiotic activity. The results described above confirmed not only the BT1583 peptide structure (with the exception of the N-terminal residue) but also the identity of the BT NRPS operon.
DISCUSSION
In silico NRPS operon identification.
Identification and isolation of an NRPS operon are essential to studies of a peptide antibiotic. However, identification of a specific NRPS operon remains a challenging task. Identification of an NRPS operon traditionally starts with identification of clones in a genomic BAC or cosmid library by hybridization with DNA probes from known NRPS genes or gene fragments amplified by PCR of genomic DNA using degenerate primers. Because the amino acid sequences of NRPS domains are usually quite similar, such approaches can be successful. However, because probes or primers are often imperfect, some NRPS operons can be missed. Moreover, microbes often contain multiple NRPS operons, so that the probes or primers may reveal some NRPS operons but not the operon sought. This often results in ill-fated efforts devoted to an incorrect gene (6).
We reasoned that the initial NRPS gene identification could be made more rationale if all NRPS operons in the genome were compared at the same time so that the best fit could be found. Such a comparison would require a draft of the genome. Fortunately, sequencing costs have decreased significantly so that academic labs can sequence microbial genomes. As we show here, twofold coverage is sufficient for accurate NRPS operon identification. In the actual BT NRPS operon selection, the following two sets of information were generated and compared to find the best candidate: the NRPS module clustering pattern according to similarities of the substrate-binding pocket constituents (Fig. 3D) and position information, such as the positions of d-form residues. In our opinion, the module clustering technique is especially powerful for establishing the candidacy of an operon that involves unknown NRPS codons (e.g., in the case of modules 5, 6, and 8 of the BT NRPS operon). The in silico strategy is particularly useful for NRPS operon identification in organisms (such as B. texasporus) that have a large number of NRPS operons in their genomes. Although the use of shotgun genomic sequencing to find NRPS genes for known peptides has been reported previously (11), using shotgun genomic sequencing to refine the structure of a peptide was not attempted before this study.
Acknowledgments
The amino acid composition experiment was performed by Jinny Johnson, and C18 reverse-phase HPLC was performed by Larry Dangott at the Texas A&M University Protein Chemistry Lab. We thank Shane Ticky (Department of Chemistry, Texas A&M University) for performing the MS-MS experiment and providing helpful insights.
This work was supported by a start-up grant from the Department of Medical Biochemistry and Genetics of Texas A&M University System Health Science Center to Y.W.J.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Challis, G. L., J. Ravel, and C. A. Townsend. 2000. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7:211-224. [DOI] [PubMed] [Google Scholar]
- 4.Conti, E., T. Stachelhaus, M. A. Marahiel, and P. Brick. 1997. Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J. 16:4174-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaitatzis, N., B. Kunze, and R. Muller. 2001. In vitro reconstitution of the myxochelin biosynthetic machinery of Stigmatella aurantiaca Sg a15: biochemical characterization of a reductive release mechanism from nonribosomal peptide synthetases. Proc. Natl. Acad. Sci. USA 98:11136-11141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hopwood, D. A. 1997. Genetic contributions to understanding polyketide synthases. Chem. Rev. 97:2465-2498. [DOI] [PubMed] [Google Scholar]
- 7.Keating, T. A., D. E. Ehmann, R. M. Kohli, C. G. Marshall, J. W. Trauger, and C. T. Walsh. 2001. Chain termination steps in nonribosomal peptide synthetase assembly lines: directed acyl-S-enzyme breakdown in antibiotic and siderophore biosynthesis. Chembiochemistry 2:99-107. [DOI] [PubMed] [Google Scholar]
- 8.Keating, T. A., and C. T. Walsh. 1999. Initiation, elongation, and termination strategies in polyketide and polypeptide antibiotic biosynthesis. Curr. Opin. Chem. Biol. 3:598-606. [DOI] [PubMed] [Google Scholar]
- 9.Kessler, N., H. Schuhmann, S. Morneweg, U. Linne, and M. A. Marahiel. 2004. The linear pentadecapeptide gramicidin is assembled by four multimodular nonribosomal peptide synthetases that comprise 16 modules with 56 catalytic domains. J. Biol. Chem. 279:7413-7419. [DOI] [PubMed] [Google Scholar]
- 10.Konz, D., S. Doekel, and M. A. Marahiel. 1999. Molecular and biochemical characterization of the protein template controlling biosynthesis of the lipopeptide lichenysin. J. Bacteriol. 181:133-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koumoutsi, A., X. H. Chen, A. Henne, H. Liesegang, G. Hitzeroth, P. Franke, J. Vater, and R. Borriss. 2004. Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J. Bacteriol. 186:1084-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linne, U., and M. A. Marahiel. 2000. Control of directionality in nonribosomal peptide synthesis: role of the condensation domain in preventing misinitiation and timing of epimerization. Biochemistry 39:10439-10447. [DOI] [PubMed] [Google Scholar]
- 13.Marahiel, M. A. 1997. Protein templates for the biosynthesis of peptide antibiotics. Chem. Biol. 4:561-567. [DOI] [PubMed] [Google Scholar]
- 14.Mootz, H. D., and M. A. Marahiel. 1997. The tyrocidine biosynthesis operon of Bacillus brevis: complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. J. Bacteriol. 179:6843-6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mootz, H. D., D. Schwarzer, and M. A. Marahiel. 2002. Ways of assembling complex natural products on modular nonribosomal peptide synthetases. Chembiochemistry 3:490-504. [DOI] [PubMed] [Google Scholar]
- 16.Offenzeller, M., G. Santer, K. Totschnig, Z. Su, H. Moser, R. Traber, and E. Schneider-Scherzer. 1996. Biosynthesis of the unusual amino acid (4R)-4-[(E)-2-butenyl]-4-methyl-l-threonine of cyclosporin A: enzymatic analysis of the reaction sequence including identification of the methylation precursor in a polyketide pathway. Biochemistry 35:8401-8412. [DOI] [PubMed] [Google Scholar]
- 17.Offenzeller, M., Z. Su, G. Santer, H. Moser, R. Traber, K. Memmert, and E. Schneider-Scherzer. 1993. Biosynthesis of the unusual amino acid (4R)-4-[(E)-2-butenyl]-4-methyl-l-threonine of cyclosporin A. Identification of 3(R)-hydroxy-4(R)-methyl-6(E)-octenoic acid as a key intermediate by enzymatic in vitro synthesis and by in vivo labeling techniques. J. Biol. Chem. 268:26127-26134. [PubMed] [Google Scholar]
- 18.Quadri, L. E., J. Sello, T. A. Keating, P. H. Weinreb, and C. T. Walsh. 1998. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem. Biol. 5:631-645. [DOI] [PubMed] [Google Scholar]
- 19.Sagisaka, S., and K. Shimura. 1959. Enzymic reduction of alpha-amino-adipic acid by yeast enzyme. Nature 184(Suppl. 22):1709-1710. [DOI] [PubMed] [Google Scholar]
- 20.Stachelhaus, T., H. D. Mootz, V. Bergendahl, and M. A. Marahiel. 1998. Peptide bond formation in nonribosomal peptide biosynthesis. Catalytic role of the condensation domain. J. Biol. Chem. 273:22773-22781. [DOI] [PubMed] [Google Scholar]
- 21.Stachelhaus, T., H. D. Mootz, and M. A. Marahiel. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493-505. [DOI] [PubMed] [Google Scholar]