Abstract
The effect of the simple and complex basic components of a fermentation medium on the surface properties of Lactobacillus acidophilus NCC2628 is studied by physicochemical methods, such as electrophoresis, interfacial adhesion, and X-ray photonelectron spectroscopy, and by transmission electron microscopy. Starting from an optimized complete medium, the effect of carbohydrates, peptones, and yeast extracts on the physicochemical properties of the cell wall is systematically investigated by consecutively omitting one of the principal components from the fermentation medium at the time. The physicochemical properties and structure of the bacterial cell wall remain largely unchanged if the carbohydrate content of the fermentation medium is strongly reduced, although the concentration of surface proteins increases slightly. Both peptone and yeast extract have a considerable influence on the bacterial cell wall, as witnessed by changes in surface charge, hydrophobicity, and the nitrogen-to-carbon ratio. Both zeta potential and the cell wall hydrophobicity show a positive correlation with the nitrogen-to-carbon ratio of the bacterial surfaces, indicative of the important role of surface proteins in the overall surface physical chemistry. The hydrophobicity of the cell wall, which is low for the cultures grown in the complete medium and in the absence of carbohydrates, becomes fairly high for the cultures grown in the medium without peptones and the medium without yeast extract. UV spectrophotometry and sodium dodecyl sulfate-polyacrylamide gel electrophoresis combined with liquid chromatography-tandem mass spectrometry are used to analyze the effect of medium composition on LiCl-extractable cell wall proteins, confirming the major change in protein composition of the cell wall for the culture fermented in the medium without peptones. In particular, it is found that expression of the S-layer protein is dependent on the protein source of the fermentation medium.
Lactobacilli are of considerable technological and commercial importance because of their role in the manufacturing and preservation of many fermented food products, but they also play an important role in the control of undesirable microorganisms in the intestinal and urogenital tract (43). Beside indigenous lactobacilli, which reside in the human gastrointestinal tract, several Lactobacillus strains from fermented food products have shown beneficial effects on gut health (19). The surface properties of lactic acid bacteria are of major importance in fermentation technology (6, 30), but they are also of importance for mediating the adhesion of the bacteria to the gastrointestinal epithelium. This is considered to be a prerequisite for the exclusion of enteropathogenic bacteria (2, 3, 26) and immunomodulation of the host (5, 22).
The gram-positive cell wall of lactobacilli consists of the sacculus, made up of peptidoglycan, which is decorated by (lipo)teichoic acids, surface proteins, and anionic and neutral polysaccharides (14). Of particular interest for the surface properties of lactobacilli are the S-layer proteins (6, 36, 39, 41), which cover the bacterial cell wall in a regular, two-dimensional array. S-layers have been found in a number of Lactobacillus species, including L. acidophilus (8, 38). The precise surface constituents of the bacterial cell wall which contribute to the surface properties of a bacterium is an issue of considerable debate (1, 11, 33, 36), but surface proteins (35), exopolysaccharides (6, 12), and lipoteichoic acids (20) have all been implicated. In general, the outer layers of the cell wall are more important for the surface properties and interactions than the inner layers because of steric effects (39a).
The surface properties of microorganisms are, in addition, dependent on the growth conditions and the composition of the fermentation medium. Consequently, the interactions of a particular strain with, for instance, the gastrointestinal epithelium or with surfaces exposed during bioprocessing may be strongly influenced by the composition of the fermentation medium and the growth conditions. However, little is known about the relationship between the fermentation conditions, changes in bacterial surface properties, and their effect on bacterial interactions.
Studies have been done of variations in microbial surface properties using various established media (15), using slight variations in the formulation of commercial media (28), using various carbon sources (31), or using variations in the concentration of a simple carbohydrate in an otherwise complex medium (10). In addition, occasional studies have been carried out using nontraditional medium components like bile salts (42).
In these studies, the composition of the medium was varied in a nonsystematic way. Ideally, one would study the effect of each individual chemical component of the medium on the properties of the microbial surface. However, bacteria generally do not thrive on highly simple synthetic media and a number of important medium components are of an intrinsically complex nature, like yeast extracts, meat extracts, dairy concentrates, wort, and peptones. Consequently, either the study of the relationship between medium composition and physicochemical properties is restricted to model systems utilizing a growth medium of an essentially theoretical interest, or the complex medium needs to be divided into classes of principal components, of which the effects on the cell wall properties are studied. Here, we opt for the latter approach.
We started with a medium containing all principal components of a full-fermentation medium. These components were either of a simple nature (e.g., chemically well-defined carbohydrates) or of a complex nature (peptones, yeast extracts). The complete medium was optimized for the microorganism of interest, in our case, L. acidophilus NCC2628. To study the effect of the principal medium compositions on the physicochemical surface properties, fermentations were carried out using medium in which one of the principal components—either chemically simple or complex—has consecutively been left out. As the cultures were able to grow in all these media, we were able to study the effect of these simple and complex medium components on the cell wall properties.
MATERIALS AND METHODS
Growth and preparation of bacterial cultures.
L. acidophilus NCC2628 was obtained from the Nestlé Culture Collection. Cultures were grown at 40°C under anaerobic conditions in test tubes containing 10 ml of medium and harvested in late stationary phase (12 to 14 h). A number of media, designated M1, M2, M3, and M4, were used. The complete medium was M1, which consisted of 1.00% Variolac 836 (MD Foods Ingredients, Denmark) by weight, 0.10% Tween 80 (Quest International, The Netherlands) by weight, 0.75% sucrose (Fluka, Switzerland) by weight, 0.75% fructose (Fluka, Switzerland) by weight, 1.00% Pisane (Cosucra, France) by weight, 3.00% yeast extract 2012 (Biospringer, France) by weight, and 1.00% Primatone RL (Quest International, The Netherlands) by weight. Variolac 836 is a whey permeate powder consisting largely of lactose but also containing about 4% proteins. Pisane is a pea protein concentrate, whereas Primatone is an enzymatic digest of meat high in amino acids and peptides. M2, M3, and M4 were each lacking one of the essential components of M1. M2 lacked sucrose and lactose, M3 lacked the Pisane and Primatone, and M4 lacked the yeast extract. After fermentation, the bacteria were harvested by centrifugation (5,000 × g, 10 min, 4°C) and washed twice with a 0.9% NaCl solution. The viable cell count was determined by serially diluting 1 ml of homogenized cell culture in 100 mM NaH2PO4 buffer. The dilutions were plated on MRS agar and anaerobically incubated for 24 h at 40°C.
Determination of electrophoretic mobility and zeta potential.
Electrophoretic mobility was measured by laser Doppler velocimetry using a ZetaSizer 4 (Malvern Instruments, Malvern, United Kingdom). A glass capillary (ZET5104; quartz capillary, 4-mm diameter) was used as the electrophoresis cell. Between 5 and 10 ml of the bacterial suspension was injected into the electrophoresis cell using a disposable syringe. Before injection of the bacterial suspension, the measurement cell was flushed with ultrapure water (MilliQ; Millipore). Electrophoretic mobilities were converted to the zeta potential using the Helmholtz-Schmoluchowski equation (18).
Bacterial hydrophobicity through interfacial adhesion.
The basic MATH (microbial adhesion to hexadecane) test was carried out essentially by following the method described in reference 34. The MATH test was recently extended by us to provide a quantitative description of the interfacial adhesion of microorganisms (36). In brief, to 10 ml of 10 mM KH2PO4 buffer at pH 7, a quantity of bacterial suspension was added such that the resulting optical density (OD) was 0.5 ± 0.05. This usually required the addition of an aliquot of bacterial suspension of 100 to 200 μl to the 10-ml buffer solution. After homogenization, 3.0 ml of the suspension was pipetted into a 15-ml sealable plastic test tube (Falcon; BD Biosciences, Allschwil, Switzerland). Subsequently, 150 μl hexadecane (purity, >98%; Fluka, Buchs, Switzerland) was added and, after the tube was hermetically closed, the mixture was vortexed at maximum speed for 30 s using a Vortex Genie 2 (Scientific Instruments, Bohemia, NY). This was repeated for 30 s after an interval of 1 min. The optical densities of both the initial and the extracted solution were determined at a λ of 600 nm using an Uvikon 810 UV/visible-light spectrophotometer (BioTek, Basel, Switzerland) and disposable polystyrene cuvettes with an effective volume of 1 ml. A blank value was determined for the phosphate buffer without added bacteria. A waiting period between 10 min and 25 min was employed in order to achieve complete phase separation between the water and hexadecane phases while ensuring that significant sedimentation of the bacteria remaining in solution did not occur.
The fraction of bacteria adhering to the hexadecane-water interface was calculated using the following equation:
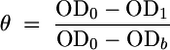 |
(1) |
where θ is the degree of interfacial adhesion and OD0, OD1, and ODb are the optical densities of the initial bacterial suspension, the extracted solution, and the blank, respectively.
We have modified the MATH assay in order to study the effects of hexadecane on bacterial interfacial adhesion (36). Instead of one adhesion value for a fixed aliquot of hexadecane, a series of adhesion values were determined by varying the amount of hexadecane between 0.5 μl and 3 ml (always on a 3-ml bacterial suspension with a cell count of 107 to 108 CFU/ml). In the interfacial-adhesion experiments, the buffer pH was kept at 7.
The interfacial-adhesion curves were quantified according to the following two-state model (36):
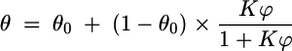 |
(2) |
where ϕ is the ratio of the volume of hexadecane to that of aqueous buffer (V [hexadecane]/V [buffer]), θ0 represents the initial plateau caused by the bacterial aggregation induced by the adsorption of very small quantities of hexadecane on hydrophobic moieties close to the outer surface layers of the bacterial cell wall model (36), and K is the interfacial-adhesion constant.
Elemental analysis of bacterial surface composition by XPS.
Because of the small penetration depth (5 to 10 nm), X-ray photonelectron spectroscopy (XPS) is highly useful for analyzing the surface compositions of sensitive biological materials. Bacterial samples for XPS analysis were prepared by freeze-drying the resuspended pellet of a 10-ml fermentation mixture. Resuspension was carried out in 100 mM NaH2PO4 buffer at pH 7. Small flakes of freeze-dried sample were adhered to a sample grid by means of double-sided sticky tape and gently compressed using a spatula in order to form a confluent layer. The samples were analyzed using an Axis Ultra XPS instrument (Kratos Scientific, Manchester, United Kingdom) for the surface concentration of the elements C, N, O, and P.
Extraction of cell wall proteins by LiCl treatment.
For the analysis of the protein content and composition of the cell wall of L. acidophilus NCC2628 fermented in the four media, cell wall proteins were extracted by a 5 M LiCl extraction (25, 38). The fermentation was carried out as described above but this time in 1-liter bottles containing 1 liter of each fermentation medium. The bacteria were harvested by centrifugation (3,500 × g, 20 min, 4°C) and washed three times with 250 ml of 10 mM KH2PO4. After being harvested and washed, the bacterial pellet of a 1-liter fermentation (about 5 ml) was resuspended in about 35 ml of KH2PO4 buffer (total, 40 ml). An equal volume of 5 M LiCl was added, and the suspension was incubated for 1 h at room temperature. Afterwards, bacteria and high-molecular-mass debris were sedimented by centrifugation (5,000 × g, 20 min, 4°C). The supernatant was removed, filtered through a 5-μm microfilter (Millex-SV; Millipore, Cork, Ireland), and dialyzed overnight against 5 liters of 50 mM Tris-HCl (pH 7.4, room temperature). Three hours after the start of the dialysis, the dialysis buffer was refreshed. Remaining high-molecular-mass debris was removed by centrifugation (5,000 × g, 20 min, 4°C). The supernatant was used undiluted for sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) analysis and was diluted 25-fold with ultrapure water for UV spectrophotometry.
UV spectrophotometry.
The diluted supernatant was analyzed on an HP 8452A diode array spectrophotometer between a λ of 200 and a λ of 400 nm. The data were collected using the HP UV/visible-light ChemStation software and imported in an Excel worksheet for further analysis. The experiments were carried out in duplicate using independently fermented cultures.
SDS-PAGE analysis.
SDS-PAGE of the supernatants of M1 to M4 was carried out using the reference molecular masses 31 kDa, 45 kDa, 66 kDa, 97.4 kDa, 116 kDa, and 200 kDa. From both the marker solution and the supernatants of M1 to M4, 20 μl was mixed with 5 μl loading buffer and denatured at 95°C for 5 min. Prior to use, 475 μl of the loading buffer (60 mM Tris at pH 6.8, 25% glycerol, 2% SDS, 0.1% bromophenol blue) was mixed with 25 μl β-mercaptoethanol. After denaturation, the samples were cooled on ice and the droplets were collected by centrifugation. The samples were run on a 10% polyacrylamide gel (Ready-gel; Bio-Rad Laboratories, Munich, Germany). Each slot was loaded with 20 μl of sample. The buffer was a 10-fold dilution in ultrapure water of a 10× Tris-glycine-SDS buffer (Bio-Rad). The gel was run for about 45 min at 200 V, washed twice in 150 ml water, and stained with a Coomassie blue stain (Bio-Safe Coomassie; Bio-Rad). The gel was destained by washing it three times with 200 ml water and dried for 1.5 h in a GelAir dryer (Bio-Rad) between cellophane sheets (Cellophane Support; Bio-Rad). Gels were run in duplicate from independently fermented cultures.
Identification of surface protein by LC-MS/MS.
Protein bands were excised from the fresh gels and in-gel digested with trypsin according to published procedures (21, 37). Extracted and dried peptides were reconstituted in 50 μl of liquid chromatography (LC) solvent A (see below) by vortexing and sonication for 15 min. Then, 25 μl of the sample was used for LC-NanoESI-tandem mass spectrometry (MS/MS) analysis. The peptides were separated and characterized using a high-performance liquid chromatography (HPLC) system consisting of a Rheos 2000 pump with a constant pressure split module (Flux Instruments, Germany) and a PAL HTC autosampler (CTC Analytics, Switzerland) coupled to an LCQ classic ion trap mass spectrometer (ThermoFinnigan, San Jose, Calif.) equipped with a NanoESI source (ThermoFinnigan). The mobile phase for LC separation was 0.1% (vol/vol) formic acid-2% (vol/vol) acetonitrile in MilliQ water (solvent A) and 0.1% (vol/vol) formic acid-80% (vol/vol) acetonitrile in MilliQ water (solvent B). Peptides were eluted with a linear gradient of 5% solvent B to 50% solvent B in 30 min, followed by a 3-min wash with 100% solvent B at an ∼800-μl/min flow rate. The NanoSpray source was operated at a potential of 2.4 kV provided by a liquid junction T configuration. The distance of the analytical column end (Magic C18, 100 μm by 10 cm; Spectronex, Basel, Switzerland) to the heated capillary (set to 140°C) was adjusted to approximately 5 mm. All spectra were obtained in positive mode and recorded at unit mass resolution. Automated MS/MS spectra were acquired with relative collision energy for collision-induced dissociation preset at 35% and an isolation width of 1 m/z unit. Full-scan MS and MS/MS data acquisition and analysis were performed with Xcalibur software V1.3 (ThermoFinnigan), including the Bioworks V3.1 software package for SEQUEST database searches (17). MS/MS spectra analysis was performed using the SEQUEST program, which correlates the uninterpreted MS/MS spectra of peptides with theoretical spectra of amino acid sequences from protein databases. In the data analysis, robust filters were employed, e.g., the SEQUEST cross-correlation factor xcorr was >2 for doubly charged ions and xcorr was >2.5 for triply charged ions. SEQUEST analysis of the acquired tandem MS spectra was performed against the latest release of the Swiss-Prot database on a local personal computer workstation. The significance of protein identification was displayed by the SEQUEST score and by the sequence coverage. The procedure is discussed in more detail in reference 32.
Transmission electron microscopy (TEM).
The bacteria were suspended in a mixture of 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer at pH 7.0 containing 0.04% Ruthenium red and incubated at 4°C. After 1 h, the sedimented part of the suspensions was microencapsulated in agar gel tubes. The samples were then fixed by incubation in 2.5% glutaraldehyde in sodium cacodylate buffer at pH 7.0 containing 0.04% Ruthenium red and incubated for 16 h at 4°C. The samples were washed three times with sodium cacodylate buffer at pH 7.0 containing 0.04% ruthenium red, followed by an incubation in 2% osmium tetroxide in sodium cacodylate buffer at pH 7.0 containing 0.04% ruthenium red for 2 h at room temperature. The samples were washed again as described above before dehydration in a series of solutions with an ethanol concentration increasing from 50% to 100%. The samples were then embedded by three successive incubations for 16 h at 4°C in 50% Spurr resin in ethanol, in 75% Spurr resin in ethanol, and finally in 100% Spurr resin. After polymerization of the resin (70°C, 48 h), ultrathin sections were cut with a Reichert OMU2 ultramicrotome. Ultrathin sections (thickness, 70 nm) stained with aqueous uranyl-acetate and lead citrate were examined by transmission electron microscopy (Philips CM12; 80 kV, magnification, ×128,000).
RESULTS
Bacterial growth.
Before analysis of the effect of the composition of the fermentation medium on the physicochemical properties of the bacterial surfaces and cell wall structure, several basic aspects of the fermentation behavior of L. acidophilus NCC2628 in the various media need to be established. It turns out that the cultures attained satisfactory cell counts between 108 and 109 CFU/ml in all media (Table 1). The lower cell density observed for the cultures grown in M2 than for the cultures grown in M1 likely reflects a carbohydrate limitation in M2. The value of the cell count for M4 is most likely on the low side, as in this medium, the strain tended to form chains of up to about 10 bacteria (micrographs not shown). The actual cell density is thus likely to be higher by a factor 2 to 5.
TABLE 1.
Viable cell countsa of L. acidophilus NCC2628 cultured in the four media
| Medium | Cell count (log CFU/ml) |
|---|---|
| M1 | 8.96 ± 0.04 |
| M2 | 8.11 ± 0.04 |
| M3 | 8.71 ± 0.01 |
| M4 | 8.08 ± 0.04 |
Values are the averages from three independently fermented cultures. The variation indicated is 1 standard deviation.
During fermentation, all cultures arrived in stationary phase after about 11 to 12 h. There was some difference in the durations of the lag phases, however. For the cultures fermented in M1 and M4, the lag time was the shortest, around 6 h. For the cultures in M2 and M3, this was about 7 h. As the cultures were harvested 12 to 14 h after inoculation, they were all in the stationary phase. From the cell counts and the growth curves, we concluded that L. acidophilus NCC2628 is able to grow in the four media, even though the variations in medium composition had some influence on the cell density attained in stationary phase.
It should be noted that, although medium components like carbohydrates and amino acid sources are omitted in the various media, they all contain both principal ingredients, albeit at strongly reduced levels (depending on the medium). For instance, the amino acid contents of the media were 2.24% (M1), 2.24% (M2), 0.74% (M3), and 1.51% (M4), all by weight, as calculated from the analysis of the amino acid composition of the Primatone, Pisane, and yeast extract (see the supplemental material). In addition, M2 contained carbohydrates, but at a very low level, mainly from the yeast extract (which contains about 10% of carbohydrates by weight) or as a carryover from the inoculant. We estimated the carbohydrate content of M2 to be about 0.3% by weight.
Zeta potential.
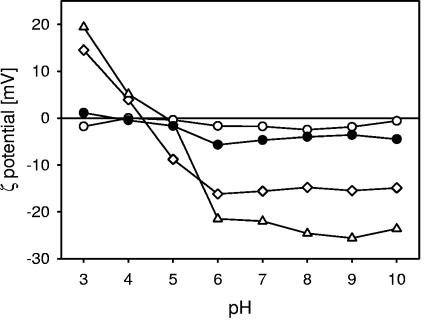
The zeta potential of L. acidophilus NCC2628cultivated in four different media as a function of the pH is shown in Fig. 1. For the bacteria grown in M1, M2, and M4, the effective charge on the bacterial surface was positive at a low pH and negative at a high pH, indicative of cell wall electrostatics determined by surface proteins and weak acids. The isoelectric point, which is defined as the pH where the zeta potential becomes zero, varied for the cultivation medium composition shown from about pH 4.3 for the culture fermented in M2 to about pH 5.2 for the culture fermented in M1 (see Table 4).
FIG. 1.
Zeta potential of L. acidophilus NCC2628 fermented in the various media as a function of pH. The bacterial cultures were suspended in a 10 mM KH2PO4 buffer. All data points are the averages of three measurements with independently fermented cultures. •, M1 (complete medium); ⋄, M2 (without sugars); ○, M3 (without peptides); ⧫, M4 (without yeast extracts). Error bars are not shown, as the standard deviations are generally smaller than the symbols.
TABLE 4.
Summary of physicochemical properties of L. acidophilus NCC2628 as fermented in the various mediaa
| Medium | Zeta potential at a pH of 3 (mV) | Minimum of zeta potential (mV) | pI | Interfacial adhesion | N/C ratio |
|---|---|---|---|---|---|
| M1 | 11.3 | −10.9 | 5.2 | Hydrophilic | 0.124 |
| M2 | 14.5 | −15.5 | 4.3 | Hydrophilic | 0.134 |
| M3 | −1.8 | −1.9 | Hydrophobic | 0.100 | |
| M4 | 19.4 | −25.6 | 4.9 | Fairly hydrophobic | 0.164 |
The standard deviation in the zeta potential data is about 1.5 mV (experiments on three independently fermented cultures) (Fig. 1). The estimated standard deviation in the isoelectric point is about 0.2 pH units.
Between the bacteria grown in M1, M2, and M4, there are some differences in cell wall composition and structure reflected in the zeta potential profile. For the bacteria grown in M2, the zeta potential as a function of pH was very close to those of the bacteria grown in M1, the values of the zeta potential being slightly more positive toward a low pH and slightly more negative toward a high pH. It is likely that, in the absence of carbohydrates in the medium, the cell wall either incorporates slightly larger amounts of protein or synthesizes slightly smaller amounts of (neutral) exopolysaccharides. Proteins are generally highly charged at both low and high pHs, whereas neutral exopolysaccharides reduce the zeta potential of the bacteria because the hydrodynamic interface is shifted outward. The bacteria grown in M4 are significantly more highly charged at both low and high pHs. This indicates that the surfaces of the bacteria grown in M4 are richer in proteins.
The major variation in the zeta potential profile was observed for the culture grown in M3, which was close to electrically neutral over the whole pH range from 3 to 10. It is clear that, with strongly reduced protein sources, the composition of the bacterial surface changes substantially. In particular, we conclude that the cell wall proteins are not expressed at the outer layers of the cell wall as the zeta potential remains close to zero even at a low pH. Normally, even relatively small amounts of cell wall proteins at the outer surface of the cell wall result in a positive zeta potential at a low pH.
In summary, the substantial differences in zeta potential observed for the strains cultured in the various media are most likely related to differences in cell wall composition, in particular, the concentration of surface proteins. This issue will be addressed later using XPS.
Interfacial adhesion assay.
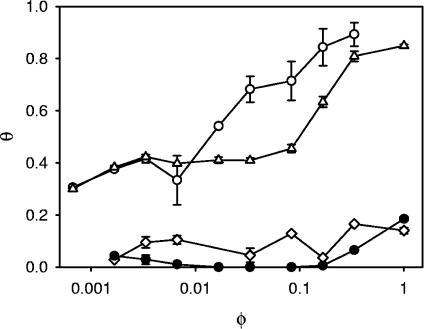
From the interfacial adhesion curves shown in Fig. 2, it is observed that L. acidophilus NCC2628 is an intrinsically hydrophilic strain, as the degree of interfacial adhesion is very low for the culture fermented in the complete medium M1. The bacterium remains hydrophilic when the carbohydrates are eliminated from the fermentation medium (M2). Again, in line with the results from the zeta potential, we conclude that the absence of carbohydrates in the medium does not lead to a cell wall which is structurally different. This changed as soon as we eliminated either the peptones or the yeast extract. For the cultures grown in M3 and M4, the bacterial cell wall became much more hydrophobic, as is witnessed by the interfacial-adhesion curves in Fig. 2 and the higher values of the adhesion constants in Table 2. Both the initial adhesion plateau, caused by the aggregation of bacteria at very low volumes of hexadecane, and the interfacial-adhesion constant, quantifying the affinity for adhesion of the bacteria to the hexadecane-water interface, are higher. The high values for both parameters imply that hydrophobic groups are present at the outer surface of the bacterial cell wall for the cultures fermented in M3 and M4. For M4, the increase in hydrophobicity is related to the protein content, as is indirectly demonstrated by the increase in zeta potential at both low and high pHs and directly using XPS (see below). For M3, an increased content of surface proteins cannot explain the increase in hydrophobicity, as this is excluded by both the zeta potential and XPS analyses.
FIG. 2.
Interfacial adhesion curves of L. acidophilus NCC2628 fermented in the various media. The bacterial cultures are suspended in a 10 mM KH2PO4 buffer at pH 7. All data points are averages from two independent analyses using separate cultures. •, medium M1 (complete medium); ⋄, M2 (lacking the sugars); ○, M3 (without peptones); ⧫, M4 (without yeast extracts). The error bars denote the standard errors.
TABLE 2.
Fitting parameters of the interfacial adhesion curvesa and MATH values
| Medium | θ0 | K | MATH valueb (%) |
|---|---|---|---|
| M1 | 0 | 0.22 | 1.1 |
| M2 | 0.05 | 0.25 | 6.2 |
| M3 | 0.33 | 25 | 70 |
| M4 | 0.37 | 4 | 48 |
Curves are fitted to the data from two experiments with independently fermented cultures (Fig. 2).
The MATH value was determined from the interfacial adhesion curves for an added volume of hexadecane of 150 μl.
In Table 2, the MATH values as determined following the standard assay in the literature (34) were included. In the standard method, the degree of interfacial adhesion was determined at 1 volume of hexadecane, 150 μl, on 3.0 ml of bacterial suspension. Although the method exhibited some significant disadvantages in particular for very hydrophilic or very hydrophobic microorganisms (36), for L. acidophilus NCC2628, it provided a good discrimination between the cultures from the various fermentation media.
Bacterial surface composition by XPS.
In Table 3, the atomic concentrations of the elements C, N, O, and P are given as a percentage of the total occurrence of these four elements. As expected, the surface concentrations of carbon and oxygen were the highest, followed by nitrogen. The surface concentration of phosphate was rather low, even if we take into account that the samples were prepared in a phosphate buffer. Consequently, the surface concentration of cell wall macromolecules containing phosphate, like lipoteichoic acids, was very low. This is in line with the results of both the zeta potential and the interfacial-adhesion assay, which do not reveal bacterial surfaces that are hydrophobic and highly charged at the same time (36). Cell wall macromolecules containing significant amounts of phosphate groups are necessarily highly charged, as phosphate is a strong acid.
TABLE 3.
Atomic concentrations of the bacterial surfaces as determined by XPS analysis of freeze-dried preparationsa
| Medium | Atomic concn (%)
|
|||
|---|---|---|---|---|
| C | N | O | P | |
| M1 | 66.9 | 8.3 | 24.2 | 0.6 |
| M2 | 58.8 | 7.9 | 32.3 | 1.1 |
| M3 | 59.7 | 6.0 | 32.7 | 1.6 |
| M4 | 67.0 | 11.0 | 21.0 | 1.0 |
The standard error in the XPS data is about 2%, as determined by analysis of two independently fermented cultures.
An important measure derived from the XPS analysis of bacterial surfaces is the nitrogen-to-carbon (N/C) ratio (Table 4). A high value of this ratio indicates the presence of large amounts of surface proteins and a low value, a reduced amount. As expected, for M3, without the two principal amino acid sources, the concentration of surface proteins is the lowest. Interestingly, if we left out the yeast extract (M4), the N/C ratio increased strongly. The absence of carbohydrates influences the N/C ratio to a lesser degree (Table 4, compare M1 and M2), indicating that differences in carbohydrate supplementation have only a limited influence on the expression of surface proteins. The values of the N/C ratio found for the bacterial cultures from the various media are within the range reported in the literature. The N/C ratio of the surfaces of lactic acid bacteria is typically between 0.09 and 0.15 (6, 16), although values as low as 0.04 have been reported previously (13).
Deconvolution of the 1s orbital of carbon (C1s) peak confirms the presence of carbohydrates at the bacterial surface, because of the significant contribution of hydroxyl- and ether-linked carbon at the bacterial surfaces. In addition, significant amounts of carbon in ester groups were detected as well as nitrogen-linked carbon, which are the sign of surface proteins. Evidence was also found for small amounts of carbon in carboxylic acid groups. The principal variation in carbon linkage between the cultures fermented in the various media is a somewhat higher value for the OH-linked carbon in the cases of M1 and M4.
Characterization of cell wall extracts.
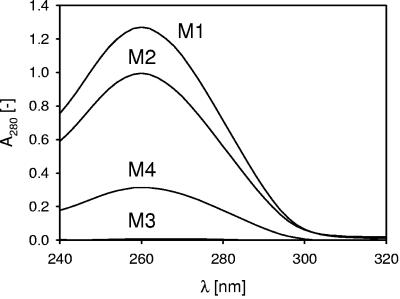
A first check on the state of the proteins in the cell walls of the cultures fermented in the various media showed a UV adsorption spectrum around a λ of 280 nm of the supernatant of the LiCl extract (Fig. 3). The spectrum in Fig. 3 shows the expected behavior. The relative protein content of the cell walls of the cultures fermented in M1 was the highest, immediately followed by the cultures fermented in M2, whereas the relative protein concentrations in the cell walls of the cultures from M4 and, in particular, M3 were the lowest. In the absence of amino acid sources (M3), the bacteria thus exhibit a strongly reduced level of proteins in their cell walls, as is corroborated by the low value for the N/C ratio reported in Table 4. In addition, if the amount of carbohydrates was reduced (M2), the microbial surface became richer in surface proteins (the relative concentration of extractable surface proteins was the highest in M2), as was confirmed by the XPS data (witness the high value for the N/C ratio for M2 compared to M1 in Table 4) and the zeta potential data (high positive charge at a low pH value) (Fig. 1).
FIG. 3.
UV spectra of the supernatant of LiCl cell wall extracts of L. acidophilus NCC2628 fermented in the various media. The supernatant was diluted 25 times with ultrapure water.
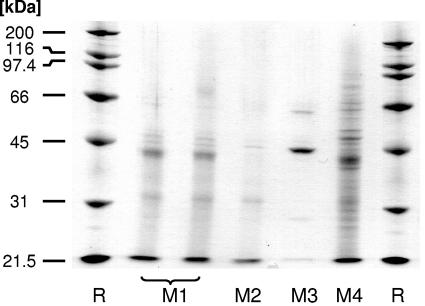
From the SDS-PAGE analysis of the supernatant from the LiCl extract (Fig. 4), more-detailed information was obtained about the presence of extractable surface proteins. The molecular masses of the extractable surface proteins varied between about 30 kDa and 70 kDa, in line with previous observations (24, 29, 38). The protein profiles of the cultures from M1 and M2 were similar, with the bands being less intense for M2. In addition, minor differences in band structure were observed between the cultures fermented in M1 and M2 (Fig. 4). The electrophoresis pattern for the culture fermented in M4 bore some similarity to the cultures fermented in M1 and M2. However, differences were observed with respect to the types of surface proteins. In addition, the intensity of the bands was higher. This is in line with the increased concentration of surface proteins, as found by XPS (Table 3).
FIG. 4.
SDS-PAGE gel of the supernatant of LiCl cell wall extracts of L. acidophilus NCC2628 fermented in M1 to M4. Levels of S-layer protein expression in the cultures fermented in M1 to M4 are shown. The level of protein expression (determined by densitometric analysis of the bands) was 342 for the extract from the cultures fermented in M1, 42.0 for M2, and 2.40 × 103 for M3 (all in arbitrary units).
Interestingly, the SDS-PAGE profile of the supernatant of M3 indicates that, in the absence of peptones, the bacterium is not able to synthesize most of the (extractable) surface proteins or only at very strongly reduced levels. However, several surface proteins were more strongly expressed in the case of fermentation in M3 than in M1 or M2 (see the bands around 45 and 55 kDa). In particular, the expression of the protein at around 45 kDa was very strongly enhanced (Fig. 4). Because of the high overall density of the bands for M4, we have not attempted quantify the intensity of the band belonging to the S-layer protein for this medium.
As S-layer proteins of Lactobacillus species vary in molecular mass between about 40 and 55 kDa (8, 24, 38), the band at ∼45 kDa is likely the S-layer protein of L. acidophilus. Indeed, LC-MS/MS analysis of the digested protein band in combination with SEQUEST database searching identified the band as belonging to the S-layer of L. acidophilus (7, 9). Six peptides (SEQUEST score 60) with high xcorr factors have been matched, yielding confident protein identification. The molecular mass of this surface protein of 444 amino acids was 46.6 kDa (Swiss-Prot entry P35829).
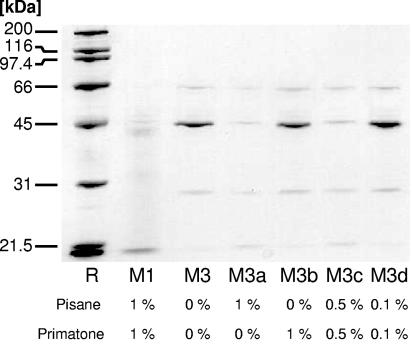
In Fig. 5, the SDS-PAGE pattern of the LiCl cell wall extract is shown for a series of media containing various concentrations of both peptones. Again, it was observed that without both peptones, the S-layer is expressed (M3) whereas with 1% of both Pisane and Primatone by weight, only a very low level of S-layer expression was seen (M1). If we leave out the Primatone while maintaining the Pisane, the S-layer expression remains at a very low level (M3a). Conversely, if 1% of Primatone by weight is added but the Pisane is left out, a high level of expression of the S-layer is again observed (M3b). The expression of the S-layer is dependent on the concentration of the Pisane: in M3c, with 0.5% of Pisane present by weight, the S-layer is expressed but fairly weakly. In M3d, containing only 0.1% Pisane by weight, S-layer expression is very strong, however.
FIG. 5.
SDS-PAGE gel of the supernatant of LiCl cell wall extracts of L. acidophilus NCC2628 fermented in M3 supplemented with various quantities of Pisane and Primatone as indicated in the figure. All concentrations are in percentages by weight. The level of protein expression (determined by densitometric analysis of the bands) was 54.6 for the extract from the cultures fermented with 1% Pisane and 1% Primatone, 828 for the cultures without Primatone and Pisane, 60.4 for the cultures with 1% Pisane, 700 for the cultures with 1% Primatone, and 101 and 1.46 × 103 for the cultures fermented with 0.5% and 0.1% Pisane and Primatone, respectively (all in arbitrary units).
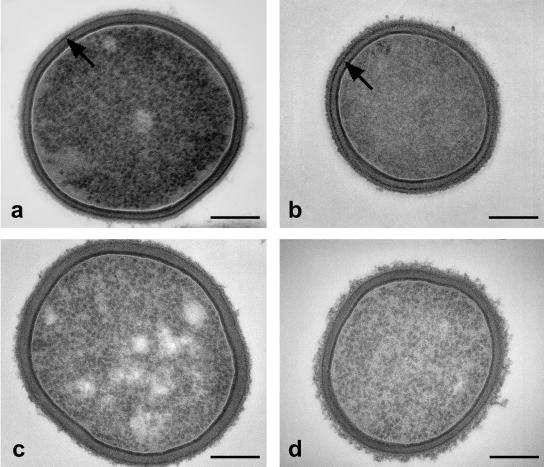
Transmission electron microscopy.
The change in protein content of the cell wall by omitting amino acid sources from the fermentation medium is confirmed by the TEM analysis (Fig. 6). In the cell wall of the cultures fermented in both M1 and M2, a dark, proteinaceous band could be observed (Fig. 6a and b). This band is reduced in intensity in the cell wall shown in Fig. 6c (fermented in M3) and Fig. 6d (fermented in M4). An S-layer on the outside of the cell wall, as found, for instance, for L. helveticus ATCC 12046 in the exponential growth phase (24, 29) and L. crispatus DSM20584 (36), was not observed. This is in agreement with the data from electrophoretic mobility analysis and the interfacial-adhesion assay, which show the bacterial surface to be both weakly charged at low pH and generally hydrophilic. An S-layer on the outside of the cell wall, conversely, would turn the surface strongly positively charged at low pH (the S-layer protein in Lactobacillus is highly basic, with an estimated pI around 10 [38]) and render it highly hydrophobic (36).
FIG. 6.
Transmission electron micrographs of L. acidophilus NCC2628 fermented in the various media. (a) M1 (complete medium); (b) M2 (without sugars); (c) M3 (without peptides); (d) M4 (without yeast extracts). The arrows in panels a and b point to a protein-rich layer within the cell wall. This layer is much less intense in panels c and d. Bars, 200 nm.
DISCUSSION
For the purpose of the present investigation, M1 is considered a reference state for investigating the effects of the medium properties on the physicochemical properties of L. acidophilus NCC2628. M1 has been optimized with respect to the cell count by variation of the type and concentration of carbohydrates, peptones, and yeast extracts (data not shown). Under these favorable growth conditions, the bacterium is hydrophilic and well dispersed in the medium and it does not form chains. L. acidophilus NCC2628 turns out to be somewhat more hydrophilic than most L. acidophilus strains reported in the literature (28, 34, 40), even when fermented in standard MRS medium (data not shown). As discussed below in more detail, hydrophobicity values for L. acidophilus show a considerable variation from strain to strain, which could in part be due to differences in growth medium and conditions.
In M2, without carbohydrates, the physicochemical properties of the bacterial surface are closely similar to those in M1, but the overall cell count is somewhat lower, reflecting the carbohydrate limitation in M2. The zeta potential of L. acidophilus NCC2628 in M2 is more positive at low pH and more negative at high pH, reflecting a higher protein content of the cell wall, as is corroborated by the XPS analysis (Table 4; see also the discussion below). This is also the case for the bacteria grown in M4, which are both highly charged and very hydrophobic.
For bacteria grown in M3, significant differences are observed in the physicochemical surface properties compared to the cultures grown in M1 and M2 (Fig. 2 and 3). In this respect, the omission of the two principal amino acid sources is having effects similar to those observed during microbial starvation, which is known to increase the microbial hydrophobicity (23). The overall protein content of the cell wall is significantly lower for cultures fermented in M3 than for the cultures fermented in M1, as is found both by XPS analysis of the freeze-dried bacterial samples and by the UV adsorption of the LiCl extract. Interestingly, the omission of the principal amino acid sources leads to significant differences in the occurrence of extractable cell wall proteins (compare the SDS-PAGE profiles in Fig. 4). In particular, in the absence of the peptones, the expression of the S-layer protein is strongly enhanced (Fig. 4). This suggests that the S-layer protein is preferentially expressed under conditions which are not optimal for bacterial growth. Of the various roles proposed for the bacterial S-layer, this is in line with a postulated protective effect whereby the S-layer is expressed in response to a stress factor (4, 7). From the experimental series in which the concentrations of Pisane and Primatone have been varied, the concentration at which the Pisane starts to have an effect may be estimated to be between 0.1 and 0.5% by weight (Fig. 5). Again, the contribution of the Pisane to the growth medium is likely that it enhances the growth conditions in the medium and it reduces the environmental stress.
We have assessed the role of the amino acids in the fermentation medium by supplementing M3 with all amino acids of which the concentration was lower than 0.1% by weight. M3 contains all 20 amino acids, but some at a rather low concentration (see the supplemental material). The supplementation of M3 with amino acids does not lead to a change in the expression of extractable surface proteins (data not shown). Consequently, the expression of the S-layer is not directly related to the lack of an amino acid in the medium. This further supports the interpretation that the S-layer is expressed in response to an environmental stress factor.
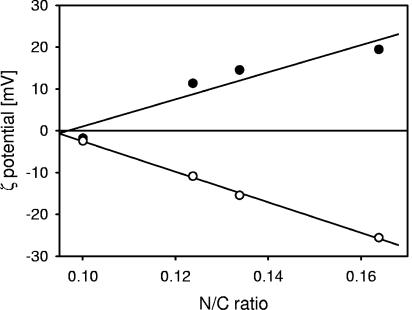
It is of interest to establish relations between the cell wall composition and the physicochemical properties of the bacterial surfaces. In Fig. 7, the zeta potential of L. acidophilus NCC2628 is plotted as a function of the N/C ratio. The data shown are for the various media, M1 to M4, and the values of the zeta potential are those at pH 3, and those at the minimum of the zeta potential in the pH range between 3 and 10. A very good linear relation between the N/C ratios of the various bacterial surfaces and their electrostatic properties was obtained. This demonstrates that the electrostatic properties of the bacterial surfaces are largely determined by their protein content and that the nature of the surface proteins does not significantly change with variations in the composition of the fermentation medium. This is in good agreement with our SDS-PAGE analysis, which shows that for the cultures fermented in M1, M2, and M4, only the concentration of surface proteins varies.
FIG. 7.
Correlation between the zeta potential and the N/C ratio of the bacterial surface as determined by XPS analysis. Filled symbols, zeta potential at pH 3; open symbols, minimum of the zeta potential in the pH range between 3 and 10. The solid lines indicate the linear regression of the experimental data. R2 = 0.89 (zeta potential at pH 3 versus N/C ratio); R2 = 1.00 (minimum value of the zeta potential versus N/C ratio).
Good correlations between the N/C ratio and the results of the interfacial-adhesion assay are also obtained, with the exception of the cultures fermented in M3 (Tables 2 and 4). This indicates that for the cultures fermented in M1, M2, and M4, the surface hydrophobicity is related to the protein content of the bacterial surfaces. The culture fermented in M3 is an exception, but this is not surprising, as the protein profile is drastically different. The contradiction between the strong expression of the S-layer and the low zeta potential and hydrophobicity of the cultures fermented in M3 is only an apparent one, as it is demonstrated that the S-layer is buried within the cell wall and therefore does not contribute to the physicochemical surface properties.
The mechanisms by which hydrophobicity of the bacterial surface is induced are most likely different between the cultures fermented in M3 and in M4, since the zeta potential profiles differ strongly between these two cultures (Fig. 2). One explanation is that other cell wall constituents like (lipo)teichoic acids or specific polysaccharides are responsible for the hydrophobicity of the bacterial surface, as these constituents have been implied in microbial hydrophobicity (36) and adhesion (1, 20). On the other hand, the far-from-optimal growth conditions in the medium without peptones might induce the bacteria to express, at low surface concentrations, a specific adhesion factor.
Variations in the surface properties of L. acidophilus have been observed before. Under standard growth conditions in MRS medium, L. acidophilus strains vary from hydrophilic to hydrophobic (28, 34, 41). Our data obtained for L. acidophilus NCC2628 fermented in standard MRS medium (Difco, France) and harvested in stationary phase support these observations (data not shown). However, the variations observed in the literature may in part be caused by variations in the composition of the MRS medium, as even small changes in the composition of a commercial MRS medium are found to lead to very significant changes in the hydrophobicity of L. acidophilus (27). Based on our results, we would infer that the change in composition of the MRS medium is related to a change in protein or peptone composition. Indeed, one of the principal changes in the composition of the MRS medium is related to its peptone content (27).
In conclusion, the composition of the fermentation medium is a major factor in determining the surface properties of microorganisms like L. acidophilus. For L. acidophilus NCC2628, the physicochemical properties and the protein composition of the bacterial cell wall remain largely unchanged if the carbohydrates are left out of the fermentation medium, although the bacterial growth is influenced. Both peptones and yeast extract have a considerable influence on the structure and physicochemical properties of the bacterial cell wall. A major change in cell wall structure is that a protein-rich band within the cell wall, as observed in transmission electron microscopy, is largely absent in the media without peptones and yeast extract. At the same time, the hydrophobicity of the cell wall, which is very low for the cultures grown in the complete medium and in the absence of carbohydrates, becomes fairly high for the cultures grown in the absence of peptone and yeast extract. The hydrophobicity and electric charge correlate well with the N/C ratio of the bacterial surfaces, indicating a major role for surface proteins in the overall physicochemical properties. The exception is formed for the cultures grown in the medium without peptones, where major changes in protein expression in the cell wall, in particular, a strongly enhanced expression of the S-layer protein, are observed. This enhanced S-layer expression is most likely stress induced.
Finally, we anticipate that our findings promote the adoption of physicochemical techniques in the microbiology of the bacterial cell wall, in particular, in relation to the development of fermentation processes and the optimization of bacterial strains for probiotic health benefits.
Supplementary Material
Acknowledgments
In the development of the fermentation media, the support of C. Cavadini, F. Praplan, and W. El-Khal is acknowledged. We thank A.-C. Pittet for technical assistance and W. Sybesma for critical reading of the manuscript. H.-J. Mathieu and N. Xanthopoulos (EPFL, Lausanne, Switzerland) are thanked for help with the XPS analysis.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abu-Lail, N. I., and T. A. Camesano. 2003. Role of lipopolysaccharides in the adhesion, retention and transport of Escherichia coli JM109. Environ. Sci. Technol. 37:2173-2183. [DOI] [PubMed] [Google Scholar]
- 2.Bernet, M. F., D. Brassart, J. R. Neeser, and A. L. Servin. 1993. Adhesion of human bifidobacterial strains to cultured human intestinal epithelial cells and inhibition of enteropathogen-cell interactions. Appl. Environ. Microbiol. 59:4121-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernet, M. F., D. Brassart, J. R. Neeser, and A. L. Servin. 1994. Lactobacillus acidophilus LA 1 binds to cultured human intestinal cell lines and inhibits cell attachment and cell invasion by enterovirulent bacteria. Gut 35:483-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beveridge, T. J., and L. L. Graham. 1991. Surface layers of bacteria. Microbiol. Rev. 55:684-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blum, S., D. Haller, A. Pfeifer, and E. J. Schiffrin. 2002. Probiotics and immune response. Clin. Rev. Allergy Immunol. 22:287-309. [DOI] [PubMed] [Google Scholar]
- 6.Boonaert, C. J. P., and P. G. Rouxhet. 2000. Surface of lactic acid bacteria: relationships between chemical composition and physicochemical properties. Appl. Environ. Microbiol. 66:2548-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boot, H. J., C. P. A. M. Kolen, J. M. van Noort, and P. H. Pouwels. 1993. S-layer protein of Lactobacillus acidophilus ATCC 4356: purification, expression in Escherichia coli, and nucleotide sequence of the corresponding gene. J. Bacteriol. 175:6089-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boot, H. J., C. P. A. M. Kolen, and P. H. Pouwels. 1995. Identification, cloning, and nucleotide sequence of a silent S-layer protein gene of Lactobacillus acidophilus ATCC 4356 which has extensive similarity with the S-layer protein of this species. J. Bacteriol. 177:7222-7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boot, H. J., C. P. A. M. Kolen, B. Pot, K. Kersters, and P. H. Pouwels. 1996. The presence of two S-layer-protein-encoding genes is conserved among species related to Lactobacillus acidophilus. Microbiology 142:2375-2384. [DOI] [PubMed] [Google Scholar]
- 10.Bowen, W. R., and R. J. Cooke. 1989. Studies of Saccharomyces cerevisiae during fermentation—an in vivo electrokinetic investigation. Biotechnol. Bioeng. 33:706-715. [DOI] [PubMed] [Google Scholar]
- 11.Busscher, H. J., R. Bos, H. C. van der Mei, and P. S. Handley. 2000. Physicochemistry of microbial adhesion from an overall approach to the limits, p. 431-458. In A. Baszkin and W. Norde (ed.), Physical chemistry of biological interfaces. Marcel Dekker, New York, N.Y.
- 12.Camesano, T. A., and N. I. Abu-Lail. 2002. Heterogeneity in bacterial surface polysaccharides, probed on single-molecule basis. Biomacromolecules 3:661-667. [DOI] [PubMed] [Google Scholar]
- 13.Cuperus, P. L., H. C. van der Mei, G. Reid, A. W. Bruce, A. H. Khoury, P. G. Rouxhet, and H. J. Busscher. 1993. Physicochemical surface properties of urogenital and poultry lactobacilli. J. Colloid Interface Sci. 156:319-324. [Google Scholar]
- 14.Delcour, J., T. Ferain, M. Deghorain, E. Palumbo, and P. Hols. 1999. The biosynthesis and functionality of the cell-wall of lactic acid bacteria. Antonie Leeuwenhoek 76:159-184. [PubMed] [Google Scholar]
- 15.Dufrêne, Y. F., and P. G. Rouxhet. 1996. Surface composition, surface properties, and adhesiveness of Azospirillum brasilense—variation during growth. Can. J. Microbiol. 42:548-556. [Google Scholar]
- 16.Dufrêne, Y. F., C. J. P. Boonaert, and P. G. Rouxhet. 1999. Surface analysis by X-ray photonelectron spectroscopy in study of bioadhesion and biofilms. Methods Enzymol. 310:375-389. [DOI] [PubMed] [Google Scholar]
- 17.Eng, J. K., A. L. McCormack, and J. R. Yates. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976-989. [DOI] [PubMed] [Google Scholar]
- 18.Evans, D. F., and H. Wennerström. 1994. The colloidal domain: where physics, chemistry, biology and technology meet. VCH Publishers, New York, N.Y.
- 19.Fuller, R. 1992. Probiotics: the scientific basis. Chapman & Hall, London, United Kingdom.
- 20.Granato, D., F. Perotti, I. Masserey, M. Rouvet, M. Golliard, A. Servin, and D. Brassart. 1999. Cell surface-associated lipoteichoic acid acts as an adhesion factor for attachment of Lactobacillus johnsonii La1 to human enterocyte-like Caco-2 cells. Appl. Environ. Microbiol. 65:1071-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hellmann, U., C. Wernstedt, J. Gonez, and C. H. Heldin. 1995. Improvement of an in-gel digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal. Biochem. 224:451-455. [DOI] [PubMed] [Google Scholar]
- 22.Isolauri, E., S. Salminen, and T. Mattila-Sandholm. 1999. New functional foods in the treatment of food allergy. Ann. Med. 31:299-302. [DOI] [PubMed] [Google Scholar]
- 23.Kjelleberg, S., and M. Hermansson. 1984. Starvation-induced effects on bacterial surface characteristics. Appl. Environ. Microbiol. 48:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lortal, S., J. van Heijenoort, K. Gruber, and U. B. Sleytr. 1992. S-layer of Lactobacillus helveticus ATCC12046: isolation, chemical characterization and re-formation after extraction with lithium chloride. J. Gen. Microbiol. 138:611-618. [Google Scholar]
- 25.Lortal, S., M. Rousseau, P. Boyaval, and J. van Heijenoort. 1991. Cell wall and autolytic system of Lactobacillus helveticus ATCC12046. J. Gen. Microbiol. 137:549-559. [Google Scholar]
- 26.Mack, D. R., S. Michail, S. Wei, L. McDougall, and M. A. Hollingsworth. 1999. Probiotics inhibit enteropathogenic E. coli adherence in vitro by inducing intestinal mucin gene expression. Am. J. Physiol. 276:G941-G950. [DOI] [PubMed] [Google Scholar]
- 27.Millsap, K. W., H. C. van der Mei, and H. J. Busscher. 1996. Physico-chemical and adhesive cell surface properties of Lactobacillus strains grown in old formula and new, standardized MRS medium. 1996. J. Microbiol. Methods 27:239-242. [Google Scholar]
- 28.Millsap, K. W., G. Reid, H. C. Van der Mei, and H. J. Busscher. 1997. Cluster analysis of genotypically characterized Lactobacillus species based on physicochemical cell surface properties and their relationship with adhesion to hexadecane. Can. J. Microbiol. 43:284-291. [Google Scholar]
- 29.Mozes, N., and S. Lortal. 1995. X-ray photoelectron spectroscopy and biochemical analysis of the surface of Lactobacillus helveticus ATCC12046. J. Gen. Microbiol. 141:11-19. [Google Scholar]
- 30.Mozes, N., and P. G. Rouxhet. 1990. Microbial hydrophobicity and fermentation technology. In R. J. Doyle and M. Rosenberg (ed.), Microbial cell surface hydrophobicity. American Society for Microbiology, Washington, D.C.
- 31.Neufeld, R. J., J. E. Zajic, and D. F. Gerson. 1980. Cell surface measurements in hydrocarbon and carbohydrate fermentations. Appl. Environ. Microbiol. 39:511-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panchaud, A., M. Kussmann, and M. Affolter. 2005. Rapid enrichment of bioactive milk proteins and iterative, consolidated protein identification by multidimensional protein identification technology. Proteomics 5:3836-3846. [DOI] [PubMed] [Google Scholar]
- 33.Razatos, A., Y. L. Ong, M. M. Sharma, and G. Georgiou. 1998. Molecular determinants of bacterial adhesion monitored by atomic force microscopy. Proc. Natl. Acad. Sci. USA 95:11059-11064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reid, G., P. L. Cuperus, A. W. Bruce, H. C. Van der Mei, L. Tomeczek, A. H. Khoury, and H. J. Busscher. 1992. Comparison of contact angles and adhesion to hexadecane of urogenital, dairy, and poulty lactobacilli: effect of serial culture passages. Appl. Environ. Microbiol. 58:1549-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schär-Zammaretti, P., and J. Ubbink. 2003. Imaging of lactic acid bacteria with AFM—elasticity and adhesion maps and their relationship to biological and structural data. Ultramicroscopy 97:199-208. [DOI] [PubMed] [Google Scholar]
- 36.Schär-Zammaretti, P., and J. Ubbink. 2003. The cell wall of lactic acid bacteria: surface constituents and macromolecular conformations. Biophys. J. 85:4076-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shevchenko, A., M. Wilm, M. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 38.Smit, E., F. Oling, R. Demel, B. Martinez, and P. H. Pouwels. 2001. The S-layer protein of lactobacillus acidophilus ATCC4356: identification and characterisation of domains responsible for S-protein assembly and cell wall binding. J. Mol. Biol. 305:245-257. [DOI] [PubMed] [Google Scholar]
- 39.Toba, T., R. Virkola, B. Westerlund, Y. Björkman, J. Sillanpää, T. Vartio, N. Kalkinnen, and T. Korhonen. 1995. 1995. A collagen-binding S-layer protein in Lactobacillus crispatus. Appl. Environ. Microbiol. 61:2467-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39a.Ubbink, J., and P. Schär-Zammaretti. 2005. Probing bacterial interactions; integrated approaches combining atomic force microscopy, electron microscopy and biophysical techniques. Micron 36:293-320. [DOI] [PubMed] [Google Scholar]
- 40.Van der Mei, H. C., R. Bos, and H. J. Busscher. 1998. A reference guide to microbial cell surface hydrophobicity based on contact angles. Colloids Surf. B 11:213-221. [Google Scholar]
- 41.Van der Mei, H. C., B. Van de Belt-Gritter, P. H. Pouwels, B. Martinez, and H. J. Busscher. 2003. Cell surface hydrophobicity is conveyed by S-layer proteins—a study in recombinant lactobacilli. Colloids Surf. B 28:127-134. [Google Scholar]
- 42.Waar, K., H. C. van der Mei, H. J. M. Harmsen, J. E. Degener, and H. J. Busscher. 2002. Adhesion to bile drain materials and physicochemical surface properties of Enterococcus faecalis strains grown in the presence of bile. Appl. Environ. Microbiol. 68:3855-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wood, B. J. B. 1992. The lactic acid bacteria in health and disease. The lactic acid bacteria, vol. 1. Elsevier Applied Science, London, United Kingdom.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.