Abstract
Degenerate primers were used to amplify 14 distinct reductive-dehalogenase-homologous (RDH) genes from the Dehalococcoides-containing mixed culture KB1. Most of the corresponding predicted proteins were highly similar (97 to >99% amino acid identity) to previously reported Dehalococcoides reductive dehalogenases. To examine the differential transcription of these RDH genes, KB1 was split into five subcultures amended with either trichloroethene, cis-1,2-dichloroethene, vinyl chloride, 1,2-dichlorethane, or no chlorinated electron acceptor. Total RNA was extracted following the onset of reductive dechlorination, and RDH transcripts were reverse transcribed and amplified using degenerate primers. The results indicate that the transcription of RDH genes requires the presence of a chlorinated electron acceptor, and for all treatments, multiple RDH genes were simultaneously transcribed, with transcripts of two of the genes being present under all four electron-accepting conditions. Two of the transcribed sequences were highly similar to reported vinyl chloride reductase genes, namely, vcrA from Dehalococcoides sp. strain VS and bvcA from Dehalococcoides sp. strain BAV1. These findings suggest that multiple RDH genes are induced by a single chlorinated substrate and that multiple reductive dehalogenases contribute to chloroethene degradation in KB1.
The industrial solvents trichloroethene (TCE) and tetrachloroethene (PCE) are pervasive and persistent groundwater contaminants. TCE- and PCE-contaminated sites frequently contain substantial quantities of the degradation products cis-1,2-dichloroethene (cDCE) and vinyl chloride (VC), which accumulate due to biotic and abiotic processes (4, 10, 23). Because of the acute and chronic toxicity of these chlorinated compounds, they pose risks to human and environmental health (13). In addition, these recalcitrant compounds tend to persist in anoxic subsurface environments (32). Fortunately, anaerobic dechlorinating microorganisms have been identified in a variety of genera. Among these, several isolates of the genus Dehalococcoides can completely reductively dechlorinate chlorinated ethenes to the nontoxic end product ethene (31). KB1 is a microbial consortium dominated by Dehalococcoides organisms (5). The successful stimulation of complete dechlorination of chlorinated ethenes in situ through bioaugmentation has been demonstrated in the field (16, 20). Molecular tools such as PCR detection of dehalogenase genes and transcripts have the potential to aid in monitoring and diagnosing the dechlorinating abilities at a site.
To date, 15 different reductive dehalogenases (RDases) have been at least partially purified from a variety of microbial genera (3, 11, 17, 19, 22-27, 30, 34, 38). These include the first RDase purified from a Dehalococcoides organism, TceA of Dehalococcoides ethenogenes strain 195 (18, 19). More recently, a VC RDase was partially purified from Dehalococcoides sp. strain VS (23). All but one of the RDases have been copurified with a corrinoid cofactor, and all contained iron-sulfur clusters. Physiological studies of whole cells and cell extracts have suggested the expression of different dechlorinating enzymes depending on the type and concentration of the initial dechlorinating substrate (8, 12, 37).
Biochemical purification has proved laborious due to the low growth yields of these organisms as well as the hydrophobic nature and oxygen sensitivity of their membrane-associated proteins. However, molecular investigations of the reductive dehalogenase genes have provided a wealth of sequence information. An analysis of the genome of Dehalococcoides strain 195 revealed 17 intact genes that appeared homologous to genes encoding biochemically purified reductive dehalogenases (31, 39). Based on these sequences, Krajmalnik-Brown et al. (15) designed degenerate primers to amplify Dehalococcoides-specific reductive-dehalogenase-homologous (RDH) genes. This allowed for the amplification of many RDH genes from different strains of Dehalococcoides, including strain BAV1 (7 homologues), strain FL2 (14 homologues), and strain CBDB1 (14 homologues). Although some strain-specific genes were identified, Hölscher et al. (14) identified 10 subclusters of genes with orthologues in two or more different Dehalococcoides strains.
The fact that each strain possesses multiple RDHs has prompted investigations of what controls the expression of these genes. Studies with the cpr gene cluster of Desulfitobacterium dehalogenans have identified two regulatory proteins, cprC and cprK, whose gene products belong to the NirI/NosR and CRP/FNR families, respectively (33). The transcription of cprA and cprB was shown to be dependent on the chlorinated substrate 3-chloro-4-hydroxyphenylacetate. A recent examination of the Dehalococcoides ethenogenes strain 195 genome revealed homologues of cprC and cprK; however, the majority of the transcriptional regulators were two-component signal transduction systems (31). What remains to be investigated is if these regulatory genes respond to specific substrates and thus cause the transcription of different dehalogenase genes based on the presence of specific chlorinated compounds used as electron acceptors.
The aims of these studies were to first identify the Dehalococcoides RDH genes in KB1 and then to understand which of these RDH genes have an active role in reductive dechlorination of TCE, cDCE, VC, and 1,2-dichloroethane (1,2-DCA). To address the latter issue, we identified RDH genes that were transcribed in KB1 during the dechlorination of these four different electron acceptors. Our results indicated that two genes were transcribed in response to all four of the chlorinated electron acceptors and that multiple genes were simultaneously transcribed during the degradation of each of the compounds. This work adds to the RDH sequence data set and further extends it by investigating the differential transcription of these genes under different electron-accepting conditions. This information will be useful in creating molecular tools to aid in the design and monitoring of bioremediation schemes. Accumulating research indicates that the 16S rRNA gene is not sufficient to distinguish the dechlorinating abilities of different strains of Dehalococcoides (5, 9, 28). However, the detection of specific RDH genes may allow for the prediction of a site's degradation potential, and the detection of RDH gene transcripts may allow estimates of in situ dechlorination activity.
MATERIALS AND METHODS
Chemicals.
All chemicals were purchased from Sigma-Aldrich, unless noted otherwise. Chlorinated ethenes were of >97% purity (Sigma-Aldrich). A 1% gas mixture of ethene and methane (Scotty II; Alltech Associates, Inc.) was used for gas chromatograph calibration.
Cultures and growth conditions.
All KB1 subcultures grown with different chloroaliphatic compounds were derived from a single, TCE-dechlorinating parent culture (6). Subcultures were maintained with different chlorinated electron acceptors in 200- to 2,000-ml volumes in glass bottles (with approximately 10% of the total volume being headspace) and amended with methanol or hydrogen plus acetate as previously described (5). The cultures were maintained in defined mineral medium (7). All KB1 cultures were replenished with 20 to 50% (vol/vol) fresh medium if degradation rates decreased, approximately every 3 to 6 months. A KB1 enrichment culture that was maintained with VC and methanol for 5 years (KB1/VC-MeOH) was the source of inoculum for the transcription experiment described herein. This ethene-producing culture was grown with weekly amendments of 445 μM (liquid concentration) VC from a 100% gas stock and of 2.2 mM methanol. The KB1/VC-MeOH culture contains two identified Dehalococcoides 16S rRNA gene sequences, KB1/VC (AY146779) and KB1/PCE (AY146780), and Dehalococcoides accounts for about 50% of the biomass, with acetogens and methanogens comprising the remainder. A second subculture (KB1/VC-H2) was also used to assay specific RDH genes. This subculture contains only one Dehalococcoides 16S rRNA gene sequence, KB1/VC, and in this culture, Dehalococcoides accounts for >90% of the biomass (5).
Transcription experiment.
The KB1/VC-MeOH subculture (2 liters) was purged with a sterile N2-CO2 (80:20 [vol/vol]) gas stream that was passed over heated copper filings until the methane, ethene, and VC concentrations were below the detection limits. Following a 70-hour incubation, 400-ml portions of the culture were dispensed into five identical 1-liter bottles (Pyrex; VWR, Mississauga, Ontario, Canada) in an anaerobic glove box (Coy Laboratory Products, Grass Lake, MI). The bottles were sealed with plastic screw-cap tops in which a hole had been drilled in order to insert a black butyl rubber septum (Geo-Microbial Technologies, Ochelata, OK) for repeated sampling. Individual chlorinated electron acceptors were added to each bottle by syringe as undiluted compounds in the following amounts: TCE, 150 μmol; cDCE, 300 μmol; VC, 150 μmol; and 1,2-DCA, 200 μmol. These amounts correspond to the following initial aqueous concentrations: TCE, 200 μM; cDCE, 475 μM; VC, 135 μM; and 1,2-DCA, 465 μM. One bottle was not amended with a chlorinated electron acceptor. Headspace samples (300 μl) were then taken (after the compounds had equilibrated) for compositional analysis, and an initial well-mixed liquid sample (50 ml) was taken from the unamended treatment for RNA analysis. Subsequently, methanol was added as the electron donor (495 μmol) to all five bottles to initiate dechlorination. In an effort to detect genes responding to the chlorinated substrate that was added to the culture and not to subsequent dechlorination products, samples for RNA extraction were taken soon after the onset of dechlorination.
Analytical procedures.
Chlorinated ethenes and ethanes, methane, and ethene were analyzed by gas chromatography of headspace samples as described previously (5).
Nucleic acid extraction.
Genomic DNAs were extracted from 15 ml of liquid culture using an UltraClean soil DNA kit (Mo Bio Laboratories Inc., Solana Beach, CA) as previously described (5). In total, DNA was extracted from six different cultures to capture the breadth of KB1 cultures. For RNA extraction, 50-ml samples were withdrawn from the culture bottles inside an anaerobic chamber (Coy Laboratory Products). To avoid creating excessive vacuum, 60 ml of N2-CO2 was injected into each culture bottle with a sterile 60-ml syringe prior to withdrawing 50 ml of culture in the same syringe. The culture sample was then dispensed into an anaerobic centrifuge tube on ice. Cells were collected by centrifugation at 2,000 × g for 40 min at 4°C. After discarding the supernatant, the pellet was resuspended in 300 μl of ice-cold lysis solution (1.4 M NaCl, 22 mM EDTA, 35 mM sodium dodecyl sulfate) and transferred to a 1.5-ml screw-cap microcentrifuge tube. Subsequently, 900 μl of ice-cold acid-phenol-chloroform-isoamyl alcohol (125:24:1, pH 4.5) (Ambion, Austin, TX) and 100 μl of zirconia/silica beads (0.5 mm; BioSpec Products Inc., Bartlesville, OK) were added to the microcentrifuge tube. The tube was then agitated horizontally on a vortex machine (VELP Scientifica, Plainview, NY) for 4 min at maximum speed followed by centrifugation at 14,000 × g for 3 min at 4°C. The aqueous supernatant was removed and transferred to a new 1.5-ml microcentrifuge tube. Ammonium acetate (0.1 volume of a 5 M solution) and isopropanol (1.1 volumes) were added, and the RNA was precipitated overnight at −20°C. The RNA solution was then purified using an RNeasy spin column (QIAGEN, Valencia, Calif.). Contaminating DNAs were removed using two successive treatments of a DNA-free kit (Ambion, Austin, TX).
Reverse transcription.
Each RNA sample was divided into two, with one half serving as a control to which no reverse transcriptase (RT) was added. Total RNA was quantified using the Ribogreen method according to the manufacturer's recommendations (Molecular Probes, Burlington, Ontario, Canada). First-strand cDNA synthesis was performed using 6 to 12 μg of RNA, 250 ng of random hexamers (Invitrogen, Carlsbad, CA), and 10 nmol of deoxynucleoside triphosphates (Invitrogen). The final volume was adjusted to 15 μl using RNase-free water. This mixture was then heated to 65°C for 5 min in a PTC-200 DNA engine thermocycler (MJ Research). The sample was cooled in an ice bath before 2 μl of a 0.1 M dithiothreitol solution (Invitrogen) and 4 μl of 5× first-strand buffer (Invitrogen) were added. The sample was then incubated at 25°C for 5 min. Finally, 1 μl of SuperScript II (Invitrogen) was added, and the thermocycler program was continued at 25°C for 10 min, 42°C for 1 h, and then 70°C for 15 min.
PCR amplification of RDH genes from DNA or cDNA.
Degenerate primers B1R and RRF2 (15) were used to amplify RDH genes from genomic DNA (for KB1 RDH gene identification) and cDNA templates (for transcription experiments). PCR mixtures (50 μl) contained 1 to 10 μl of cDNA or 50 ng of genomic DNA, a 0.5 μM concentration of each primer, 2.5 mM MgCl2, a 0.25 mM concentration of each deoxynucleotide (MBI Fermentas, Burlington, Ontario, Canada), 0.13 mg/ml of bovine serum albumin, and 1.25 U of Taq DNA polymerase (New England Biolabs, Beverly, MA) in 1× PCR buffer (New England Biolabs). PCRs were carried out with the following parameters: 130 s at 94°C; 30 cycles of 30 s at 94°C, 45 s at 48°C, and 130 s at 72°C; and a final extension of 6 min at 72°C. Agarose gels (1%) were run to verify amplification and amplicon sizes. To detect contamination of the cDNA with genomic DNA, the PCR products from cDNAs generated without RT were also analyzed in agarose gels.
RDH-specific primers.
Specific PCR primers were designed for each of the RDH genes found in the KB1 subcultures (Table 1). For specific RDH genes, PCRs were carried out as described above, except that the annealing and extension steps were performed for only 30 s, and the annealing temperature was 60°C. The specificity of these primers was assessed by attempting PCR amplification of nontarget DNA from Escherichia coli, a mixed toluene-degrading consortium, and clones containing nontarget RDH genes. In addition, PCRs were performed with genomic DNA from three different KB1 subcultures at three different annealing temperatures. The resulting amplicons were sequenced, the chromatograms were visually inspected, and the sequences were aligned.
TABLE 1.
Specific primer sequences designed for KB1 RDH genes
| KB1 gene target | Primer namea | Sequence (5′-3′) |
|---|---|---|
| rdhA1 | rdhA1_246f | ATCGGAGCTGCACAAGTAGG |
| rdhA1_336r | TCTTGTGAGCGGTGTCTTTG | |
| rdhA2 | rdhA2_720f | CAAAGGAGATGTTCCGGTGT |
| rdhA2_985f | CAGGTGGAAAAGACCGGTTA | |
| rdhA3 | rdhA3_1149f | CATTCTCCGGGAAGAAAACA |
| rdhA3_1379r | CCAGGCTTCCTTGTCTTCAG | |
| rdhA4 | rdhA4_754f | TTGTTATGCCGCCAATATGA |
| rdhA4_925r | TCTATCCATTTCGCCCAGAC | |
| rdhA5 | rdhA5_1017f | GATGCAGGCATTTACCGTTT |
| rdhA5_1137r | GTCTCTTTGCCTTCGGTCAG | |
| rdhA6 | rdhA6_318f | ATTTAGCGTGGGCAAAACAG |
| rdhA6_555r | CCTTCCCACCTTGGGTATTT | |
| rdhA7 | rdhA7_1391f | GCTAAAGAGCCGTCATCCTG |
| rdhA7_1539r | GCAGTAACAACAGCCCCAAT | |
| rdhA8 | rdhA8_845f | CCCAAGGTAGGTGTGCAGAT |
| rdhA8_1016r | CCCGGTTAGTTACCCCGTAT | |
| rdhA9 | rdhA9_251f | CTGACCTTGAAACCCCTGAA |
| rdhA9_425r | TTGCCACCCATTTCCATATT | |
| rdhA10 | rdhA10_710f | GCTGAAACACCCACCAAACT |
| rdhA10_860r | CGACAAAGGGGAATCTTTGA | |
| rdhA11 | rdhA11_429f | TAATGGCAACCGGAGGTAAG |
| rdhA11_609r | TCTACCGGTATGGCCTGAAC | |
| rdhA12 | rdhA12_864f | AGGAGTTCCTGTGGGGACTT |
| rdhA12_994r | TTTGGGGGTCATAACTGCTC | |
| rdhA13 | rdhA13_1356f | CAGGGTACCTGTCCCTTCAA |
| rdhA13_1493r | AGGGTTCTTCCGTCCGTACT | |
| rdhA14 | rdhA14_642f | GAAAGCTCAGCCGATGACTC |
| rdhA14_846r | TGGTTGAGGTAGGGTGAAGG |
The position numbers are shown within the names with respect to the first nucleotide of each gene following the RRF2 primer binding site. f, forward; r, reverse.
Cloning.
For each cloning reaction, amplicons from three to five PCRs were pooled, ligated into the pCR2.1-TOPO vector, and used to transform One Shot TOP10 competent E. coli cells (Invitrogen, Carlsbad, California). The success of the cloning reaction was determined by using an overnight E. coli culture as the template for PCR amplification of the insert with T7f and M13r primers and subsequent product visualization on a 1% agarose gel. For cultures in which the plasmids had appropriate insert sizes (1.4 to 1.7 kb), plasmid DNA was extracted using a GeneElute plasmid miniprep kit (Sigma-Aldrich, Oakville, Ontario, Canada).
Sequence determination.
Sequencing of the plasmid inserts was carried out by the Ontario Cancer Institute's sequencing facility (Princess Margaret Hospital, Toronto, Ontario, Canada), using a Beckman Coulter CEQ 2000 automatic sequencer. Inserts were sequenced using vector primers T7 forward and M13 reverse (Invitrogen).
Sequence assembly and analysis.
A total of 87 inserts, derived from both DNA and cDNA templates, were sequenced with both the T7f and M13r primers. For each unique insert, 3 to 10 times sequence coverage was obtained over the entire length. Internal sequencing primers were designed using Primer3 software (http://www.genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). The sequences were assembled into contigs using the EditSeq and SeqMan programs of the DNAStar software package (DNASTAR Inc., Madison, WI). Conceptual translations were made using the translate tool of Expasy (http://us.expasy.org/tools/dna.html). These amino acid sequences were aligned and compared to known sequences using ClustalX (36). Manual editing of the alignment was performed using Genedoc (http://www.psc.edu/biomed/genedoc). Additionally, an identity/similarity matrix was constructed using MatGAT, a program that creates pairwise alignments (2). Phylogenetic trees were constructed using PAUP* (Sinauer Associates, Inc., Sunderland, MA).
Nucleotide sequence accession numbers.
The sequences of the RDH genes and their associated B gene fragments have been deposited in GenBank under the following accession numbers: DQ177506 to DQ177519 (KB1 rdhA1 to rdhA14) and DQ115513 and DQ115514 (FL2 rdhA12 and rdhA13).
RESULTS
RDH genes in KB1 culture.
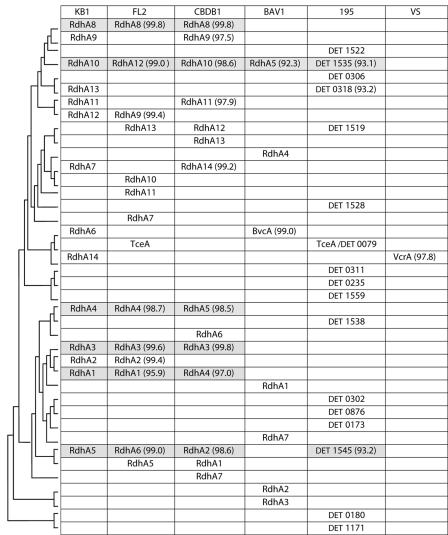
Fourteen distinct RDH gene sequences were identified in DNA samples from different KB1 subcultures (Fig. 1). All of these genes were also amplified from DNAs from the culture used in the transcription study (KB1/VC-MeOH). Furthermore, fragments of seven of these RDH genes (KB1 rdhA1, rdhA2, rdhA3, rdhA8, rdhA10, rdhA12, and rdhA14) were identified in a random genomic fragment library of KB1/VC-MeOH (E. A. Edwards, unpublished data). Each of the KB1 RDH genes identified is highly similar (93 to >99% identity at the protein level) to a previously identified RDH gene in one or more Dehalococcoides strains. Sequences with >99% identity differ by only one or two amino acids. KB1 shares nine highly similar RDH genes with strain CBDB1, eight with strain FL2, three with strain 195, and two with strain BAV1. Of the three Dehalococcoides RDH genes with ascribed functions, KB1 contains genes that are highly similar to both reported VC RDase genes (VS vcrA and BAV1 bvcA) but no genes similar to Dehalococcoides TCE RDase genes (195 tceA and FL2 tceA) (11). Although the primer set RRF2/B1R does not amplify tceA-like genes, even specific tceA-targeted primers (18) failed to amplify tceA from different KB1 subcultures (data not shown). Phylogenetic analyses of the KB1 and other Dehalococcoides RDH genes using both parsimony and neighbor-joining methods produced trees that were similar in topology to the tree presented by Hölscher et al. (14).
FIG. 1.
Comparison of translated sequences of RDH genes detected in cultures KB1 and VS and strains FL2, CBDB1, and 195. Highly similar sequences (∼93 to 100% amino acid identity) in different strains are grouped in the same row. The percent identity compared to the sequence in KB1 is shown in parentheses. The phylogenetic relationship among the proteins is indicated by the most parsimonious cladogram to the left of the table. The KB1 sequences that were detected in cDNA are listed in Table 3. Highly similar homologues found in three or more cultures are shaded. No extensive RDH gene survey has been conducted with culture VS, and vcrA is the only reported sequence.
A similarity analysis performed with conceptual protein translations of the KB1 RDH genes indicated that the majority share about 30% amino acid identity with each other (Table 2). The two most similar proteins are KB1 RdhA8 and RdhA9, which share 91.4% amino acid identity. Although the KB1 RDH protein sequences are not similar (<25% amino acid identity) to those of RDHs in other dechlorinating organisms, such as Sulfurospirillum, Dehalobacter, and Desulfitobacterium (data not shown), they still share common conserved motifs such as the two iron-sulfur cluster binding motifs and the twin arginine leader peptide.
TABLE 2.
Identity/similarity matrix of RDH gene translations in KB1
| Protein | % Identity or similarity with KB1 proteina
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RdhA8 | RdhA9 | RdhA10 | RdhA13 | RdhA11 | RdhA12 | RdhA7 | RdhA6 | RdhA14 | RdhA4 | RdhA3 | RdhA2 | RdhA1 | RdhA5 | |
| RdhA8 | 91.4 | 59.3 | 46.1 | 35.8 | 37.3 | 31.7 | 34.2 | 34.6 | 35.0 | 33.4 | 32.1 | 31.8 | 32.5 | |
| RdhA9 | 95.6 | 59.3 | 47.4 | 36.6 | 37.3 | 31.6 | 34.7 | 35.2 | 34.8 | 34.5 | 32.1 | 33.2 | 32.9 | |
| RdhA10 | 75.8 | 75.6 | 46.5 | 35.5 | 35.0 | 34.3 | 33.3 | 32.4 | 32.4 | 32.5 | 32.4 | 32.5 | 30.2 | |
| RdhA13 | 63.5 | 64.9 | 64.3 | 39.5 | 40.6 | 34.5 | 33.0 | 35.1 | 32.3 | 31.0 | 32.0 | 31.2 | 30.8 | |
| RdhA11 | 53.0 | 54.1 | 53.9 | 58.2 | 61.0 | 31.2 | 31.2 | 31.1 | 31.8 | 28.9 | 30.7 | 29.2 | 29.8 | |
| RdhA12 | 54.5 | 55.7 | 55.0 | 59.9 | 74.9 | 29.9 | 31.8 | 30.9 | 30.6 | 30.5 | 32.2 | 30.4 | 30.1 | |
| RdhA7 | 51.3 | 52.1 | 50.7 | 53.8 | 49.1 | 50.1 | 31.0 | 31.5 | 28.5 | 30.9 | 30.4 | 33.2 | 31.5 | |
| RdhA6 | 49.8 | 50.0 | 52.6 | 52.4 | 47.6 | 49.6 | 51.6 | 39.0 | 31.1 | 29.8 | 29.7 | 30.2 | 28.4 | |
| RdhA14 | 51.9 | 50.9 | 50.3 | 50.7 | 48.9 | 49.9 | 49.9 | 55.5 | 30.3 | 29.2 | 31.6 | 31.1 | 28.2 | |
| RdhA4 | 53.2 | 52.4 | 51.9 | 49.7 | 49.9 | 50.2 | 48.9 | 47.4 | 47.3 | 38.0 | 38.5 | 38.7 | 35.6 | |
| RdhA3 | 51.0 | 52.7 | 51.7 | 52.1 | 47.3 | 49.0 | 53.2 | 48.6 | 49.1 | 56.2 | 55.4 | 54.1 | 36.5 | |
| RdhA2 | 50.1 | 49.1 | 48.7 | 49.1 | 49.3 | 49.5 | 50.3 | 50.0 | 48.5 | 54.0 | 69.6 | 58.3 | 40.0 | |
| RdhA1 | 50.3 | 51.6 | 51.5 | 49.9 | 48.0 | 48.1 | 53.2 | 49.4 | 46.3 | 56.4 | 67.4 | 72.7 | 39.9 | |
| RdhA5 | 51.9 | 52.7 | 49.0 | 51.9 | 47.3 | 48.8 | 50.3 | 47.4 | 43.3 | 53.1 | 56.4 | 56.9 | 57.7 | |
The upper triangle of data represents the percent identity between two amino acid sequences in a pairwise alignment, and the lower triangle of data represents percent similarity. Similarity was calculated using the BLOSUM50 scoring matrix and a modified Smith-Waterman algorithm (2). Nearly complete translations of the rdhA genes starting after the RRF2 primer proteins were used to generate the identity and similarity matrixes.
Identification of specific RDH genes in KB1 Dehalococcoides strains.
Two strains of Dehalococcoides have been identified in KB1, based on differences in 16S rRNA gene sequence and substrate range (5). The corresponding 16S rRNA gene sequences have been designated KB1/PCE and KB1/VC, because cultures that harbor only the latter do not dechlorinate PCE (5). Genomic DNA from the mixed culture KB1/VC-MeOH (which contains both KB1/PCE and KB1/VC sequences) and genomic DNA from the KB1/VC-H2 culture (which contains only the KB1/VC sequence) were assayed for RDH genes, using the 14 primer pairs for each specific KB1 RDH gene. All 14 KB1 RDH genes were detected in culture KB1/VC-MeOH, while all except the sequence corresponding to KB1 rdhA6 (bvcA-like gene) were detected in culture KB1/VC-H2 (Fig. 2). These data suggest that the BAV1 bvcA-like gene originated from the Dehalococcoides strain with the KB1/PCE sequence and not from the strain containing the KB1/VC sequence. These data reinforce that these two 16S rRNA gene sequences (which differ by only 1 bp) do indeed correspond to two distinct strains of Dehalococcoides.
FIG. 2.
PCR amplification of DNA from different KB1 subcultures using primers specific to frequently encountered RDH genes and the Dehalococcoides 16S rRNA gene. Four different specific primer sets (rdhA14, rdhA6, rdhA5, and 1f/259r [5]) were tested on six different DNA templates. Template 1, genomic DNA from KB1/VC-H2 (only contains Dehalococcoides sequence KB1/VC); template 2, genomic DNA from a KB1/TCE-MeOH culture (contains both KB1/VC and KB1/PCE sequences); template 3, genomic DNA from KB1/VC-MeOH (contains both KB1/VC and KB1/PCE sequences); template 4, positive control (plasmid DNA containing the corresponding RDH gene or 16S rRNA gene); template 5, negative control (plasmid DNA containing a nontarget RDH gene); and template 6, negative control (no template). L, 100-bp ladder. The white arrow points to the result showing that KB1 rdhA6 (bvcA-like gene) was not detected in KB1/VC-H2.
Transcription experiment.
Complete dechlorination to ethene was observed in all cultures amended with chlorinated electron acceptors (Fig. 3). RNA extraction and cDNA synthesis were successful for each sample taken. Furthermore, the RNA preparations were not contaminated with genomic DNA because no visible amplicons were generated from PCRs of samples processed without RT (Fig. 4).
FIG. 3.
Dechlorination profiles of four treatments amended with chlorinated substrates. The arrows indicate when RNAs were extracted. The TCE-amended culture was sampled 7 h after electron donor addition; at this point, 30 μmol of TCE had been dechlorinated to cDCE, and only traces of VC were present. The cDCE-amended culture was sampled after 24 h, during the transformation of cDCE to VC before ethene production began. The VC-amended culture was also sampled after 24 h. The 1,2-DCA-amended culture was sampled after 300 h (12.5 days) owing to a long lag time of 7 days before appreciable degradation took place.
FIG. 4.
PCR amplification of cDNAs with degenerate primers targeting RDH genes (primers RRF2 and BR1). For cDNA samples from each treatment group (TCE, cDCE, VC, and 1,2-DCA), two PCRs were performed: “+” indicates that RT was used for cDNA synthesis, and “−” indicates that no RT was used. L, 1,000-bp ladder (New England Biolabs); N, PCR product from RNA extracted from the unamended culture.
RDH genes identified in transcription experiments.
In total, seven different RDH sequences were identified in cDNA samples from cultures amended with different substrates (Table 3). In each cDNA sample, multiple distinct RDH transcripts were detected, with up to six being simultaneously detected in the cDCE treatment. Two genes, i.e., KB1 rdhA6 and KB1 rdhA5, those most similar to BAV1 bvcA and FL2 rdhA6, were transcribed regardless of which chlorinated electron acceptor was provided to the culture. No RDH sequences were amplified from cDNAs generated from the unamended culture.
TABLE 3.
Comparison of the different RDH genes identified in RNA samples
| KB1 RDH gene | Most similar RDH gene | No. of clones containing each RDH gene transcript per treatmenta
|
|||
|---|---|---|---|---|---|
| TCE | cDCE | VC | 1,2-DCA | ||
| rdhA6 | BAV1 bvcA | 5 | 3 | 4 | 3 |
| rdhA14 | VS vcrA | 2 | 0 | 5 | 0 |
| rdhA5 | FL2 rdhA6 | 3 | 7 | 1 | 12 |
| rdhA3 | CBDB1 rdhA3 | 0 | 1 | 0 | 0 |
| rdhA7 | CBDB1 rdhA14 | 0 | 1 | 0 | 0 |
| rdhA1 | CBDB1 rdhA4 | 0 | 1 | 1 | 0 |
| rdhA8 | FL2 rdhA8 | 0 | 1 | 0 | 0 |
| Total | 10 | 14 | 11 | 15 | |
For each chlorinated substrate, RNA was extracted at one time point soon after dechlorination began; RDH genes were amplified from cDNA samples, cloned, and sequenced. The number of occurrences of each RDH gene transcript from the different treatments is reported.
DISCUSSION
KB1/VC contains RDH genes that are highly similar to genes in strains CBDB1, FL2, BAV1, 195, and VS. In combination with previously reported RDH gene sequences, these results suggest that a pool of Dehalococcoides RDH genes exists and that each strain possesses a different complement of these genes. Phylogenetic analyses by Hölscher et al. (14) delineated 10 subclusters of highly similar genes in strains CBDB1, FL2, BAV1, and 195. With new sequence information from the KB1 culture, six additional subclusters based on sequence similarity are evident (Fig. 1, with each row being a subcluster), as genes that seemed to be unique now have highly similar genes in KB1. Dehalococcoides strains 195 and BAV1 still have the largest proportions of unique RDH genes, with 12 out of 17 (71%) and 5 out of 7 (71%), respectively. Without more information on the evolutionary relationship between these genes, it is not possible to tell if these subclusters of highly similar homologues represent true orthologues or not; however, their close identity suggests that they share function, perhaps even to the level of substrate specificity. For example, the KB1 mixed culture contains highly similar homologues of both reported VC RDase genes, bvcA (15) and vcrA (23), consistent with the observation that this culture dechlorinates VC efficiently.
A comparison of the dechlorination conditions under which particular RDH genes were transcribed in KB1 and the substrate range of Dehalococcoides strains that possess highly similar homologues to those genes suggests that, in contrast to expectations, these highly similar genes do not necessarily encode RDases that share substrate specificity. For example, KB1 rdhA5 was transcribed under all four electron-accepting conditions. This gene has highly similar homologues in three other Dehalococcoides strains, namely, FL2 (rdhA6), CBDB1 (rdhA2), and 195 (DET1545). No chlorinated substrate has been reported to be common among these three organisms and KB1, suggesting a broad substrate range, an unidentified common substrate, different regulatory mechanisms in different strains, or a nonspecific regulatory mechanism for this putative dehalogenase. KB1 rdhA6, which is highly similar to BAV1 bvcA, was transcribed during the degradation of all four compounds (TCE, cDCE, VC, and 1,2-DCA); however, TCE is not a growth substrate for Dehalococcoides strain BAV1 (10). It could be that transcription of KB1 rdhA6 was induced by the presence of small amounts of cDCE that were formed from TCE dechlorination. KB1 rdhA14, which is highly similar to VS vcrA, was transcribed during TCE degradation, in contrast to the tested substrate range of partially purified protein fractions containing VcrA. These partially purified fractions dechlorinated all three DCE isomers at rates similar to the VC dechlorination rate; however, TCE was dechlorinated at 5% of this rate (23). Although vcrA of strain VS is closely related to KB1 rdh14, there are still significant sequence differences between these genes. There are 18 nucleotide differences in the first 72 base pairs, corresponding to six amino acid substitutions at the N terminus of the protein, which could explain differences in the substrate specificity of the enzyme. The identification of a transcript's function is complicated in sequential reactions, where dechlorination of parent and intermediate products often occurs simultaneously. However, even during VC and 1,2-DCA dechlorination reactions, each of which only involves a single dechlorination step, multiple genes were transcribed. Sequencing of Dehalococcoides genomes will provide additional insight into regulatory networks and allow more detailed experimentation to explore RDase gene transcription.
A recent analysis of the Dehalococcoides strain 195 genome described a close association between most of the RDH genes and genes coding for transcription regulators (31). Furthermore, many of these regulatory genes are homologous to those belonging to two-component signal transduction systems, whose associated histidine kinase sensors appear to reside in the cytoplasm rather than the typical membrane location (29, 31, 35). Because of the presence of PAS/PAC motifs implicated in sensing cytosolic changes in redox potential, oxygen, and light, it has been suggested that the histidine kinase sensors respond to intracellular stimuli to modulate the energy level of the cell (31, 35). In addition, an analysis of the genome of strain 195 also revealed that one RDase gene, tceA, is not situated near a transcriptional regulator. This is interesting because the corresponding RDase, TceA, was partially purified from a mixed TCE-dechlorinating culture containing strain 195. It may be that within one Dehalococcoides organism, individual RDase genes are regulated by distinct transcriptional control mechanisms, and that these controls respond to multiple stimuli.
An alternative explanation for the different substrate ranges of the Dehalococcoides strains that transcribe highly similar RDH genes is that although similar apoenzymes may be produced from these genes, perhaps different corrinoid cofactors modulate substrate specificity. All but one of the purified RDases are dependent on the corrinoid cofactors that are associated with the enzyme. The genome sequence of Dehalococcoides strain 195 suggests that these organisms cannot synthesize the corrinoid ring structure and must salvage them from the environment (31), which in the case of pure cultures must be the medium, but in the case of a mixed culture or the environment, might also be other organisms. Therefore, different cofactors may be associated with the RDases when strains are grown in mixed or pure cultures. Strain CBDB1 was able to degrade pentachlorobenzene and hexachlorobenzene when grown in a mixed culture, but not when grown in isolation (1). This was attributed to the state in which the compound was administered (i.e., in crystal form, as opposed to a hexadecane phase); however, this different substrate range could be due to the availability of different cofactors associated with the RDase(s). Dehalococcoides strain 195 exhibited different rates and extents of dechlorination depending on what amendments were made to the medium. For example, when mixed-culture supernatant was added to the pure culture, TCE was degraded to ethene, but when yeast extract or E. coli extract was added, no degradation beyond cDCE occurred (21). The possibility that the corrinoid cofactors affect the substrate range and specificity of the holoenzyme would explain observations made with mixed and pure cultures, and this hypothesis should be explored further.
Understanding the transcriptional controls of reductive dechlorination is relevant for bioremediation applications. Mixed consortia like KB1 have been used successfully in bioaugmentation field studies (20). The sequence information described in this paper will be useful in designing probes or primers to detect the presence of these genes or transcripts in situ. Dehalogenase genes offer more resolution than 16S rRNA gene-based probes, which cannot distinguish between Dehalococcoides strains with differing dechlorinating activities. The detection of a specific complement of RDH genes may provide more reliable strain identification. Furthermore, the detection of transcription of certain RDH genes may provide quantitative information about the metabolic processes occurring in situ. However, more research is required to determine if the identification of certain transcripts at a site correlates with a particular compound being degraded or if site geochemical parameters such as available corrinoid cofactors modulate substrate degradation. These approaches will lead to the development of refined molecular tools to predict and monitor in situ dechlorination processes.
ADDENDUM IN PROOF
Since the acceptance of this paper, the publication of a study analyzing the genome sequence of Dehalococcoides strain CBDB1 revealed the presence of 32 RDH genes (M. Kube, A. Beck, S. H. Zinder, H. Kuhl, R. Reinhardt, and L. Adrian, Nat. Biotechnol. 23:1269-1273, 2005). This is significantly more than the 14 RDH genes detected by Hölscher et al. using the same degenerate primers that were used to amplify the RDH genes in KB1. However, this does not change the topology of the tree in Fig. 1, nor does it change the conclusions of this paper.
Acknowledgments
This research was supported by grants from the Natural Science and Engineering Research Council of Canada (to E.A.E.) and the U.S. Department of Energy Office of Cleanup Technologies, administered by the Savannah River Operations Office (contract no. DE-AC09-96SR18500) (to E.A.E. and F.E.L.). A.S.W. was supported by an Ontario graduate scholarship.
REFERENCES
- 1.Adrian, L., U. Szewzyk, J. Wecke, and H. Görisch. 2000. Bacterial dehalorespiration with chlorinated benzenes. Nature 408:580-583. [DOI] [PubMed] [Google Scholar]
- 2.Campanella, J. J., L. Bitincks, and J. Smalley. 2003. MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinformatics 4:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christiansen, N., B. K. Ahring, G. Wohlfarth, and G. Diekert. 1998. Purification and characterization of the 3-chloro-4-hydroxy-pheny acetate reductive dehalogenase of Desulfitobacterium hafniense. FEBS Lett. 436:159-162. [DOI] [PubMed] [Google Scholar]
- 4.Cupples, A. M., A. M. Spormann, and P. L. McCarty. 2003. Growth of a Dehalococcoides-like microorganism on vinyl chloride and cis-dichloroethene as electron acceptors, as determined by competitive PCR. Appl. Environ. Microbiol. 69:953-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duhamel, M., K. Mo, and E. A. Edwards. 2004. Characterization of a highly enriched Dehalococcoides-containing culture that grows on vinyl chloride and trichloroethene. Appl. Environ. Microbiol. 70:5538-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duhamel, M., S. Wehr, L. Y. Yu, H. Rizvi, D. Seepersad, S. Dworatzek, E. E. Cox, and E. A. Edwards. 2002. Comparison of anaerobic dechlorinating enrichment cultures maintained on tetrachloroethene, trichloroethene, cis-1,2-dichloroethene and vinyl chloride. Water Res. 36:4193-4202. [DOI] [PubMed] [Google Scholar]
- 7.Edwards, E. A., and D. Grbic-Galic. 1994. Anaerobic degradation of toluene and o-xylene by a methanogenic consortium. Appl. Environ. Microbiol. 60:313-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerritse, J., O. Drzyzga, G. Kloetstra, M. Keijmel, L. P. Wiersum, R. Hutson, M. D. Collins, and J. C. Gottschal. 1999. Influence of different electron donors and accepters on dehalorespiration of tetrachloroethene by Desulfitobacterium frappieri TCE1. Appl. Environ. Microbiol. 65:5212-5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He, J., K. M. Ritalahti, M. R. Aiello, and F. E. Löffler. 2003. Complete detoxification of vinyl chloride by an anaerobic enrichment culture and identification of the reductively dechlorinating population as a Dehalococcoides species. Appl. Environ. Microbiol. 69:996-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He, J., K. M. Ritalahti, K.-L. Yang, S. S. Koenigsberg, and F. E. Löffler. 2003. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424:62-65. [DOI] [PubMed] [Google Scholar]
- 11.He, J., Y. Sung, R. Krajmalnik-Brown, K. M. Ritalahti, and F. E. Löffler. 2005. Isolation and characterization of Dehalococcoides sp. strain FL2, a trichloroethene (TCE)- and 1,2-dichloroethene-respiring anaerobe. Environ. Microbiol. 7:1442-1450. [DOI] [PubMed] [Google Scholar]
- 12.He, Q., and R. A. Sanford. 2002. Induction characteristics of reductive dehalogenation in the ortho-halophenol-respiring bacterium, Anaeromyxobacter dehalogenans. Biodegradation 13:307-316. [DOI] [PubMed] [Google Scholar]
- 13.Henschler, D. 1994. Toxicity of chlorinated organic compounds: effects of the introduction of a chlorine in organic molecules. Angew. Chem. Int. 33:1920-1935. [Google Scholar]
- 14.Hölscher, T., R. Krajmalnik-Brown, K. M. Ritalahti, F. von Wintzingerode, H. Gorisch, F. E. Löffler, and L. Adrian. 2004. Multiple nonidentical reductive-dehalogenase-homologous genes are common in Dehalococcoides. Appl. Environ. Microbiol. 70:5290-5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krajmalnik-Brown, R., T. Hölscher, I. N. Thomson, F. M. Saunders, K. M. Ritalahti, and F. E. Löffler. 2004. Genetic identification of a putative vinyl chloride reductase in Dehalococcoides sp. strain BAV1. Appl. Environ. Microbiol. 70:6347-6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lendvay, J. M., F. E. Löffler, M. Dollhopf, M. R. Aiello, G. Daniels, B. Z. Fathepure, M. Gebhard, R. Heine, R. Helton, J. Shi, R. Krajmalnik-Brown, C. L. Major, Jr., M. J. Barcelona, E. Petrovskis, J. M. Tiedje, and P. Adriaens. 2003. Bioreactive barriers: bioaugmentation and biostimulation for chlorinated solvent remediation. Environ. Sci. Technol. 37:1422-1431. [Google Scholar]
- 17.Löffler, F. E., R. A. Sanford, and J. M. Tiedje. 1996. Initial characterization of a reductive dehalogenase from Desulfitobacterium chlororespirans Co23. Appl. Environ. Microbiol. 62:3809-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magnuson, J. K., M. F. Romine, D. R. Burris, and M. T. Kingsley. 2000. Trichloroethene reductive dehalogenase from Dehalococcoides ethenogenes: sequence of tceA and substrate range characterization. Appl. Environ. Microbiol. 66:5141-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magnuson, J. K., R. V. Stern, J. M. Gossett, S. H. Zinder, and D. R. Burris. 1998. Reductive dechlorination of tetrachloroethene to ethene by a two-component enzyme pathway. Appl. Environ. Microbiol. 64:1270-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Major, D. W., M. L. McMaster, E. E. Cox, E. A. Edwards, S. M. Dworatzek, E. R. Hendrickson, M. G. Starr, J. A. Payne, and L. W. Buonamici. 2002. Field demonstration of successful bioaugmentation to achieve dechlorination of tetrachloroethene to ethene. Environ. Sci. Technol. 36:5106-5116. [DOI] [PubMed] [Google Scholar]
- 21.Maymó-Gatell, X., Y.-T. Chien, J. Gossett, and S. Zinder. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568-1571. [DOI] [PubMed] [Google Scholar]
- 22.Miller, E., G. Wohlfarth, and G. Diekert. 1998. Purification and characterization of the tetrachloroethene reductive dehalogenase of strain PCE-S. Arch. Microbiol. 169:497-502. [DOI] [PubMed] [Google Scholar]
- 23.Müller, J. A., B. M. Rosner, G. Von Abendroth, G. Meshulam-Simon, P. L. McCarty, and A. M. Spormann. 2004. Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distribution. Appl. Environ. Microbiol. 70:4880-4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neumann, A., G. Wohlfarth, and G. Diekert. 1995. Properties of tetrachloroethene and trichloroethene dehalogenase of Dehalospirillum multivorans. Arch. Microbiol. 163:276-281. [Google Scholar]
- 25.Neumann, A., G. Wohlfarth, and G. Diekert. 1996. Purification and characterization of tetrachloroethene reductive dehalogenase from Dehalospirillum multivorans. J. Biol. Chem. 271:16515-16519. [DOI] [PubMed] [Google Scholar]
- 26.Ni, S., J. K. Fredrickson, and L. Xun. 1995. Purification and characterization of a novel 3-chlorobenzoate-reductive dehalogenase from the cytoplasmic membrane of Desulfomonile tiedjei DCB-1. J. Bacteriol. 177:5135-5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okeke, B. C., Y. C. Chang, M. Hatsu, T. Suzuki, and K. Takamizawa. 2001. Purification, cloning and sequencing of an enzyme mediating the reductive dechlorination of tetrachloroethylene (PCE) from Clostridium biferentans DPH-1. Can. J. Microbiol. 47:448-456. [PubMed] [Google Scholar]
- 28.Ritalahti, K. M., and F. E. Löffler. 2004. Populations implicated in the anaerobic reductive dechlorination of 1,2-dichloropropane in highly enriched bacterial communities. Appl. Environ. Microbiol. 70:4088-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schobert, M., and H. Görisch. 2001. A soluble two-component regulatory system controls expression of quinoprotein ethanol dehydrogenase (QEDH) but not expression of cytochrome c550 of the ethanol-oxidation system in Pseudomonas aeruginosa. Microbiology 147:363-372. [DOI] [PubMed] [Google Scholar]
- 30.Schumacher, W., C. Holliger, A. J. B. Zehnder, and W. R. Hagen. 1997. Redox chemistry of cobalamin and iron-sulfur cofactors in the tetrachloroethene reductase of Dehalobacter restrictus. FEBS Lett. 409:421-425. [DOI] [PubMed] [Google Scholar]
- 31.Seshadri, R., L. Adrian, D. E. Fouts, J. A. Eisen, A. M. Phillippy, B. A. Methe, N. L. Ward, W. C. Nelson, R. J. Deboy, S. C. Daugherty, L. M. Brinkac, S. A. Sullivan, R. Madupu, K. E. Nelson, K. H. Kang, M. Impraim, K. Tran, J. M. Robinson, H. A. Forberger, C. M. Fraser, S. H. Zinder, and J. F. Heidelberg. 2005. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307:105-108. [DOI] [PubMed] [Google Scholar]
- 32.Smidt, H., and W. M. de Vos. 2004. Anaerobic microbial dehalogenation. Annu. Rev. Microbiol. 58:43-73. [DOI] [PubMed] [Google Scholar]
- 33.Smidt, H., M. van Leest, J. van der Oost, and W. M. de Vos. 2000. Transcriptional regulation of the cpr gene cluster in ortho-chlorophenol-respiring Desulfitobacterium dehalogenans. J. Bacteriol. 182:5683-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suyama, A., M. Yamashita, S. Yoshino, and K. Furukawa. 2002. Molecular characterization of the PceA reductive dehalogenase of Desulfitobacterium sp. strain Y51. J. Bacteriol. 184:3419-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van de Pas, B. A., J. Gerritse, W. M. de Vos, G. Schraa, and A. J. M. Stams. 2001. Two distinct enzyme systems are responsible for tetrachloroethene and chlorophenol reductive dehalogenation in Desulfitobacterium strain PCE1. Arch. Microbiol. 176:165-169. [DOI] [PubMed] [Google Scholar]
- 38.van de Pas, B. A., H. Smidt, R. W. Hagens, J. van der Oost, G. Schraa, A. J. M. Stams, and W. M. de Vos. 1999. Purification and molecular characterization of ortho-chlorophenol reductive dehalogenase, a key enzyme of halorespiration in Desulfitobacterium dehalogenans. J. Biol. Chem. 274:20287-20292. [DOI] [PubMed] [Google Scholar]
- 39.Villemur, R., M. Saucier, A. Gauthier, and R. Beaudet. 2002. Occurrence of several genes encoding putative reductive dehalogenases in Desulfitobacterium hafniense/frappieri and Dehalococcoides ethenogenes. Can. J. Microbiol. 48:697-706. [DOI] [PubMed] [Google Scholar]