Abstract
The abundant microbial population in a 3,043-m-deep Greenland glacier ice core was dominated by ultrasmall cells (<0.1 μm3) that may represent intrinsically small organisms or starved, minute forms of normal-sized microbes. In order to examine their diversity and obtain isolates, we enriched for ultrasmall psychrophiles by filtering melted ice through filters with different pore sizes, inoculating anaerobic low-nutrient liquid media, and performing successive rounds of filtrations and recultivations at 5°C. Melted ice filtrates, cultures, and isolates were analyzed by scanning electron microscopy, flow cytometry, cultivation, and molecular methods. The results confirmed that numerous cells passed through 0.4-μm, 0.2-μm, and even 0.1-μm filters. Interestingly, filtration increased cell culturability from the melted ice, yielding many isolates related to high-G+C gram-positive bacteria. Comparisons between parallel filtered and nonfiltered cultures showed that (i) the proportion of 0.2-μm-filterable cells was higher in the filtered cultures after short incubations but this difference diminished after several months, (ii) more isolates were obtained from filtered (1,290 isolates) than from nonfiltered (447 isolates) cultures, and (iii) the filtration and liquid medium cultivation increased isolate diversity (Proteobacteria; Cytophaga-Flavobacteria-Bacteroides; high-G+C gram-positive; and spore-forming, low-G+C gram-positive bacteria). Many isolates maintained their small cell sizes after recultivation and were phylogenetically novel or related to other ultramicrobacteria. Our filtration-cultivation procedure, combined with long incubations, enriched for novel ultrasmall-cell isolates, which is useful for studies of their metabolic properties and mechanisms for long-term survival under extreme conditions.
The prevalence of ultrasmall microorganisms in natural environments has been known for 30 years. However, ultrasmall cells have been difficult to cultivate, and because there are few isolates, our knowledge of their diversity, physiology, and ecological role remains limited (3, 17). Initial studies found that more than 70% of the soil microorganisms, named “dwarf,” had diameters of less than 0.3 μm, and only 0.2% of these were culturable (1, 2). More recent studies have further explored the abundance and diversity of ultrasmall Bacteria and Archaea in different soil, subsurface, marine, and freshwater habitats (10, 12, 13, 17, 23, 28). These small cells have been reported using different names, including the term “ultramicrobacteria” (38), which was used to describe cells with volumes of less than 0.1 μm3 (31). Small microbial cell size is considered to be advantageous for more efficient nutrient uptake in oligotrophic conditions due to a larger surface-to-volume ratio, protection against predators, and occupation of microenvironments (3). Members of this ultrasmall population either may represent a distinct class of intrinsically small organisms or may be starved, dormant, minute forms of normal-sized microbes (10, 13, 28). An important problem in environmental microbiology is to distinguish between these two possibilities. This is especially relevant with the increased interest in understanding cellular stress responses, adaptation to extreme environments, and the viable-but-not-culturable phenomenon and for exploring the size limitations for life.
One of the few cultivated and characterized ultramicrobacteria is Sphingopyxis alaskensis, which is an abundant oligotrophic organism in the bay waters of Alaska (4, 30). Recently, improved cultivation strategies resulted in the isolation of some previously uncultured “C-shaped” ultramicrobacteria, such as Pelagibacter ubique (SAR11) (Alphaproteobacteria) (27), Polynucleobacter necessarius (Betaproteobacteria) (9), and gram-positive Actinobacteria from the Luna 1 and 2 clusters (10). Studies with these few model ultrasmall microorganisms have been important for establishing their existence and examining their unique characteristics. The phylogenetic and physiological diversity of these initial isolates illustrates the potential variety of small-celled organisms and demonstrate the need to develop better detection and cultivation strategies. Further investigations of ultrasmall cells are needed to determine their abundance and diversity in different environments and to answer questions about their distinctive features, metabolic activities, and roles in global cycling. An important step in this process is to determine whether these cells pass though filters with different pore sizes, because many sampling protocols include a filtration step and analyze only the cells trapped on the filter. If the ultrasmall cells pass through these filters, a prominent portion of the microbial diversity may be lost in the filtrates.
Glacial ice is an intriguing habitat for studying ultrasmall prokaryotes, because it represents an extreme environment with low nutrient concentrations and subzero temperatures. Studies of frozen environments not only will expand our knowledge of microbial diversity but will provide insights into microbial survival for extended times, help define the limits of life on Earth, and serve as analogues for extraterrestrial cold habitats such as Mars. Previously, we found an abundant and diverse microbial population in a 3,043-m-deep Greenland ice core (22, 34). During that study, we observed many cells smaller than 1 μm that might belong to the group of ultramicrobacteria. Small cell sizes had been detected in other ice samples (16, 26), but no reports have focused on the distribution and diversity of ultrasmall microorganisms in permanently frozen environments. In the present study, we investigated the abundance and diversity of ultrasmall cells in the 120,000-year-old “silty” portion of the Greenland ice. We enriched for ultrasmall psychrophiles by selectively filtering melted ice through filters with different pore sizes and incubating the samples in anaerobic, low-nutrient media at low temperatures. The objectives were to (i) examine methods suitable for the observation, enumeration, size estimation, cultivation, and identification of ultrasmall microorganisms; (ii) compare the cell size distributions and rates of isolate recovery from populations in filtered and nonfiltered parallel cultures; and (iii) obtain isolates to determine their phylogenetic diversity and test whether they retain their small-cell-size characteristics.
MATERIALS AND METHODS
Ice core and sampling.
The ice core sample studied originated from the Greenland Ice Sheet Project (GISP2) and corresponded to depths of 3,042.67 to 3,042.80 m below the surface. The aseptic sampling procedure was performed as described previously (34).
Media and cultivation.
Anaerobic liquid cultures were incubated at −2°C or 5°C in 160-ml serum bottles containing 50 ml of one of the following media: MM1 (a low-salt medium supplemented with 0.1 M formate or acetate as a carbon source), full strength R2B (1×), 1/4 strength R2B (1/4×), or 1/10 strength R2B (1/10×) (22). Agar media containing 1×, 1/10×, or 1/100× R2A or tryptic soy agar (TSA) were incubated aerobically at 5°C. Plates containing similar media were also incubated anaerobically with the enzyme additive Oxyrase (Oxyrase, Inc., Mansfield, OH). Growth of isolates was further tested on the same media at 2°C, 10°C, 18°C, 25°C, 30°C, and 37°C.
Selective filtrations of melted ice.
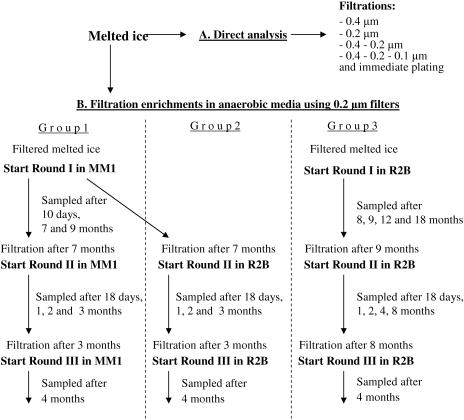
Ice core samples were aseptically melted and subjected to either one or multiple successive filtrations through polypropylene filters with different pore sizes (Acrodisc; Pall Corporation, Ann Arbor, MI), as follows: (i) 0.4-μm filtration; (ii) 0.4- and 0.2-μm filtrations; (iii) 0.4-, 0.2-, and 0.1-μm filtrations; and (iv) 0.2-μm filtration (Fig. 1A). Filtrates from each step were analyzed by flow cytometry, scanning electron microscopy (SEM), cultivation on agar media, and DNA analyses.
FIG. 1.
Flow chart of the filtration and enrichment cultivation procedure and analyses. Similar nonfiltered cultures were started and analyzed in parallel.
Enrichments for ultrasmall microorganisms.
Samples from the original melted ice were passed through 0.2-μm filters to enrich for ultrasmall cells, and the filtrates were inoculated into low-nutrient media (MM1 and R2B of different strengths). After anaerobic incubation for 2 to 8 months at −2°C or 5°C, samples were taken from each culture and filtered again, and the filtrates were used for the next round of cultivation as diagrammed in Fig. 1B. Parallel cultures were also started using nonfiltered inocula. Cultures inoculated with filtrates are designated filtered cultures, whereas those inoculated without filtration are referred to as nonfiltered cultures. The cultures were divided into three groups (Fig. 1B). The first group included first, second, and third rounds of filtered cultures in minimal medium MM1 containing either acetate or formate. The second group was inoculated from the first-round cultures in MM1, but further rounds were cultured in R2B of different strengths. All rounds of the third group were in R2B broth (Fig. 1B). Samples, taken periodically from each filtered and nonfiltered culture, were examined by flow cytometry, scanning electron microscopy, cultivation on agar media, and DNA analyses.
Flow cytometry.
Samples (0.5 ml) were fixed with glutaraldehyde (1% final concentration) for 18 h at 4°C in order to preserve the cells and permit dye penetration. The cells were stained with 2 μl 0.5 mM green fluorescent SYTO13 (Molecular Probes, Eugene, OR) for 30 min in the dark and analyzed on an XL-MCL Beckman-Coulter (Miami Lakes, FL) flow cytometer by the following optimized protocol. In addition to careful setting of the flow cytometry parameters to avoid the instrument noise, we used autoclaved H2O that had been filtered through a 0.1-μm Millipore filter as a sheath solution, filtered dimethyl sulfoxide for dilution of SYTO13, a small quantity of beads added to samples with low cell density, and extended times (20,000 events or 300 s) for each analysis at a low rate. For cell enumeration, a solution containing 104 precounted 1-μm red fluorescent beads (Polysciences, Inc.) was added as an internal standard. The number of ultrasmall filterable cells was estimated by passing a portion of each culture sample through a 0.2-μm filter and performing comparative flow cytometry analysis of the filtrate and the original sample. We also used different gate settings to enumerate only the ultrasmall cells and confirm their relative abundances in different enrichments.
SEM.
All samples from enrichments and filtrates were applied on Poretics polycarbonate 0.2-μm filters (Osmonics, Inc., Minnetonka, MN). Filters were kept in the filter holders, and a syringe was used for all successive processing and washing as described previously (33). SEM images were taken on a JEOL 5400 electron microscope at 20 kV. Cell volumes were calculated as described by Jansen et al. (15) and are presented as average values for at least 10 cells.
Genomic DNA extraction from isolates and 16S rRNA gene sequence analyses.
Genomic DNA was extracted from cells by using the PureGene kit (Gentra Systems, Inc., Minneapolis, MN). Cells resistant to lysis were disrupted by bead beating for 5 min, using a MiniBeadbeater-8 cell disrupter (Biospec Products, Inc.). The 16S rRNA genes were amplified with 63F-1387R bacterial primers (20) or 515F-1492R universal primers, using Ready-to-go PCR Beads (Amersham Biosciences, NJ). The PCR profile was as follows: initial denaturation at 95°C for 5 min; 35 cycles consisting of 95°C for 1 min, 55°C for 1 min, and 72°C for 1.5 min; and a final elongation step at 72°C for 7 min. Amplification ribosomal DNA restriction analysis (ARDRA) of the PCR products was performed with RsaI or MspI (Promega, Madison, WI) to group the isolates. The bacterial 16S rRNA gene products representing each distinct pattern were further purified using a PCR purification kit (QIAGEN, MA) and sequenced at the Penn State Nucleic Acid Facility on an ABI 370 sequencer, using the 63F and 704F primers. Sequences were compared with those from the GenBank database and the Ribosomal Database Project II (19) and aligned with reference sequences by using ClustalX. PAUP 4.0 Beta 10 (37) was used to generate rooted phylogenetic trees.
Genomic DNA extraction from cells in filtrates and cultures.
Total genomic DNA was extracted from 3- to 5-ml samples by using the Ultra Clean microbial DNA kit (MoBio Laboratories Inc., Solana Beach, CA), applying 3 min of bead beating. The DNA was used for PCR amplification of the 16S rRNA genes and the 16S rRNA-23S rRNA intergenic spacer (IGS) region and for enterobacterial repetitive intergenic consensus (ERIC) PCR fingerprinting.
Cloning of 16S rRNA gene and sequence analyses.
PCR products obtained from total genomic DNA extracted from filtrates of enrichment cultures by using bacterial primers 63F-1387R were ligated into the PCR-Script Amp vector (Stratagene, La Jolla, CA) according to the manufacturer's protocol. Ligation products were transformed into Z-Competent Escherichia coli DH5α cells (Zymo Research, Orange, CA). For initial screening, transformants containing plasmids with inserts were grown in 1.5 ml of LB broth with ampicillin (100 μg/ml) at 37°C, and plasmid DNA was extracted by the boiling procedure (29). Plasmid DNAs were restricted with PstI and NotI to determine which ones contained inserts of the appropriate size. Candidate plasmids were then used for reamplification of the 16S rRNA gene with T7-T3 primers. ARDRA was performed with either RsaI or MspI to group the amplified products. Individual PCR-amplified 16S rRNA gene products were purified and sequenced as described above.
Ribosomal IGS DNA analysis and genomic fingerprinting.
The IGS regions were PCR amplified as described for the 16S rRNA gene amplification, using bacterial (16S-1406F and 23S-115R) primers and Ready-to-go PCR beads. Products were analyzed in 3% agarose (OmniPur; Merck) gels in Tris-acetate buffer, pH 8. Comparative genomic fingerprinting was performed with a single primer, ERIC (5′-AAGTAAGTGACTGGGTGAGCG-3′), according to the protocol previously described (21). Electrophoretic separation of the products was in 1% or 2.5% Tris-acetate agarose at 5 V/cm.
Nucleotide sequence accession numbers.
GenBank accession numbers for the 16S rRNA reference gene sequences are given in Fig. 3 and Fig. 8. GenBank 16S rRNA gene sequence accession numbers for each of the isolates used in the alignments in Fig. 3 are as follows: SO3-1, DQ145736; SO3-2, DQ145737; SO3-3N, DQ148293; SO3-4b, DQ145738; SO3-5, DQ145739; SO3-6, DQ145740; SO3-7r, DQ145741; SO3-7w, DQ145742; SO3-8, DQ145743; SO3-9, DQ145744; SO3-10-4, DQ145745; SO3-11-3, DQ145746; SO3-12, DQ145747; SO3-13, DQ145748; SO3-14, DQ145749; SO3-16, DQ145750; and SO3-17a, DQ145751. GenBank 16S rRNA gene sequence accession numbers for each of the isolates used in the alignments in Fig. 8 are as follows: UMB 2, DQ147559; UMB 3, DQ147560; UMB 4, DQ147561; UMB 5, DQ147562; UMB 8, DQ147563; UMB 10, DQ147564; UMB 11, DQ147565; UMB 12, DQ147566; UMB 13, DQ147567; UMB 14, DQ147568; UMB 15, DQ147569; UMB 17, DQ147570; UMB 19, DQ147571; UMB 20, DQ147572; UMB 24y, DQ147573; UMB 25or, DQ147574; UMB 26, DQ147575; UMB 27y, DQ147576; UMB 28, DQ147577; UMB 29, DQ147578; UMB 32, DQ147579; UMB 33p, DQ147580; UMB 33y, DQ147581; UMB 34, DQ147582; UMB 37, DQ147583; UMB 38, DQ147584; UMB 40, DQ147585; UMB 42, DQ147586; UMB 45, DQ147587; UMB 46, DQ147588; UMB 49, DQ147589; UMB 51, DQ147590; UMB 62, DQ147591; UMB 64, DQ147592; UMB 69, DQ147593; UMB 70, DQ147594; UMB 71, DQ147595; UMB 71aN, DQ147596; UMB 72, DQ147597; UMB 74w, DQ147598; UMB 79, DQ147599; UMB 80, DQ147600; UMB 81, DQ147601; UMB 82a, DQ147602; and UMB 82b, DQ147603.
FIG. 3.
Phylogenetic relationships of 17 rRNA gene sequences obtained from isolates recovered from melted ice and filtrates and of 13 closely related sequences, based on a distance analysis (neighbor-joining algorithm with Juke-Cantor model; 1,000 bootstrap replicates were performed).
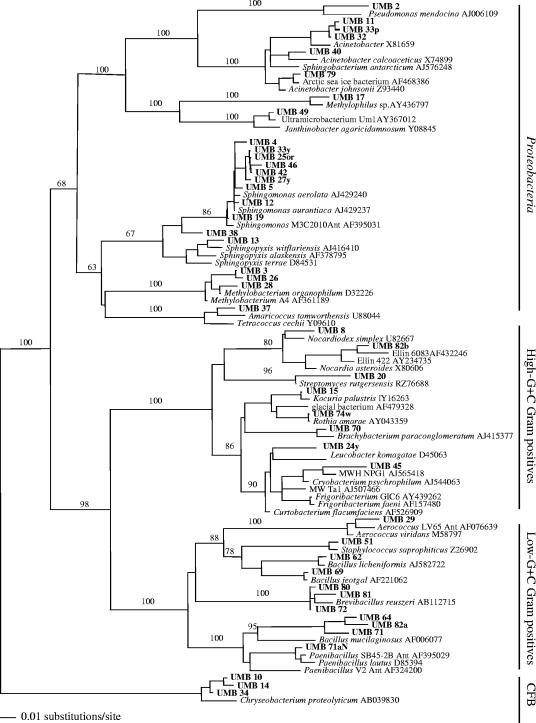
FIG. 8.
Phylogenetic relationships of the 45 rRNA gene sequences obtained from the isolates and 46 closely related sequences, based on a distance analysis (neighbor-joining algorithm with Juke-Cantor model; 1,000 bootstrap replicates were performed).
RESULTS
Direct cultivation of filterable microorganisms from melted ice.
In order to study the abundance of the potentially ultrasmall cells in the deep Greenland ice core, we first compared the cells in melted ice versus the same volume of melted ice passed through either 0.4-μm or 0.2 μm filters or through a series of successive filtrations using either 0.4- and 0.2-μm or 0.4-, 0.2-, and 0.1-μm pore sizes (Fig. 1A). SEM and flow cytometry analyses showed that a single filtration through a 0.4-μm filter successfully separated ultrasmall cells and that the successive filtrations through 0.4-, 0.2-, and 0.1-μm filters further reduced the cell sizes and numbers, due to removal of larger cells (data not shown).
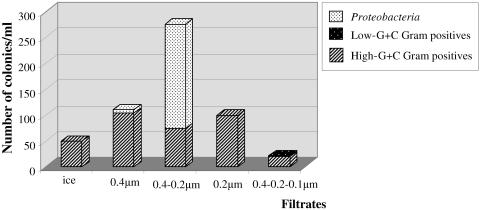
Equal volumes of both nonfiltered and filtered ice core samples were plated onto 1/10× and 1/100× TSA or R2A agar media to determine whether the separation provided by filtration would enhance recovery and growth of ultrasmall microorganisms. Colonies appeared after 2 to 6 months of incubation at 5°C. The number of colonies obtained was 50 per ml of melted nonfiltered ice and ranged from 22/ml for the sample filtered through the 0.4-, 0.2-, and 0.1-μm-pore-size series to 276/ml for the sample filtered through 0.4- and 0.2-μm pore sizes (Fig. 2). The recovery of culturable organisms from the melted ice was extremely low (0.00035%, based on the originally estimated 1.4 × 107 cells/ml) and increased to 2 to 6% recovery for the filtrates which contained 4 × 103 to 7 × 103 cells/ml. The morphological diversity of isolates was limited and was dominated by white and yellowish-pigmented colonies of non-spore-forming gram-positive or red or yellow colonies of gram-negative ultrasmall cells. Approximately 100 fungal colonies of different morphologies also grew, but these were not studied further.
FIG. 2.
Total numbers of colonies obtained after plating samples of either melted ice or liquid from filtrates that had passed through filters with different pore sizes. The phylogenetic distribution of isolated bacteria was determined by ARDRA and 16S rRNA gene sequence analysis.
Phylogenetic analysis of isolates from filtered melted ice.
Seventeen bacterial isolates (15 from filtrates and 2 from the melted ice) were selected to represent the observed different colony and cell morphotypes. Results from ARDRA and sequence analysis of 16S rRNA genes from these isolates were used to represent the distribution of these and similar colonies belonging to the same morphotype into the broad groups of high-G+C gram-positive bacteria, low-G+C gram-positive bacteria, and Proteobacteria (Fig. 2). The most abundant group of high-G+C gram-positive bacteria originated predominantly from the filtrates, indicating that their cells had been sufficiently small to pass through 0.4-, 0.2-, and even 0.1-μm filters. Interestingly, an increase in the number and diversity of isolates was obtained from the filtered samples versus the directly plated melted ice (Fig. 2), possibly because some inhibitors had been removed or clumped cells were detached from debris during filtration. Dissociation of cell aggregates could account for the more than 200 colonies of similar-appearing Proteobacteria recovered from the 0.4-μm-0.2-μm filtrate.
Further phylogenetic comparisons (Fig. 3), revealed that 14 of the 17 isolates were related to the genera Arthrobacter and Microbacterium within the families Micrococcineae and Microbacteriaceae, one isolate was affiliated with the genus Paenibacillus, and two were related to psychrophilic representatives of Sphingomonas. The 16S rRNA gene sequences of the two isolates from the melted ice, SO3-8 and SO3-9, were highly similar to those of other Arthrobacter isolates from filtrates. Particularly interesting as novel isolates were SO3-5, SO3-6, SO3-7r, and SO3-12. These had 16S rRNA gene sequence distances to the closest validated species of 6.7% to Arthrobacter globiformis, 4.0% to the Antarctic species Paenibacillus wynnii, 4.8% to Sphingomonas faeni, and 2.2% to Arthrobacter chlorophenolicus, respectively (Table 1).
TABLE 1.
Characteristics of representative isolates with ultrasmall cells recovered from melted ice filtrates, filtered cultures, and nonfiltered cultures
| Origin | Isolate | Isolation conditionsa | Cell morphology | Avg cell vol (μm3) | Growth temp range (°C) | Closest validated relative species (accession number) | Distance (%) |
|---|---|---|---|---|---|---|---|
| Melted ice filtrate | SO3-2 | 0.4-0.2-0.1 μm | Coccoid | 0.065 | 10-37 | Microbacterium aurum (Y17229) | 1.0 |
| SO3-5 | 0.4-0.2-0.1 μm | Short rods | 0.070 | 2-37 | Arthrobacter globiformis (M23411) | 6.7 | |
| SO3-6 | 0.4-0.2-0.1 μm | Rods, spores | 2-37 | Paenibacillus wynnii (AJ633647) | 4.0 | ||
| SO3-7r | 0.4 μm | Thin rods | 0.086 | 2-30 | Sphingomonas faeni (AJ429239) | 4.8 | |
| SO3-7w | 0.4 μm | Short rods | 0.070 | 2-30 | Arthrobacter sulfonivorans (AF235091) | 1.6 | |
| SO3-10-4 | 0.4 μm | Short rods | 0.070 | 2-30 | Arthrobacter globiformis (M23411) | 1.5 | |
| SO3-12 | 0.4 μm | Coccoid | 0.062 | 2-37 | Arthrobacter chlorophenolicus (AF102267) | 2.2 | |
| SO3-17a | 0.2 μm | Short rods | 2-37 | Arthrobacter globiformis (M23411) | 2.1 | ||
| Cultures Filtered | UMB 13 | Round II, R2B, 18 days | Thin rods | 0.054 | 2-30 | Sphingopyxis witflariensis (AJ416410) | 2.0 |
| UMB 14 | Round II, R2B, 18 days | Short rods | 2-37 | Chryseobacterium proteolyticum (AB039830) | 3.8 | ||
| UMB 38 | Round II, 1/100 × R2B, 3 mo | Small ovoid | 0.043 | 18-30 | Sphingomonas aerolata (AJ429240) | 4.8 | |
| UMB 40 | Round II, 1/100 × R2B, 3 mo | Short rods | 10-37 | Acinetobacter calcoaceticus (X74899) | 2.9 | ||
| UMB 24y | Round I, MM1 + formate, 18 days | Coccoid | 0.100 | 2-37 | Curtobacterium flaccumfaciens (AF526909) | 0.7 | |
| UMB 49 | Round I, R2B, 7 mo | Thin rods | 0.043 | 2-30 | Janthinobacter agaricidamnosum (Y08845) | 2.7 | |
| UMB 82a | Round II, R2B, 8 mo | Short rods, small spores | 2-37 | Bacillus mucilaginosus (AF006077) | 9.5 | ||
| Nonfiltered | UMB 10 | Round II, MM1 + formate, 18 days | Short rods | 0.080 | 10-37 | Chryseobacterium proteolyticum (AB039830) | 3.7 |
| UMB 19 | Round II, R2B, 30 days | Short rods | 0.080 | 2-30 | Sphingomonas aurantiaca (AJ429236) | 1.1 | |
| UMB 34 | Round II, MM1 + formate, 18 days | Short rods | 10-37 | Chryseobacterium proteolyticum (AB039830) | 3.4 | ||
| UMB 45 | Round I, MM1 + formate, 7 mo | Pleomorphic rods | 0.080 | 2-18 | Cryobacterium psychrophilum (AJ544063) | 1.0 |
See Fig. 1.
Because some bacterial isolates showed nearly identical 16S rRNA gene sequences, we performed ERIC PCR genomic fingerprinting in order to either confirm identity or detect differences. Our results showed that all isolates, with the exception of the highly similar, SO3-2 and SO3-3N, had unique strain-specific profiles, demonstrating that we were not reisolating identical strains (data not shown).
Comparisons of filtered and nonfiltered cultures.
Even though we obtained small-celled and potentially novel organisms following the direct plating of the melted ice and filtrates onto agar media, their low number and restricted diversity prompted further attempts to improve culturability. Our previous results (22) suggested that long-term incubation in anaerobic, low-nutrient liquid media prior to plating increased recovery of both aerobic and facultatively anaerobic heterotrophs. Thus, we designed cultivation and sampling strategy to incorporate incubation in anaerobic media at low temperature while addressing the question of whether successive filtrations would enrich for ultrasmall-celled organisms (Fig. 1B). Samples from each round of cultivation started from filtered or nonfiltered inocula were examined by flow cytometry and SEM to determine whether the proportion of cells in the filtered inoculum remained small or represented organisms that were small only during a portion of their life cycle and thus would be excluded during subsequent filtrations. The IGS analyses of the rRNA genes were used to follow the dynamic population changes in the samples. Three rounds of parallel filtered and nonfiltered cultures in different media (Fig. 1B) were examined periodically for 20 months.
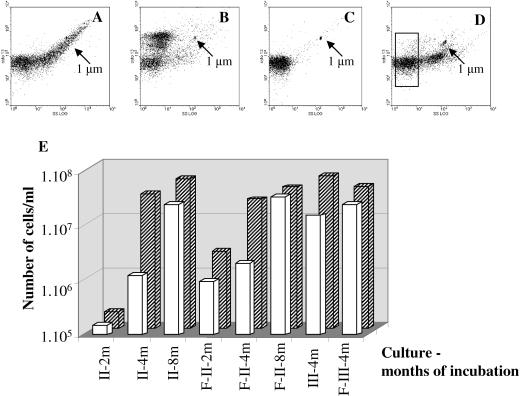
Flow cytometry analyses monitored the cell numbers and size distributions in each culture after different incubation times. Typical results showed a wide distribution of cell sizes in the original melted ice sample (Fig. 4A). An examination of an R2B culture started with a filtrate and incubated for several months showed an enrichment for small cells that clustered into groups with different fluorescence intensity, which is related to the amount of DNA (Fig. 4B). The cluster of cells with increased fluorescence indicates higher DNA content and probable cell growth. Filtration of samples of the enrichment culture prior to flow cytometry analysis usually resulted in the appearance of only small cells in the filtrate (Fig. 4C). In contrast, a sample from a nonfiltered culture typically had a wider distribution of cell sizes (Fig. 4D). Similar flow cytometry results with other cultures verified the effectiveness of the filtration step as an enrichment approach for small-celled populations.
FIG. 4.
Examples of flow cytometry plots of side scatter (SS) versus green fluorescence of different enrichment populations. Cells were stained with SYTO 13. Arrows show the position of 1-μm beads. (A) Melted ice sample; (B) round II filtered R2B culture showing three clusters of small cells with different fluorescence intensities; (C) filtrate of an enrichment culture showing small cells (see white bars in panel E); (D) round II nonfiltered culture in R2B with two cell clusters with different sizes and a gate for ultrasmall cells; (E) flow cytometry results showing the number of filterable cells (white bars) that pass through a 0.2-μm filter related to the total number of cells (hatched bars) in rounds II and III of nonfiltered and filtered R2B cultures (designated F-II and F-III) after 2, 4, and 8 months.
Having established the utility of flow cytometry for monitoring cell size distributions, we used the numerical flow cytometry data to determine the proportion of ultrasmall cells in an aliquot passed through a 0.2-μm filter in comparison to the total cell numbers in each culture. The results for the cultures from round II incubated in anaerobic R2B and sampled at 2, 4, and 8 months and from round III after 4 months (Fig. 4E) are representative of those obtained for several cultures. The comparisons of filtered and nonfiltered cultures showed that (i) the total number of cells increased during the 8 months of incubation, demonstrating growth; (ii) the number of filterable small cells was higher in the filtered enrichments than in the nonfiltered cultures, especially after 2 months; and (iii) the differences in cell sizes between the nonfiltered and filtered cultures were less pronounced after prolonged incubations of 4 and 8 months in round II and after 4 months in round III.
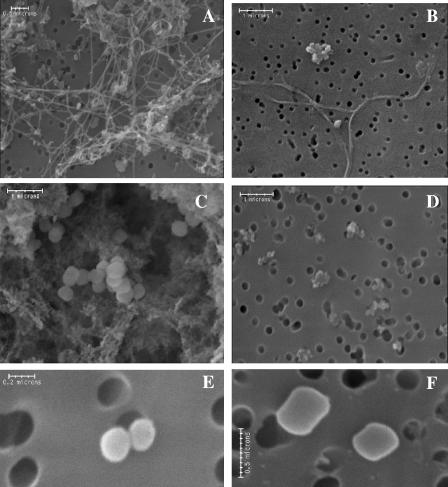
The morphological diversity in different cultures was examined by electron microscopy (Fig. 5). Filamentous structures were observed in all MM1 cultures, and their presence in filtered enrichments indicated that at least some portion of these thin structures passed through the filter pores (Fig. 5A and B). The R2B cultures had a significant amount of exopolymeric substance in addition to the cells (Fig. 5C and D). Ultrasmall cells were detected in all cultures. Examples of these cells (Fig. 5E and F) had calculated volumes of 0.011, 0.05, and 0.07 μm3.
FIG. 5.
Scanning electron micrographs showing morphology of cells from filtered enrichment cultures in different media. (A) Round II in MM1 plus formate, 2 months; (B) 0.2-μm filtrate of the same culture; (C) round II in R2B, 8 months; (D) 0.2-μm filtrate of the same culture; (E and F) individual cells from rounds I and II R2B cultures, respectively.
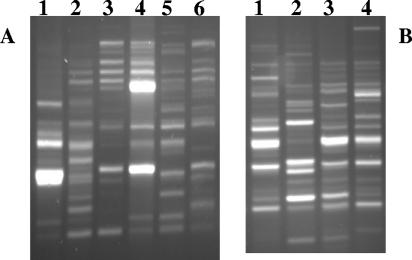
Changes in the population composition also were monitored at different times by using bacterial 16S-23S rRNA IGS PCR profiles (Fig. 6). The profiles obtained from DNA extracted from the round I MM1-plus-formate cultures inoculated with filtered melted ice (Fig. 6A, lanes 4 to 6) differed from those of similarly grown cultures inoculated with nonfiltered melted ice (lanes 1 to 3). Both cultures showed remarkably different profiles after only 10 days (lanes 1 and 4), while the profiles from filtered and nonfiltered cultures had fewer differences after longer incubation times of 7 months and 9 months (lanes 2, 5, 3, and 6). A similar pattern was observed for filtered and nonfiltered MM1-plus-acetate cultures (not shown). Comparisons of profiles obtained from round II anaerobic incubations in R2B of different strengths also showed differences in size and intensity of PCR fragments, particularly those from cells in 1/100× R2B (Fig. 6B). Purification and partial sequencing of representative bands showed relationships to novel psychrophilic Actinobacteria and Cytophaga. The IGS profiles illustrate that changes in the abundance and diversity of organisms during cultivation depended on the medium, filtration status, and incubation time. These results were consistent with the observations (Fig. 4E) that the differences between the filtered and nonfiltered cultures were more pronounced during the initial incubations and that the populations became more uniform upon subsequent longer cultivation.
FIG. 6.
Results from PCR amplification of 16S-23S rRNA gene IGS regions, using DNAs extracted from anaerobic enrichment cultures. (A) Round I enrichments in MM1 plus acetate inoculated with either nonfiltered (lanes 1 to 3) or filtered (lanes 4 to 6) ice after 10 days (lanes 1 and 4), 7 months (lanes 2 and 5), and 9 months (lanes 3 and 6). (B) Round II filtered enrichments after 18 days in 1/10× R2B (lane1), 1/100× R2B (lane 2), 1× R2B (lane 4), and nonfiltered 1× R2B (lane 3).
In addition, the diversity of the ultrasmall microbial populations of some enrichments was examined by constructing 16S rRNA gene clone libraries. The sequence analysis of 180 clones containing the 16S rRNA gene amplified from DNA of filtrates of three enrichment cultures in R2B showed relatedness to only a few organisms, including Sphingopyxis alaskensis, uncultured Betaproteobacteria, and Methylobacterium organophilum.
Recovery of isolates from filtered and nonfiltered cultures.
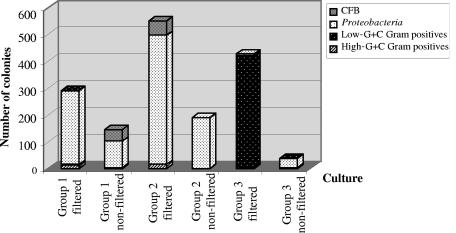
Because one goal of this study was to examine methods for cultivating ultrasmall cells and to obtain isolates for future characterization, samples from different cultures were plated and colonies examined. A total of 1,737 colonies were obtained on agar media from different cultures during a 20-month period (Fig. 7). The majority of the isolates (1,290) were recovered from successively filtered enrichments, compared to 447 isolates from similar cultures started from nonfiltered inocula. The three groups of cultures (outlined in Fig. 1B) yielded 443, 797, and 497 isolates, respectively. Despite the long-term incubation in liquid media, colonies did not appear until after 2 to 4 months of incubation at 5°C; most colonies were on low-strength R2A, with only a few on 1/10× and 1/100× TSA. A number of fungi grew as well but were not studied.
FIG. 7.
Total number of isolates from the three groups of filtered and nonfiltered cultures and their phylogenetic distribution as determined by morphotypes, ARDRA grouping, and 16S rRNA gene sequence analysis. CFB, Cytophaga-Flavobacteria-Bacteroides.
An examination of the morphotypes based on colony and cell morphology of selected isolates relative to their phylogenetic relationships (presented below) allowed grouping of all recovered colonies in broad categories based on origin (Fig. 7). The total number of isolates from the filtered enrichments was three times higher than that from nonfiltered enrichments. This statistically significant difference was best shown for Proteobacteria representatives, which dominated the isolates from both filtered and nonfiltered enrichments in groups 1 and 2. A striking difference was found in filtered group 3 enrichments incubated in R2B for 18 months, which resulted in the isolation of low-G+C gram-positive spore-forming bacteria that probably originated from filterable spores.
Phylogenetic diversity of isolates from cultures.
A total of 82 purified isolates were used for DNA extraction, 16S rRNA gene PCR amplification, and ARDRA comparisons. Phylogenetic analyses based on BLAST searches of closest validly described species were performed on 45 of the 16S rRNA gene sequences (Fig. 8). Isolates were affiliated with several major bacterial phyla: high-G+C gram-positive bacteria, low-G+C gram-positive bacteria, Proteobacteria, and Cytophaga-Flavobacteria-Bacteroides. The majority of those (24 isolates, representing more than 1,000 colonies, recovered from different enrichments) belonged to the Proteobacteria phylum, whereas each of the remaining three phyla included 2 to 11 isolates. The highest number of initially isolated colonies was related to spore-forming, low-G+C gram-positive bacteria. About 50% of the isolates could be assigned to validly described species, but at least 14 isolates may represent novel taxa, with similarity to known species of below 98%. Of significance for our study was finding diverse and novel isolates with ultrasmall cells from all major phyla.
The most numerous group of Alphaproteobacteria included 23 small-celled isolates, 9 of which formed a tight cluster closely related to Sphingomonas aurantiaca and Sphingomonas aerolata. One isolate (UMB 38) also clustered within this group but may represent a novel genus or higher taxon, based on a significant phylogenetic distance to Sphingomonas aerolata of 4.8% (Table 1). Isolate UMB 13 clustered within the genus Sphingopyxis and was related to Sphingopyxis witflariensis and the model ultramicrobacterium Sphingopyxis alaskensis, with distances of 2.0% and 2.2%, respectively. A Betaproteobacteria small-celled isolate (UMB 49), recovered on anaerobic agar with Oxyrase, was closely related to previously uncultured ultramicrobacteria Um1 (unpublished) and ND5 (13), and its closest described species, Janthinobacterium agaricidamnosum (18), was at a distance of 2.7% (Table 1).
Isolates phylogenetically related to ultramicrobacteria also were found among the high-G+C gram-positive bacteria. Based on 16S rRNA gene sequence analysis, UMB 45 clustered with the recently recovered nonculturable, ultrasmall, C-shaped bacteria MWH-NPG1 and MWTa1 from the Luna 1 cluster (10, 11) and was related to the Antarctic isolate Cryobacterium psychrophilum (36). Three ultrasmall isolates, UMB 10, UMB 14, and UMB 34, obtained after 18 days of incubation from either filtered R2B or nonfiltered MM1-plus-formate cultures, had 16S rRNA gene sequences significantly different from that of the closest validated species, Chryseobacterium proteolyticum, at distances of 3.7, 3.8, and 3.4%, respectively. These isolates may represent a new genus.
The relatively large group of low-G+C gram-positive bacteria represented more than 400 spore-forming bacilli related to Bacillus licheniformis, Bacillus jeotgal, Paenibacillus lautus, and Brevibacillus reuszeri. Some were also related to previously reported Antarctic Paenibacillus isolates (6). A numerically significant and phylogenetically novel group (UMB 64, UMB 82a, and UMB 71) showed very distant relatedness (7.5 to 9.5% distance) to a polysaccharide-degrading bacterium, Bacillus mucilaginosus (32) (Fig. 8; Table 1). Alignments of our isolates' 16S rRNA gene sequences with that of B. mucilaginosus showed that the region of greatest divergence was between positions 175 and 235 according to the E. coli numbering. These spore-forming Bacillus isolates were cultivated from the group 3, round I and II filtered enrichments in R2B after 18 months and 8 months, respectively.
The DNAs from all 82 isolates was subjected to ERIC PCR analyses, and in the majority of cases they were shown to represent different strains (data not shown). One exception was a group of isolates within the Proteobacteria and related to Sphingomonas species that had nearly identical 16S rRNA gene sequences (Fig. 8) and highly similar ERIC PCR fingerprints. Interestingly, these isolates originated from different cultivations and incubation times. Nevertheless, several isolates (UMB 25or, UMB 5, UMB 27y, UMB 4, and UMB 19) from this same group had different ERIC profiles (data not presented). Isolate UMB 38, which was related to but different at 4.8% from the above-mentioned group by 16S rRNA gene sequence, also had a distinct ERIC-PCR pattern, consistent with its 16S rRNA gene differences.
Morphology, size, and growth characteristics of the ultrasmall isolates.
Isolates initially recovered from filtrates of melted ice and from filtered and nonfiltered cultures were characterized after subsequent cultivation on different media. Most isolates grew faster when recultivated on low- and high-nutrient media (different strengths of R2A or TSA), and some changed their colony morphology. One group that initially formed yellowish colonies was subsequently separated into yellow and white colonies, which were later identified as Chryseobacterium and Acinetobacter, respectively. One small-celled Curtobacterium-related bacterial isolate, UMB 24y, was originally isolated as a coculture with a fungus belonging to Ascomycota (Fig. 9) and was eventually separated. In other cases the colony morphology varied with different growth conditions. For example, isolates UMB 19, UMB 24y, and UMB 25or formed more exopolymer on R2A at 2 or 10°C than at 18 or 25°C. Others, such as UMB 4, UMB 8, and UMB 25or, exhibited stronger pigmentation at 2°C than at higher temperatures. Many isolates grew within the tested temperatures of between 2 and 30°C, but a few grew at 37°C (Table 1).
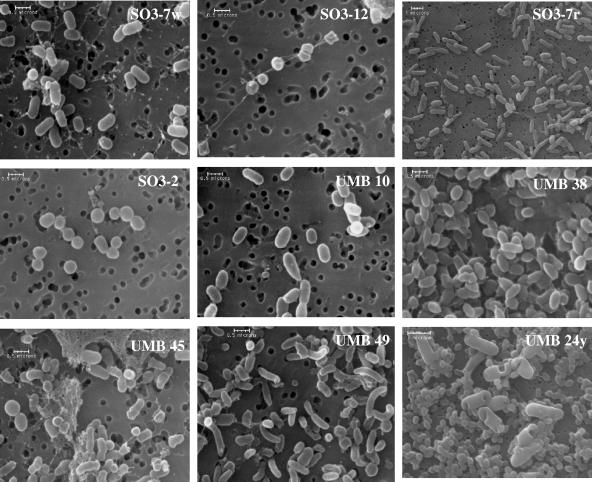
FIG. 9.
Scanning electron micrographs of newly isolated representative small-celled strains from filtered ice and from enrichment cultures with most closely related genera: isolates SO3-7w (Arthrobacter), SO3-12 (Arthrobacter), SO3-7r (Sphingomonas), SO3-2 (Microbacterium), UMB 10 (Chryseobacterium), UMB 38 (Sphingomonas), UMB 45 (Frigoribacterium), UMB 49 (Janthinobacter), and UMB 24y (Curtobacterium, also showing the Ascomycota fungus that had been present originally).
The cells were examined microscopically in order to determine whether initially recovered isolates continued to form small cells when recultivated on R2A or TSA at 18°C. Cell morphologies included rod, ellipsoid, and spherical shapes. A significant number of the isolates (95% of the isolates from melted ice in Fig. 3 and 85% of the isolates from cultures in Fig. 8) retained their small cell or spore sizes. SEM images (Fig. 9) of selected isolates showed that many had coccoidal morphology with diameters of less than 0.5 μm (SO3-2 and SO3-12) or were rod shaped (1 by 0.2 μm or 0.6 by 0.4 μm in size) (SO3-5, SO3-7r, SO3-7w, SO3-10-4, UMB 10, UMB 13, and UMB 19). The ultrasmall cells of isolate UMB 49 exhibited polymorphic shapes, and the rods had a slightly curved form (Fig. 9), similar to the “C shape” of some recently isolated ultramicrobacteria (9). The cells of isolates UMB 38 and UMB 45 (Fig. 9) were embedded in copious exopolymeric matrix, whereas SO3-7w and SO3-12 (Fig. 9) formed very thin extracellular structures. The calculated cell volumes averaged between 0.04 and 0.10 μm3, which was within the accepted volume limits of <0.1 μm3 for ultramicrobacteria (Table 1). Many of the small-celled isolates presented in Fig. 9 are phylogenetically novel and may represent new, previously uncultured and undescribed taxa.
DISCUSSION
Isolating and studying environmental ultrasmall microorganisms with possible novel metabolic activities is important because it may help define their functional diversity and ecological role. This study was based on a previous observation that very small cells dominated the microbial population found in a deep Greenland ice core (22). It has been suggested that ultrasmall microbial cells are capable of occupying the narrow liquid veins in glacier ice and possibly metabolizing at very low rates (25). Because small cells may be well adapted to the multiple stress conditions in glacier ice, the ultrasmall isolates obtained from glacier ice will be valuable for future experiments designed to test their survival and metabolic capabilities in ice and the factors needed for their recovery and cultivation.
In this study we examined a variety of methods for analyzing ultrasmall cell populations and for cultivating isolates. We found that carefully controlled flow cytometry and SEM analyses confirmed the presence of numerous small cells capable of passing through 0.2-μm, as well as 0.1-μm, filters. Thus, the traditional method of collecting cells by filtering and examining only those captured on the filters excludes many ultrasmall cells and small spores. Occasionally, cells larger than the filter pore sizes were observed in some filtrates. These cells either may have passed through the filters because of variations in pore sizes or flexibility of the cells during filtration (12, 28) or may have originally been smaller, starved dormant cells that increased their size following cultivation. According to DeLong (8), this physiological strategy appears to be common, but its actual distribution among microbial phyla is poorly understood. It is still unknown what fraction of the naturally occurring small cells represents physiologically induced dwarf forms versus stable diminutive phenotypes. Limited studies of several cultivated, intrinsically small bacteria suggest that they keep their small size independent of the growth conditions. It also is assumed that these organisms are slowly but constantly growing and thus can become a dominant population in their habitat. This is true for Sphingopyxis alaskensis (4), but it is not clear whether similar characteristics will be found for other cultured ultrasmall organisms, such as our isolates.
Other reports indicated that a large portion of the nonculturable cells in environmental samples actually corresponds to the ultrasmall cells (17). Strategies and procedures for cultivating these organisms differ, but general principles are to mimic the natural environment, use filtration or extinctive dilutions in very-low-nutrient media, and use gradual adaptation to laboratory cultivation conditions (5, 7, 9, 11, 28, 30, 35). Our enrichment strategy combined a selective filtration step with low-temperature incubation in oligotrophic, anaerobic media for several months. The 0.2-μm filtration excluded the larger cells and allowed only the ultrasmall and very thin filamentous cells to be transferred to the next round of enrichment, thus preventing overgrowth by larger, fast-growing organisms and the possible accumulation of substances inhibitory to the smaller cells. The anaerobic conditions possibly prevented oxidative stress and allowed cells that could have been damaged or dormant to resuscitate. Finally, the long-term incubations at low temperature and in low-nutrient media mimicked some conditions in the glacier environment.
One goal of this work was to answer the question whether isolates obtained after filtration and cultivation differed morphologically and phylogenetically from those not specifically enriched for ultrasmall cells. Our comparisons of the numbers and diversity of isolates from filtered and nonfiltered melted ice and filtered and nonfiltered, low-temperature cultures lead to several conclusions. First, we found that the filtration step increased the number of colonies obtained from both the melted ice and liquid cultures. The increase in culturability of cells directly from filtered melted ice to 2 to 6% may be caused partially by the physical separation and detachment of cell aggregates or by the removal of inhibitory substances during filtration. In addition, there was a significant increase in the number of isolates (1,290) obtained from the filtration enrichments compared to the number of isolates (447) obtained from unfiltered cultures. It is possible that the successive rounds of filtration and incubation provided the acclimation and recovery conditions that ultrasmall cells required to form colonies.
A second conclusion is that the filtration-cultivation steps not only increased the number of colonies but could also alter the diversity of isolates. For example, members of the high-G+C group were isolated mostly from the directly plated melted ice and its filtrates, whereas Proteobacteria were dominant among isolates from enrichment cultures. A third observation is that the proportions of 0.2-μm-filterable cells in filtered and nonfiltered liquid cultures differed after short incubations but these differences diminished after several months. This is consistent with our report (22) that long-term cultivation in liquid oligotrophic media at low temperature significantly improved the recovery and/or growth of previously nonculturable organisms, including ultrasmall isolates. These results suggest that the time of incubation is an important factor that may allow damaged or dormant cells to resuscitate and that after sufficient time, ultrasmall cells can become dominant. Other authors also pointed out that long incubation periods at low temperature during subcultivation of ultrasmall environmental microorganisms may initiate an unknown mechanism allowing colony formation on rich media (3, 30).
An important question is whether the small-celled population in a specific environment is composed of only a few species or whether it is highly diverse. Different studies have shown that direct cultivation of 0.2-μm-filterable cells from different environments resulted in obtaining remarkably low numbers of isolates of restricted diversity (39). Our results also showed that despite the relative increase in culturability from filtered melted ice, the isolates obtained directly from the melted ice had limited diversity. The dominance of Arthrobacter spp. may be explained by pleomorphism (rod-coccus cycle) and their ability to reduce their cell sizes up to 10 times in response to starvation (17). Some isolates (SO3-1, SO3-5, SO3-12, SO3-16, and SO3-17a), however, were distantly related to known Arthrobacter species and may represent novel species that form intrinsically smaller cells.
Based on the phylogenetic analysis of the isolates obtained from filtered and nonfiltered cultures, we conclude that diversity increased after the long-term cultivation, and a large number of ultrasmall cells were found in all major phylogenetic groups. Some isolates were related to organisms known for their small sizes, such as Microbacterium and Sphingomonas. Representatives of the genus Sphingomonas were the most numerous Proteobacteria from all filtered and nonfiltered group 1 and 2 cultures. However, the increased number of isolates obtained from the filtered enrichments suggests that the filtration-cultivation procedure provided favorable growth conditions for this bacterial group, including previously uncultured organisms and some that are related to known ultramicrobacteria, such as UMB 13 and UMB 38. Other Proteobacteria isolates were related to Acinetobacter, the characterized species of which also undergo cell size reduction in response to starvation (14). Interestingly, phylogenetically novel ultrasmall bacteria belonging to the Cytophaga-Flavobacteria-Bacteroides group were isolated from short-term enrichments in different media, suggesting that their cells may have predominated prior to being outcompeted or have been relatively fast recovering and/or growing during the initial incubation.
The cultivation of spore-forming representatives is interesting because we had previously noted that the absence of spore-forming isolates was surprising (22), since spores could survive for thousands of years. One explanation had been the inability of spores in the sample to germinate. In contrast, the filtration and liquid incubation in the present investigation resulted in the isolation of a significant number of diverse spore-forming bacilli after 18 months of incubation. Because little is known about spore germination from environmental samples (24), it is not clear whether the filtration step enhanced germination, perhaps by eliminating inhibitory factors or other competing cells, or whether the longer incubation permitted germination and subsequent cell growth. Several isolates, distantly related to Bacillus mucilaginosus (9.5%), possessed small cells and spores with similar small sizes and deserve further attention as a possible novel ultrasmall-cell taxon at the family or order level.
An important result from this work is that our procedure of successive filtration-cultivation steps led to a significant increase in the total number and diversity of cultured isolates with a parallel increase of the small-celled organisms compared to direct plating of filtered melted ice. The extended low-temperature cultivation in liquid oligotrophic medium also appeared to be a factor in the recovery of ultrasmall microorganisms and should be considered in the development of strategies for their isolation. In conclusion, the filtration-cultivation procedure combined with long cultivation times allowed the enrichment of ultrasmall microbes from the deep Greenland glacier ice core sample, improved their culturability, and resulted in the isolation of diverse ultramicrobacteria, including phylogenetically novel ones. Further characterization of our collection of ultrasmall isolates may provide insight into novel metabolic properties and the mechanisms for long-term survival under extreme cold conditions.
Acknowledgments
This research was supported by Department of Energy grant DE-FG02-93ER20117, NASA Astrobiology Institute grant NNA04CC06A and NSF grant MO-0347475.
We thank Todd Sowers and members of our laboratory for helpful discussions and Elaine Kunze and Missy Hazen for help with the flow cytometry and electron microscopy work.
REFERENCES
- 1.Bae, H., E. Cota-Robles, and L. Casida. 1972. Microflora of soil as viewed by transmission electron microscopy. Appl. Microbiol. 23:637-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakken, L., and R. Olsen. 1987. The relationship between cell size and viability of soil bacteria. Microb. Ecol. 13:103-114. [DOI] [PubMed] [Google Scholar]
- 3.Cavicchioli, R., and M. Ostrowski. 2003. Ultramicrobacteria. In Encyclopedia of life sciences. [Online.] McMillan Publishers Ltd., Nature Publishing Group, London, United Kingdom. http://www.els.net.
- 4.Cavicchioli, R., M. Ostrowski, F. Fegatella, A. Goodchild, and N. Guixa-Boixereu. 2003. Life under nutrient limitations in oligotrophic marine environments: an eco/physiological perspective of Sphingopyxis alaskensis. Microb. Ecol. 46:1-12. [DOI] [PubMed] [Google Scholar]
- 5.Cho, J.-C., and S. J. Goiovannoni. 2004. Cultivation and growth characteristics of a diverse group of oligotrophic marine Gammaproteobacteria. Appl. Environ. Microbiol. 70:432-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christner, B. C., E. Mosley-Thompson, L. G. Thompson, and J. N. Reeve. 2003. Bacterial recovery from ancient glacial ice. Environ. Microbiol. 5:433-436. [DOI] [PubMed] [Google Scholar]
- 7.Connon, S. A., and S. J. Giovannoni. 2002. High-thoughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl. Environ. Microbiol. 68:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLong, E. F. 2000. Diminutive cells in the ocean—unanswered questions, p. 81-84. In Proceedings of a workshop on size limits of very small microorganisms. National Academy Press, Washington, D.C.
- 9.Hahn, M. W. 2003. Isolation of strains belonging to the cosmopolitan Polynucleobacter necessarius cluster from freshwater habitats located in three climatic zones. Appl. Environ. Microbiol. 69:5248-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hahn, M. W., H. Lunsdorf, Q. Wu, M. Schauer, and M. G. Hofle. 2003. Isolation of novel ultramicrobacteria classified as actinobacteria from five freshwater habitats in Europe and Asia. Appl. Environ. Microbiol. 69:1442-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hahn, M. W., P. Stadler, Q. Wu, and M. Pockl. 2004. The filtration-acclimation method for isolation of an important fraction of the not readily culturable bacteria. J. Microbiol. Methods 57:379-390. [DOI] [PubMed] [Google Scholar]
- 12.Haller, C. M., S. Rolleke, D. Vybiral, A. Witte, and B. Velimirov. 1999. Investigation of 0.2 μm filterable bacteria from the Western Mediterranean Sea using a molecular approach: dominance of potential starvation forms. FEMS Microbiol. Ecol. 31:153-161. [DOI] [PubMed] [Google Scholar]
- 13.Iizuka, T., S. Yamanaka, T. Nishiyama, and A. Hiraishi. 1998. Isolation and phylogenetic analysis of aerobic copiotrophic ultramicrobacteria from urban soil. J. Gen. Appl. Microbiol. 44:75-84. [DOI] [PubMed] [Google Scholar]
- 14.James, G. A., D. R. Korber, D. E. Caldwell, and J. W. Costerton. 2005. Digital image analysis of growth and starvation responses of a surface-colonizing Acinetobacter sp. J. Bacteriol. 177:907-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janssen, P. H., A. Schuhmann, E. Morschel, and F. A. Rainey. 1997. Novel anaerobic ultramicrobacteria belonging to the Verrucomicrobiales lineage of bacterial descent isolated by dilution culture from anoxic rice paddy soil. Appl. Environ. Microbiol. 63:1382-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karl, D. M., D. F. Bird, K. Bjõrkman, T. Houlihan, R. Shackelford, and L. Tupas. 1999. Microorganisms in the accreted ice of Lake Vostok, Antarctica. Science 286:2144-2147. [DOI] [PubMed] [Google Scholar]
- 17.Kief, T. L. 2000. Size matters: Dwarf cells in soil and subsurface terrestrial environments, p. 19-47. In R. R. Colwell and D. J. Grimes (ed.), Nonculturable microorganisms in the environment. ASM Press, Washington, D.C.
- 18.Lincoln, S. P., T. R. Fermor, and B. J. Tindall. 1999. Janthinobacterium agaricidamnosum sp. nov., a soft rot pathogen of Agaricus bisporus. Int. J. Syst. Evol. Microbiol. 49:1577-1589. [DOI] [PubMed] [Google Scholar]
- 19.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, P. R. Saxman, J. M. Stredwick, G. M. Garrity, B. Li, G. J. Olsen, S. Pramanik, T. M. Schmidt, and J. M. Tiedje. 2000. The RDP (Ribosome Database Project) continues. Nucleic Acids Res. 28:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchesi, J. R., T. Sato, A. J. Weightman, T. A. Martin, J. C. Fry, S. J. Hiom, D. Dymock, and W. G. Wade. 1998. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 64:795-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miteva, V. I., S. Selenska-Pobell, and V. S. Mitev. 1999. Random and repetitive primer amplified polymorphic DNA analysis of Bacillus sphaericus. J. Appl. Microbiol. 86:928-936. [Google Scholar]
- 22.Miteva, V. I., P. P. Sheridan, and J. B. Brenchley. 2004. Phylogenetic and physiological diversity of microorganisms isolated from a deep Greenland ice core. Appl. Environ. Microbiol. 70:202-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyoshi, T., T. Iwatsuki, and T. Naganuma. 2005. Phylogenetic characterization of 16S rRNA gene clones from deep-groundwater microorganisms that pass through 0.2-micrometer-pore-size filters. Appl. Environ. Microbiol. 71:1084-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicholson, W. L. 2002. Roles of Bacillus endospores in the environment. Cell. Mol. Life Sci. 59:410-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price, P. B., and T. Sowers. 2004. Temperature dependence of metabolic rates for microbial growth, survival and maintenance. Proc. Natl. Acad. Sci. USA 101:4631-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Priscu, J. C., E. E. Adams, W. B. Lyons, M. A. Voytek, D. W. Mogk, R. L. Brown, C. P. McKay, C. D. Takacs, K. A. Welch, C. F. Wolf, J. D. Kirshtein, and R. Avci. 1999. Geomicrobiology of subglacial ice above Lake Vostok, Antarctica. Science 286:2141-2143. [DOI] [PubMed] [Google Scholar]
- 27.Rappe, M. S., S. A. Connon, K. L. Vergin, and S. J. Giovannoni. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplancton clade. Nature 418:630-633. [DOI] [PubMed] [Google Scholar]
- 28.Rutz, B. A., and T. L. Kief. 2004. Phylogenetic characterization of dwarf archaea and bacteria from a semiarid soil. Soil Biol. Biochem. 36:825-833. [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Schut, F., E. de Vries, J. Gottschal, B. Robertson, W. Harder, R. Prins, and D. Button. 1993. Isolation of typical marine bacteria by dilution culture: growth, maintenance, and characteristics of isolates under laboratory conditions. Appl. Environ. Microbiol. 59:2150-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schut, F., R. Prins, and J. Gottschal. 1997. Oligotrophy and pelagic marine bacteria: facts and fiction. Aquat. Microb. Ecol. 12:177-202. [Google Scholar]
- 32.Shelobolina, E. S., Z. A. Avakyan, E. E. Boulygina, T. T. Tourova, A. M. Lysenko, G. A. Osipov, and G. I. Karavaiko. 1997. Description of a new species of mucilaginosus bacteria, Bacillus edaphicus sp. nov., and conformation of the taxonomic status of Bacillus mucilaginosus based on data of phenotypic and genotypic analysis. Mikrobiologiia 66:813-822. [Google Scholar]
- 33.Sheridan, P. P., J. Loveland-Curtze, V. I. Miteva, and J. E. Brenchley. 2003. Rhodoglobus vestali gen. nov. sp. nov, a novel psychrophilic organism isolated from an Antarctic Dry Valley lake. Int. J. Syst. Evol. Microbiol. 53:985-994. [DOI] [PubMed] [Google Scholar]
- 34.Sheridan, P. P., V. I. Miteva, and J. B. Brenchley. 2003. Phylogenetic analysis of anaerobic psychrophilic enrichment cultures obtained from a Greenland ice core. Appl. Environ. Microbiol. 69:2153-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stevenson, B. S., S. A. Eichorst, J. T. Wertz, T. M. Schmidt, and J. T. Breznak. 2004. New strategies for cultivation and detection of previously uncultured microbes. Appl. Environ. Microbiol. 70:4748-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki, K., J. Sasaki, M. Uramoto, T. Nakase, and K. Komagata. 1997. Cryobacterium psychrophilum gen. nov., sp. nov., nom. rev., comb. nov., an obligately psychrophilic actinomycete to accomodate “Curtobacterium psychrophilum” Inoue and Komagata 1976. Int. J. Syst. Evol. Microbiol. 47:474-478. [DOI] [PubMed] [Google Scholar]
- 37.Swofford, D. L. 2002. PAUP (Phylogenetic Analysis Using Parsimony.), version 4. Sinauer Associates, Sunderland, Mass.
- 38.Torrella, F., and R. Morita. 1981. Microcultural study of bacterial size changes and microcolony and ultramicrocolony formation by heterotrophic bacteria in seawater. Appl. Environ. Microbiol. 41:518-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vybiral, D., E. Denner, C. M. Haller, H.-J. Busse, A. Witte, M. G. Hofle, and B. Velimirov. 1999. Polyphasic classification of 0.2 μm filterable bacteria from the Western Mediterranean sea. Syst. Appl. Microbiol. 22:635-646. [DOI] [PubMed] [Google Scholar]