Abstract
Genes for two enzymes of the tetrahydromethanopterin-linked C1 transfer pathway (fae and fhcD) were detected in hypersaline, hyperalkaline Mono Lake (California), via PCR amplification and analysis. Low diversity for fae and fhcD was noted, in contrast to the diversity previously detected in a freshwater lake, Lake Washington (Washington).
Methylotrophic bacteria are a group of organisms that consume a wide range of C1 compounds, such as methane, methanol, methylated amines, methylated glycines, halomethanes, and methylated sulfur species (1, 17). They are found in a variety of environments, such as freshwater, marine, and terrestrial habitats, as well as habitats characterized by extreme conditions, such as highly saline, alkaline, or acidic habitats (2, 4-6, 8, 11, 18, 21, 24, 25, 28). Along with the classic cultivation approaches, molecular tools have been in use for culture-independent detection and characterization of natural methylotroph populations. The traditional molecular tools include oligonucleotide probes and PCR primer sets targeting genes conserved among specific groups of methylotrophic bacteria, such as 16S rRNA genes (3, 8, 23, 24, 26, 27), or functional genes encoding specific methylotrophic functions, such as particulate and soluble methane monooxygenases (5, 8, 18, 20), methanol dehydrogenase (8, 19, 24), corrinoid-linked methyltransferase (21), or methanesulfonic acid monooxygenase (15). The phylogenetic probes have been used successfully to uncover the diversity of α- and γ-proteobacterial methylotrophs (3, 8, 23, 24, 25, 27), and the functional probes have been used for detecting a range of methylotrophs possessing respective primary oxidation genes (5, 8, 15, 18-21, 24). Recently, Kalyuzhnaya and colleagues have developed a suite of novel primer sets designed for a broader detection of C1-oxidizing capacity in the environment (12, 13). These primer sets target four genes in the tetrahydromethanopterin (H4MPT)-linked pathway, the pathway widespread in methylotrophic bacteria (30) but also found in nonmethylotrophs: fae, mtdB, mch, and fhcD. These new tools, tested on microbial populations in a freshwater lake (Lake Washington, Washington), uncovered a broad diversity of fae, mtdB, mch, and fhcD phylotypes belonging to α-, β-, and γ-proteobacteria, including methanotrophs, nonmethanotrophic methylotrophs, and bacteria not known for methylotrophic ability, such as Burkholderia spp. (12, 13). In addition, sequences belonging to highly divergent bacterial groups, such as Planctomycetes, and yet unaffiliated divergent species have been uncovered (12, 13). The broadest range of divergent sequences was detected using two of the primer sets, those targeting fae and fhcD. Most of the phylotypes detected in Lake Washington were not closely related to known organisms, suggesting that the majority of the population potentially involved in C1 metabolism in this environment remain unidentified (12, 13). Mono Lake (California) is an extreme environment characterized by high rates of methane production and methane oxidation (10). Initial insights into the methane-oxidizing bacterial population in the site were recently obtained via fluorescence in situ hybridization and denaturing gradient gel electrophoresis employing oligonucleotide primers specific for known methanotroph groups (4). The goal of this work was the assessment of the diversity of C1-utilizing bacteria in Mono Lake by use of tools with a broader detection range, i.e., PCR primers targeting fae and fhcD.
Water samples from Mono Lake were collected from 20 discrete depths, between 5 and 38 m, near a permanently moored buoy in the central basin of Mono Lake (station 6; 37°57.822′ N, 119°01.305′ W) by using 5-liter Niskin bottles deployed from a small boat in August 2002. Dissolved methane concentrations were determined using headspace extraction followed by gas chromatography (10). Dissolved oxygen concentrations were determined using an O2 sensor (Yellow Spring Instruments) (4). Rates of methane oxidation were determined using a [3H]CH4 tracer technique (4). Oxygen concentrations in the upper water column were high (>100 μM), while methane concentrations were low (Table 1). Rates of methane oxidation increased at the base of the oxycline as methane concentrations increased and oxygen concentrations decreased (Table 1).
TABLE 1.
Data for water samples from different depthsa
| Depth (m) | Concn (μM) of:
|
Oxidation rate (nmol liter−1 day−1) | Recovery of:
|
||
|---|---|---|---|---|---|
| O2 | CH4 | fae | fhcD | ||
| 5 | 137.0 | 0.3 | 0.0 | + | + |
| 9 | 128.9 | 0.2 | 0.0 | + | + |
| 10 | 115.8 | 0.4 | 0.1 | + | + |
| 11 | 82.8 | 0.5 | 0.2 | + | + |
| 12 | 71.3 | 0.7 | 1.4 | + | + |
| 13 | 60.2 | 0.6 | 1.5 | + | + |
| 13.5 | 36.9 | 0.4 | 0.0 | + | + |
| 14 | 30.2 | 0.5 | 11.3 | + | + |
| 15 | 24.9 | 0.6 | 11.7 | + | + |
| 16 | 9.9 | 1.3 | 32.5 | − | + |
| 17 | 9.2 | 2.4 | 8.1 | − | + |
| 18 | 0.0 | 3.5 | 5.4 | − | + |
| 20 | 0.0 | 7.1 | 2.0 | − | + |
Depth of water column versus dissolved oxygen (O2) and methane (CH4) concentrations, biological methane oxidation rate, and recovery of fae- and fhcD-specific PCR products.
Water samples for DNA extraction were stored in clean sample-rinsed plastic cubitainers at 4°C until filtration through a Sterivex filter cartridge (0.22 μm; Millipore). Excess water was expelled, and the cartridge was filled with 1.8 ml of lysis buffer (22). Total community DNA was extracted from 20 discrete samples from different depths by using a previously described method (9), and these were used as templates to PCR amplify fae and fhcD, as described previously (12, 13). Nine samples (between the depths of 5 and 15 m) were positive for fae, and 13 samples (between the depths of 5 and 20 m) were positive for fhcD (Table 1). PCR products were then pooled, cloned into the pCR2.1 vector (Invitrogen), and analyzed, as described below.
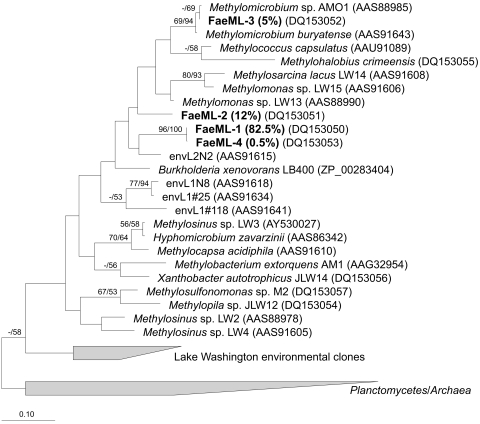
A total of 189 plasmids containing fae inserts were analyzed based on their restriction fragment length polymorphism (RFLP) patterns, as described before (12). A total of five RFLP patterns were identified. Two to five representatives of each pattern were sequenced, and the sequences were categorized into phylotypes, based on a 94% DNA similarity cutoff value, a value recently suggested for discriminating between microbial species (16, 29), based on extensive comparisons between closely related bacterial strains (16). A total of four unique phylotypes were identified, and these were distributed as follows: phylotype FaeML1, 155 clones (82.5%); phylotype FaeML2, 23 clones (12%); phylotype FaeML3, 10 clones (5%); and phylotype FaeML4, 1 clone (0.5%). The homologous coverage value (7) for this library was calculated at 0.99, suggesting that the sampling effort was adequate and covered the major phylotypes present in the library. Comparisons with the sequences deposited with GenBank as well as with our proprietary databases showed that only one phylotype, FaeML3, showed over 94% identity at the DNA level with known fae sequences, those belonging to Methylomicrobium species that are γ-proteobacterial methanotrophs. A representative of each unique phylotype was included in the phylogenetic analyses, along with the sequences from a variety of cultured bacteria, as well as the sequences previously recovered from Lake Washington by use of the same primer set, as previously described (12). Phylogenetic analyses (Fig. 1) revealed that, while phylotype FaeML3, as expected, tightly clustered with Methylomicrobium sequences, the three remaining phylotypes loosely clustered with known sequences belonging to γ-proteobacteria.
FIG. 1.
Phylogenetic tree reflecting relationships of Fae sequences detected in Mono Lake water column. Analyses were performed using neighbor-joining (NJ) and maximum parsimony (MP) methods, using inferred amino acid sequences (92 positions). The scale bar indicates the number of expected amino acid substitutions per site per unit of branch length. Bootstrap values above 50% for NJ/MP analyses are shown above branches. Note that two types of fae are found in Methylosinus strains (represented here by Methylosinus sp. LW2 and Methylosinus sp. LW3) that cluster separately on phylogenetic trees (12).
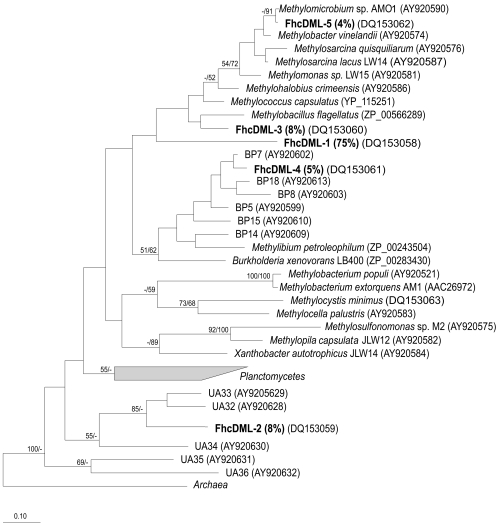
A total of 186 plasmids containing fhcD inserts were analyzed in a similar fashion, as previously described (13). A total of five RFLP patterns and a total of five phylotypes were identified, based on a 94% cutoff at the DNA level, and these were distributed as follows: phylotype FhcDML1, 139 clones (75%); phylotype FhcDML2, 15 clones (8%); phylotype FhcDML3, 14 clones (8%); phylotype FhcDML4, 10 clones (5%); and phylotype FhcDML5, 8 clones (4%). The homologous coverage value (7) for this library was calculated at 1. Comparisons with the sequences deposited with GenBank as well as with our proprietary databases revealed that only one phylotype, FhcDML5, showed significant identity at the DNA level (90%) with known fhcD sequences that belonged to Methylomicrobium. The remaining four phylotypes were only distantly related to known fhcD sequences (67 to 87% identity at the amino acid level). Phylogenetic analyses (Fig. 2) revealed that, as expected, phylotype FhcDML5 tightly clustered with Methylomicrobium sequences, while phylotypes FhcDML1 and FhcDML3 loosely clustered with known γ-proteobacterial sequences, phylotype FhcDML4 clustered with the sequences of uncultured β-proteobacteria, and phylotype FhcDML2 clustered with sequences of unaffiliated uncultured bacteria previously identified in Lake Washington (13).
FIG. 2.
Phylogenetic tree reflecting relationships of FhcD sequences detected in Mono Lake water column. Analyses were performed using neighbor-joining (NJ) and maximum parsimony (MP) methods, using inferred amino acid sequences (130 positions). The scale bar indicates the number of expected amino acid substitutions per site per unit of branch length. Bootstrap values above 50% for NJ/MP analyses are shown above branches. Note that sequences of Methylobacillus flagellatus genes for the H4MPT-linked reactions cluster with γ-proteobacterial rather than β-proteobacterial sequences (14). Sequences designated with BP and UA represent yet-uncultured β-proteobacteria and unaffiliated bacteria detected in Lake Washington (13), respectively.
Overall, our data suggest that only a few species possessing the genes for the H4MPT-linked C1 transfer pathway are present in Mono Lake and that of these, only one group, the Methylomicrobium group, is identifiable. Sequences highly similar to the 16S rRNA sequences from Methylomicrobium strains isolated from soda lakes (11, 18, 28) have recently been detected in Mono Lake (4), pointing toward the ubiquitous nature of these species in environments characterized by high salinity and high alkalinity. The remaining sequences detected in this work, including the most abundant fae and fhcD phylotypes, only loosely clustered with sequences of known γ-proteobacterial methanotrophs. Based on the abundance of these phylotypes in PCR-amplified libraries, they likely represent species with an ecologically important function, which may be in methane oxidation or in oxidation of other C1 compounds. Carini et al. have recently reported on the presence of α-proteobacterial 16S rRNA gene sequences, including those closely related to Methylosinus sequences (4). However, no α-proteobacterial fae or fhcD sequences were recovered in this work. This is unlikely to be due to primer bias, as the same primers have readily detected α-proteobacterial fae or fhcD sequences from Lake Washington (12, 13), or to insufficient sampling in the clone libraries (see above) but is likely due to the low abundance of these sequences in the samples. The low diversity of detected fae and fhcD sequences in Mono Lake is in contrast with the diversity previously uncovered in a freshwater lake, Lake Washington, where a variety of sequences belonging to α-, β-, and γ-proteobacteria have been identified using the same molecular tools (12, 13). In addition, sequences clustering with planctomycete-related sequences have been identified, as have sequences deeply diverging from the sequences affiliated with known organisms, suggesting the presence of novel phyla with no cultured representatives (12, 13).
The presence of a phylotype in the fhcD Mono Lake library (FhcDML2) that is related to this latter group of sequences is intriguing, but nothing is known about the phylogenetic position of the organisms possessing these sequences, their physiological properties, or their role in the environment. Work is under way to address these questions, including metagenomic analysis of the Lake Washington microbial community and specific cell separation using flow cytometry.
In conclusion, we provided evidence that the pathway involved in C1 transfers mediated by H4MPT is present in microbial populations inhabiting an extreme environment, a soda lake, but that the diversity of the detected genes is very limited compared to the diversity previously found in a freshwater lake. The observed lower diversity may result from special adaptations required for C1 microbes to survive in the unique geochemical environment present in Mono Lake. The dominant groups of fae and fhcD sequences recovered in PCR-based libraries belong to unknown bacterial species likely to be involved in oxidation of C1 compounds, while about 5% of the sequences belong to the well characterized genus Methylomicrobium.
Acknowledgments
This work, part of the Microbial Observatories program, was funded by the National Science Foundation (MCB-0131957 and MCB 99-77886) and in part by CRDF grant RBI-2509-MO-03.
We gratefully acknowledge P. Dunfield for providing DNA of Methylohalobius crimeensis, D. Stahl for sharing a Unix server, and J. T. Hollibaugh for providing DNA extracts.
REFERENCES
- 1.Anthony, C. 1982. The biochemistry of methylotrophs. Academic Press, Inc., London, United Kingdom.
- 2.Auman, A. J., S. Stolyar, A. M. Costello, and M. E. Lidstrom. 2000. Molecular characterization of methanotrophic isolates from freshwater lake sediment. Appl. Environ. Microbiol. 66:5259-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodrossy, L., N. Stralis-Pavese, J. C. Murrell, S. Radajewski, A. Weilharter, and A. Sessitsch. 2003. Development and validation of a diagnostic microbial microarray for methanotrophs. Environ. Microbiol. 5:566-582. [DOI] [PubMed] [Google Scholar]
- 4.Carini, S., N. Bano, G. LeCleir, and S. B. Joye. 2005. Aerobic methane oxidation and methanotroph community composition during seasonal stratification in Mono Lake, California (USA). Environ. Microbiol. 7:1127-1138. [DOI] [PubMed] [Google Scholar]
- 5.Costello, A. M., and M. E. Lidstrom. 1999. Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl. Environ. Microbiol. 65:5066-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dedysh, S. N., W. Liesack, V. N. Khmelenina, N. E. Suzina, Y. A. Trotsenko, J. D. Semrau, A. M. Bares, N. S. Panikov, and J. M. Tiedje. 2000. Methylocella palustris gen. nov., sp. nov., a new methane-oxidizing acidophilic bacterium from peat bogs, representing a novel subtype of serine-pathway methanotrophs. Int. J. Syst. Evol. Microbiol. 50:955-969. [DOI] [PubMed] [Google Scholar]
- 7.Good, I. J. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237-262. [Google Scholar]
- 8.Horz, H. P., M. T. Yimga, and W. Liesack. 2001. Detection of methanotroph diversity on roots of submerged rice plants by molecular retrieval of pmoA, mmoX, mxaF, and 16S rRNA and ribosomal DNA, including pmoA-based terminal restriction fragment length polymorphism profiling. Appl. Environ. Microbiol. 67:4177-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Humayoun, S. B., N. Bano, and J. T. Hollibaugh. 2003. Depth distribution of microbial diversity in Mono Lake, a meromictic soda lake in California. Appl. Environ. Microbiol. 69:1030-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joye, S. B., T. L. Connell, L. G. Miller, R. S. Oremland, and R. S. Jellison. 1999. Oxidation of ammonia and methane in an alkaline, saline lake. Limnol. Oceanogr. 44:178-188. [Google Scholar]
- 11.Kaluzhnaya, M., V. Khmelenina, B. Eshinimaev, N. Suzina, D. Nikitin, A. Solonin, J. L. Lin, I. McDonald., C. Murrell, and Y. Trotsenko. 2001. Taxonomic characterization of new alkaliphilic and alkalitolerant methanotrophs from soda lakes of the Southeastern Transbaikal region and description of Methylomicrobium buryatense sp. nov. Syst. Appl. Microbiol. 24:166-176. [DOI] [PubMed] [Google Scholar]
- 12.Kalyuzhnaya, M. G., M. Lidstrom, and L. Chistoserdova. 2004. Utility of environmental primers targeting ancient enzymes: methylotroph detection in Lake Washington. Microb. Ecol. 48:463-472. [DOI] [PubMed] [Google Scholar]
- 13.Kalyuzhnaya, M. G., O. G. Nercessian, M. E. Lidstrom, and L. Chistoserdova. 2005. Development and application of polymerase chain reaction primers based on fhcD for environmental detection of methanopterin-linked C1-metabolism in bacteria. Environ. Microbiol. 7:1269-1274. [DOI] [PubMed] [Google Scholar]
- 14.Kalyuzhnaya, M. G., N. Korotkova, G. Crowther, C. J. Marx, M. E. Lidstrom, and L. Chistoserdova. 2005. Analysis of gene islands involved in methanopterin-linked C1 transfer reactions reveals new functions and provides evolutionary insights. J. Bacteriol. 187:4607-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelly, D. P., and J. C. Murrell. 1999. Microbial metabolism of methanesulfonic acid. Arch. Microbiol. 172:341-348. [DOI] [PubMed] [Google Scholar]
- 16.Konstantinidis, K. T., and J. M. Tiedje. 2005. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. USA 102:2567-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lidstrom, M. E. 2001. The aerobic methylotrophic bacteria. In M. Dworkin (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed. Springer-Verlag, New York, N.Y. http://link.springer-ny.com/service/books/10125.
- 18.Lin, J. L., S. Radajewski, B. T. Eshinimaev, Y. A. Trotsenko, I. R. McDonald, and J. C. Murrell. 2004. Molecular diversity of methanotrophs in Transbaikal soda lake sediments and identification of potentially active populations by stable isotope probing. Environ. Microbiol. 6:1049-1060. [DOI] [PubMed] [Google Scholar]
- 19.McDonald, I. R., and J. C. Murrell. 1997. The methanol dehydrogenase structural gene mxaF and its use as a functional gene probe for methanotrophs and methylotrophs. Appl. Environ. Microbiol. 63:3218-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDonald, I. R., and J. C. Murrell. 1997. The particulate methane monooxygenase gene pmoA and its use as a functional gene probe for methanotrophs. FEMS Microbiol. Lett. 156:205-210. [DOI] [PubMed] [Google Scholar]
- 21.McDonald, I. R., K. L. Warner, C. McAnulla, C. A. Woodall, R. S. Oremland, and J. C. Murrell. 2002. A review of bacterial methyl halide degradation: biochemistry, genetics and molecular ecology. Environ. Microbiol. 4:193-203. [DOI] [PubMed] [Google Scholar]
- 22.Murray, A. E., J. T. Hollibaugh, and C. Orrego. 1996. Phylogenetic compositions of bacterioplankton from two California estuaries compared by denaturing gradient gel electrophoresis of 16S rDNA fragments. Appl. Environ. Microbiol. 62:2676-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murrell, J. C., I. R. McDonald, and D. G. Bourne. 1998. Molecular methods for the study of methanotroph ecology. FEMS Microbiol. Ecol. 27:103-114. [Google Scholar]
- 24.Murrell, J. C., and S. Radajewski. 2000. Cultivation-independent techniques for studying methanotroph ecology. Res. Microbiol. 151:807-814. [DOI] [PubMed] [Google Scholar]
- 25.Pacheco-Oliver, M., I. R. McDonald, D. Groleau, J. C. Murrell, and C. B. Miguez. 2002. Detection of methanotrophs with highly divergent pmoA genes from Arctic soils. FEMS Microbiol. Lett. 209:313-319. [DOI] [PubMed] [Google Scholar]
- 26.Radajewski, S., P. Ineson, N. R. Parekh, and J. C. Murrell. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646-649. [DOI] [PubMed] [Google Scholar]
- 27.Radajewski, S., G. Webster, D. S. Reay, S. Morris, P. Ineson, D. B. Nedwell, J. I. Prosser, and J. C. Murrell. 2002. Identification of active methylotroph populations in an acidic forest soil by stable-isotope probing. Microbiology 148:2331-2342. [DOI] [PubMed] [Google Scholar]
- 28.Sorokin, D. Y., B. E. Jones, and J. G. Kuenen. 2000. An obligate methylotrophic, methane-oxidizing Methylomicrobium species from a highly alkaline environment. Extremophiles 4:145-155. [DOI] [PubMed] [Google Scholar]
- 29.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. C. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y.-H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 30.Vorholt, J. A., L. Chistoserdova, S. M. Stolyar, R. K. Thauer, and M. E. Lidstrom. 1999. Distribution of tetrahydromethanopterin-dependent enzymes in methylotrophic bacteria and phylogeny of methenyl tetrahydromethanopterin cyclohydrolases. J. Bacteriol. 181:5750-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]