Abstract
Eight Enterobacteriaceae strains that produce zeaxanthin and derivatives of this compound were isolated from a variety of environmental samples. Phylogenetic analysis showed that these strains grouped with different clusters of Erwinia type strains. Four strains representing the phylogenetic diversity were chosen for further characterization, which revealed their genetic diversity as well as their biochemical diversity. The carotenoid synthesis gene clusters cloned from the four strains had three different gene organizations. Two of the gene clusters, those from strains DC416 and DC260, had the classical organization crtEXYIBZ; the gene cluster from DC413 had the rare organization crtE-idi-XYIBZ; and the gene cluster from DC404 had the unique organization crtE-idi-YIBZ. Besides the diversity in genetic organization, these genes also exhibited considerable sequence diversity. On average, they exhibited 60 to 70% identity with each other, as well as with the corresponding genes of the Pantoea type strains. The four different clusters were individually expressed in Escherichia coli, and the two idi-containing clusters gave more than fivefold-higher carotenoid titers than the two clusters lacking idi. Expression of the crtEYIB genes with and without idi confirmed the effect of increasing carotenoid titer by the type II idi gene linked with the carotenoid synthesis gene clusters.
The carotenoids represent one of the most widely distributed and structurally diverse classes of natural pigments, producing light yellow to orange to deep red colors. Eye-catching examples include lycopene from tomatoes, β-carotene from carrots, and lutein from marigolds. In addition to synthesis in photosynthetic organisms, carotenoids are also synthesized in some bacteria and fungi (15). These pigments have important functions in photosynthesis, nutrition, and protection against photooxidative damage. Many studies have reported health benefits of carotenoids, including prevention of cancer (5), enhancement of immune responses (20), and improvement of visual function (6, 11). Currently, carotenoids are used as nutritional supplements, pharmaceuticals, food colorants, and animal feed additives.
Most naturally occurring carotenoids are hydrophobic tetraterpenoids containing a C40 methyl-branched hydrocarbon backbone derived from successive condensation of eight C5 isoprene units. The C5 isoprene unit, isopentenyl pyrophosphate (IPP), can be generated from an acetyl coenzyme A precursor by the mevalonate pathway (21) or from the pyruvate and glyceraldehyde-3-phosphate precursors by the nonmevalonate pathway (14). IPP is isomerized to dimethylallyl pyrophosphate (DMAPP) by IPP isomerase encoded by the idi gene. IPP is then condensed with DMAPP to form the C10 compound geranyl pyrophosphate and elongated to the C15 compound farnesyl pyrophosphate (FPP). FPP is present in both carotenogenic and noncarotenogenic bacteria. The carotenogenic pathway to extend FPP to common C40 carotenoids, such as β-carotene, includes geranylgeranyl pyrophosphate synthase (CrtE), phytoene synthase (CrtB), phytoene dehydrogenase (CrtI), and lycopene cyclase (CrtY). Sometimes additional enzymes, including β-carotene ketolases (CrtW), β-carotene hydroxylases (CrtZ), and zeaxanthin glycosylases (CrtX), carry out subsequent modifications of β-carotene to generate a variety of C40 carotenoids.
Multiple carotenoid synthesis gene clusters have been isolated for Erwinia species (now classified as Pantoea species). Most of these gene clusters (GenBank accession numbers AY166713, D90087, AB076662, and M90698) have the classic gene organization crtEXYIBZ, with the crtEXYIB genes transcribed as an operon and the crtZ gene transcribed in the opposite orientation. In a rare gene cluster from Pantoea agglomerans (GenBank accession number M87280), an unknown gene (ORF6) is present between the crtE and crtX genes. We cloned several new carotenoid synthesis gene clusters from environmental Enterobacteriaceae strains. We discovered more diversity in the carotenoid synthesis gene clusters and demonstrated the function of the unknown gene (identified as idi), which increases the carotenoid titer.
MATERIALS AND METHODS
Strain isolation.
A variety of environmental samples were collected, and eight yellow-pigmented strains were isolated for growth on Luria-Bertani (LB) plates at 30°C. DC404 was isolated from soil collected from a residential garden in Wilmington, Del.; DC409, DC413, and DC414 were isolated from a Florida soil sample; DC416 and DC519 were isolated from tree bark collected in Florida; DC260 was isolated from the surface of a brick on a west-facing wall in a suburb of Wilmington, Del.; and DC266 was isolated from elm tree bark collected in Wilmington, Del. Samples were resuspended and streaked at least twice on LB plates to homogeneity. The 16S rRNA genes of the strains were amplified and sequenced as described previously (4). Fatty acid profiles of a subset of the strains were determined by gas chromatography by Microbial ID, Inc. (Newark, Del.). Biochemical tests were also performed on the strains by Microbial ID, Inc., using the Enterotube II system (Becton Dickinson, Cockeysville, MD).
Carotenoid analysis.
The environmental strains or recombinant Escherichia coli strains were grown in 100 ml LB at 30°C overnight. The cells were harvested by centrifugation at 4,000 × g for 15 min. The cell pellets were extracted twice with 10 ml of an acetone-methanol mixture (50:50, vol/vol), which removed all the pigments in the cell pellets. The solvent was evaporated under nitrogen, and the carotenoids were resuspended in 1 ml of the same solvent. The resulting suspension was filtered with an Acrodisc CR 25-mm syringe filter (Pall Corporation, Ann Arbor, MI) and was analyzed using an Agilent series 1100 LC/MSD (Agilent, Foster City, CA) as described previously (17).
To determine the β-carotene titers, parts of the overnight cultures were used to measure dry cell weight and parts were used to measure carotenoids. Authentic β-carotene from Sigma (St. Louis, MO) was used to construct a standard curve for carotenoid measurement. Zeaxanthin and β-cryptoxanthin standards were purchased from CaroteNature (Lupsingen, Switzerland) for pigment identification.
Cosmid library construction.
A cosmid library of each strain was constructed using a pWEB cosmid cloning kit from Epicenter Technologies (Madison, WI). Genomic DNA was sheared by passing it through a syringe needle. The sheared DNA was end repaired and size selected on a low-melting-point agarose gel. DNA fragments that were approximately 40 kb were purified and ligated into the blunt-ended pWEB cosmid vector. The library was packaged using ultra-high-efficiency MaxPlax lambda packaging extracts and titrated with E. coli EPI100 cells. Approximately 600 cosmid clones were grown in LB with 100 μg/ml ampicillin and screened by color. A positive yellow cosmid clone was sequenced using an EZ-TN<TET-1> kit (Epicenter Technologies). The sequences were assembled with the Sequencher program (Gene Codes Corp., Ann Arbor, MI).
Cloning of β-carotene synthesis gene clusters.
Four β-carotene synthesis plasmids (pDCQ329, pDCQ330, pDCQ331, and pDCQ332) were constructed by amplifying the carotenoid synthesis gene clusters without the crtZ gene from four strains, DC260, DC404, DC416, and DC413. The PCR fragments containing the β-carotene synthesis genes were cloned into the EcoRI site of the pBHR1 vector (MoBiTec GmbH, Goettingen, Germany). Two other β-carotene synthesis plasmids (pDCQ350 and pDCQ380) were also constructed in pBHR1 by SOEing (gene splicing by overlapping extension) PCR of the genes from the DC413 gene cluster. Plasmid pDCQ350 contained the crtE gene linked with the crtYIB genes from DC413, and plasmid pDCQ380 contained the crtE and idi genes linked with the crtYIB genes from DC413.
Nucleotide sequence accession numbers.
The nucleotide sequences of the carotenoid synthesis gene clusters isolated from DC260, DC404, DC416, and DC413 have been deposited in the GenBank database under accession numbers DQ090833 to DQ090836.
RESULTS AND DISCUSSION
Isolation of pigmented strains from the environment.
Eight yellow-pigmented strains were isolated from a variety of environmental samples. Both 16S rRNA gene analysis and fatty acid profile analysis indicated that these strains belong to the family Enterobacteriaceae. The 16S rRNA genes of these strains exhibited more than 97% identity to the 16S rRNA gene sequences of Enterobacteriaceae strains, including Pantoea strains, which are known to produce zeaxanthin and derivatives of this compound. A phylogenetic analysis of 16S rRNA gene sequences of these strains and several Pantoea type strains was performed. The phylogenetic tree (Fig. 1) showed that DC260, DC413, and DC414 were closely related to cluster I of Pantoea species (10), DC409, DC416, and DC519 were closely related to cluster II of Pantoea species, and DC266 and DC404 were more distantly related and grouped neither with the cluster I Pantoea species nor with the cluster II Pantoea species.
FIG. 1.
Phylogenetic relationship based on a comparison of nearly complete 16S rRNA gene sequences of the isolated Enterobacteriaceae strains with the sequences of Erwinia species. The following Erwinia strains were used in the analysis: Erwinia carotovora ATCC 15713 (accession no. U80197), Erwinia stewartii (reclassified as Pantoea stewartii) ATCC 8199 (accession no. U80208), Erwinia uredovora (reclassified as Pantoea ananatis) ATCC 33244 (accession no. U80196), Pantoea ananatis ATCC 19321 (accession no. U80209), Erwinia amylovora ATCC 15580 (accession no. U80195), and Erwinia cypripedii ATCC 29267 (accession no. U80201). Clusters I, II, and III of the Erwinia type strains were based on the classification of Kwon et al. (10). The phylogenetic tree was generated using the average distance method of the ClustalW program (18) and was constructed using TreeView, version 1.6.6 (12). The scale bar indicates the fraction of nucleotide substitutions.
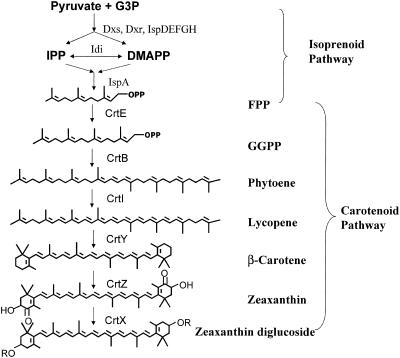
High-performance liquid chromatography (HPLC) analysis of the pigments indicated that all eight strains produced zeaxanthin, β-cryptoxanthin, and β-carotene based on a comparison with authentic standards. Mass spectrometry analysis confirmed that the molecular weight (MH+) of the zeaxanthin peak was 569, the molecular weight of the β-cryptoxanthin peak was 553, and the molecular weight of the β-carotene peak was 537. The peaks that eluted earlier than zeaxanthin were likely zeaxanthin derivatives (e.g., zeaxanthin monoglucoside and zeaxanthin diglucoside), as suggested by liquid chromatography-mass spectrometry. A biochemical pathway for synthesis of zeaxanthin and derivatives of this compound in these Enterobacteriaceae strains is proposed in Fig. 2. The nonmevalonate pathway from pyruvate and glyceraldehyde-3-phosphate precursors was proposed for IPP synthesis in these Enterobacteriaceae strains since the nonmevalonate pathway was reported previously for the closely related Erwinia strains (13).
FIG. 2.
Pathway for synthesis of zeaxanthin diglucoside proposed for the isolated Enterobacteriaceae strains. Multiple steps in the nonmevalonate isoprenoid pathway catalyzed by Dxs, Dxr, IspD, IspE, IspF, IspG, and IspH are not shown in detail. Idi, isopentenyl pyrophosphate isomerase; IspA, fanesyl pyrophosphate synthase; CrtE, geranylgeranyl pyrophosphate synthase; CrtB, phytoene synthase; CrtI, phytoene desaturase; CrtY, lycopene β-cyclase; CrtZ, 3,(3′)-β-ionone hydroxylase; CrtX, zeaxanthin glucosyl transferase. The abbreviations for compounds are as follows: G3P, glyceraldehyde-3-phosphate; IPP, isopentenyl pyrophosphate; DMAPP, dimethylallyl pyrophophate; FPP, farnesyl pyrophosphate; GGPP, geranylgeranyl pyrophosphate.
From the eight zeaxanthin-producing strains, DC413, DC260, DC416, and DC404 were selected for further characterization to represent phylogenetic diversity. A panel of biochemical tests using the Enterotube II system (Table 1) showed that DC413 and DC416 had the same metabolic profile. DC260 differed from these two strains in adonitol fermentation and citrate utilization, and DC404 differed from these two strains in gas production and citrate utilization.
TABLE 1.
Biochemical tests for strains using the Enterotube II systema
| Test | Results
|
|||
|---|---|---|---|---|
| DC260 | DC404 | DC416 | DC413 | |
| Glucoseb | + | + | + | + |
| Gasc | − | + | − | − |
| Lysined | − | − | − | − |
| Ornithinee | − | − | − | − |
| H2Sf | − | − | − | − |
| Indoleg | − | − | − | − |
| Adonitolh | + | − | − | − |
| Lactosei | + | + | + | + |
| Arabinosej | + | + | + | + |
| Sorbitolk | − | − | − | − |
| Dulcitoll | − | − | − | − |
| Phenylalaninem | − | − | − | − |
| Urean | − | − | − | − |
| Citrateo | − | − | + | + |
The Enterotube II system was obtained from Becton Dickinson (catalog number 211832).
Ability to ferment glucose.
Gas production from growth on glucose medium.
Ability to decarboxylate lysine.
Ability to decarboxylate ornithine.
H2S production by reducing sulfur-containing compounds in the medium.
Indole production from metabolism of tryptophan.
Ability to ferment adonitol.
Ability to ferment lactose.
Ability to ferment arabinose.
Ability to ferment sorbitol.
Ability to ferment dulcitol.
Ability to deaminate phenylalanine.
Ability to hydrolyze urea.
Ability to utilize sodium citrate as the sole carbon source.
Comparison of carotenoid synthesis gene clusters.
Cosmid libraries were constructed in E. coli for the four strains selected, and yellow positive E. coli clones were obtained for each of the strains. HPLC analysis showed that the cosmid clones from DC260 and DC413 produced zeaxanthin and glucosides. The cosmid clone from DC416 produced β-carotene and β-cryptoxanthin intermediates in addition to zeaxanthin and glucosides, and the cosmid clone from DC404 produced only zeaxanthin and no zeaxanthin glucosides. The data suggested that the cosmid clones isolated from DC260, DC413, and DC416 contained all the genes necessary for synthesis of zeaxanthin glucosides. The cosmid clone isolated from DC404 contained the genes necessary for synthesis of zeaxanthin but lacked the crtX gene for synthesis of zeaxanthin glucosides. Since the DC404 native strain had the ability to synthesize zeaxanthin glucosides, there is most likely a crtX gene located elsewhere in the chromosome.
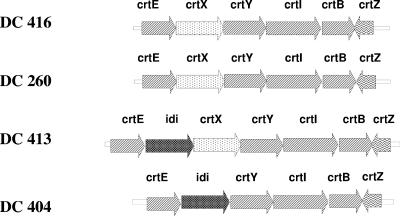
The yellow positive cosmid clones were sequenced, and the carotenoid synthesis gene clusters that were determined from sequence assembly are shown in Fig. 3. Consistent with the HPLC results for the cosmid clones, all the carotenoid synthesis genes for zeaxanthin glucosides (crtEXYIBZ) were present in the clusters cloned from DC260, DC413, and DC416. The DC404 gene cluster contained the crtEYIBZ genes and lacked the crtX gene. In addition, two of the clusters, those from DC413 and DC404, contained an idi gene, encoding isopentenyl pyrophosphate isomerase (7), in the isoprenoid pathway. The organization of the carotenoid synthesis gene cluster also indicated that DC404 was more distantly related to the other strains, as suggested by phylogenetic analysis. DC404 was the first zeaxanthin-producing strain described in which the crtX gene was separated from the carotenoid synthesis gene cluster. The unique organization of the DC404 carotenoid synthesis gene cluster offers advantages for heterologous production of high-value carotenoids, such as astaxanthin. The presence of the isoprenoid idi gene in the cluster should increase the carotenoid titer (see below). The absence of the crtX gene in the cluster should prevent formation of by-products, such as zeaxanthin glucosides.
FIG. 3.
Organization of carotenoid synthesis gene clusters in the Enterobacteriaceae strains isolated. The genes required for β-carotene synthesis are indicated by arrows with diagonal stripes; the crtX genes are indicated by arrows with dots; the crtZ genes are indicated by arrows with wavy lines; and the idi genes are indicated by arrows with grids. The directions of the arrows indicate the directions of transcription of the genes.
Besides the diversity in genetic organization, these genes also exhibited considerable sequence diversity. Table 2 summarizes the pairwise comparison of amino acid identities for the carotenoid synthesis genes from different strains. The carotenoid synthesis genes that we cloned from the environmental Enterobacteriaceae strains exhibited on average 60 to 70% identity with each other, as well as with the corresponding genes from the Pantoea type strains. This was also the case for the isoprenoid idi gene located in the carotenoid synthesis gene cluster. The idi genes from DC404 and DC413 exhibited 70% identity with each other and 66% identity with the ORF6 protein in Pantoea agglomerans. The ORF6 protein was annotated as having an unknown function in the GenBank database (accession number M87280) and was identified in an updated annotation as a type II Idi in the Swiss-Prot database (accession number Q01335). For the strains isolated, the carotenoid synthesis genes from DC416 and DC260 were most similar to each other. Among the different carotenoid synthesis genes, the CrtI gene appeared to be the most conserved gene.
TABLE 2.
Pairwise comparison of the products of carotenoid synthesis genes
| Gene product | Organisma | % Amino acid identity
|
||||||
|---|---|---|---|---|---|---|---|---|
| DC260 | DC416 | DC404 | DC413 | P. ananatis | P. agglomerans | P. stewartii | ||
| CrtE | DC260 | 100 | 88 | 64 | 60 | 58 | 58 | 51 |
| DC416 | 100 | 64 | 61 | 59 | 58 | 50 | ||
| DC404 | 100 | 64 | 63 | 63 | 53 | |||
| DC413 | 100 | 69 | 69 | 51 | ||||
| P. ananatis | 100 | 89 | 52 | |||||
| P. agglomerans | 100 | 52 | ||||||
| P. stewartii | 100 | |||||||
| CrtX | DC260 | 100 | 88 | NAb | 58 | 52 | 49 | 55 |
| DC416 | 100 | NA | 58 | 53 | 50 | 55 | ||
| DC404 | NA | NA | NA | NA | NA | |||
| DC413 | 100 | 62 | 48 | 61 | ||||
| P. ananatis | 100 | 47 | 80 | |||||
| P. agglomerans | 100 | 48 | ||||||
| P. stewartii | 100 | |||||||
| CrtY | DC260 | 100 | 77 | 58 | 58 | 57 | 57 | 58 |
| DC416 | 100 | 60 | 58 | 58 | 59 | 58 | ||
| DC404 | 100 | 56 | 57 | 60 | 58 | |||
| DC413 | 100 | 65 | 57 | 63 | ||||
| P. ananatis | 100 | 56 | 82 | |||||
| P. agglomerans | 100 | 56 | ||||||
| P. stewartii | 100 | |||||||
| CrtI | DC260 | 100 | 91 | 79 | 80 | 81 | 76 | 80 |
| DC416 | 100 | 79 | 81 | 81 | 76 | 82 | ||
| DC404 | 100 | 82 | 80 | 77 | 82 | |||
| DC413 | 100 | 87 | 75 | 88 | ||||
| P. ananatis | 100 | 76 | 94 | |||||
| P. agglomerans | 100 | 76 | ||||||
| P. stewartii | 100 | |||||||
| CrtB | DC260 | 100 | 88 | 67 | 68 | 66 | 64 | 64 |
| DC416 | 100 | 65 | 67 | 64 | 63 | 63 | ||
| DC404 | 100 | 68 | 67 | 61 | 66 | |||
| DC413 | 100 | 77 | 65 | 74 | ||||
| P. ananatis | 100 | 64 | 85 | |||||
| P. agglomerans | 100 | 62 | ||||||
| P. stewartii | 100 | |||||||
| CrtZ | DC260 | 100 | 88 | 66 | 79 | 77 | 67 | 77 |
| DC416 | 100 | 66 | 80 | 78 | 66 | 77 | ||
| DC404 | 100 | 69 | 70 | 65 | 69 | |||
| DC413 | 100 | 76 | 69 | 75 | ||||
| P. ananatis | 100 | 66 | 92 | |||||
| P. agglomerans | 100 | 67 | ||||||
| P. stewartii | 100 | |||||||
Effect of type II Idi on carotenoid production in E. coli.
Two types of isopentenyl pyrophosphate isomerases (Idi) have been characterized so far. The type I Idi from various organisms, including E. coli (7), Saccharomyces cerevisiae (2), and Arabidopsis thaliana (3), requires only divalent metals for activity. In contrast, the nonhomologous type II Idi recently found in Streptomyces (9) and Bacillus subtilis (16) requires flavin mononucleotide, NAD(P)H, and divalent metal ions. The idi genes in some native carotenoid-producing organisms (Phaffia rhodozyma and Hematococcus pluvialis) exhibit homology with the type I idi genes (8). The idi genes identified in the carotenoid synthesis gene clusters from DC413 and DC404 exhibit homology with the type II idi genes. Although insertion of the isoprenoid idi gene into the carotenoid synthesis gene cluster was observed previously in P. agglomerans, many of the crtEXYIBZ gene clusters in Pantoea species (GenBank accession numbers AY166713, D90087, AB076662, and M90698), as well as the clusters from DC260 and DC416, do not contain the idi gene. The idi insertion into the carotenoid synthesis gene cluster might have occurred through a rare event during evolution.
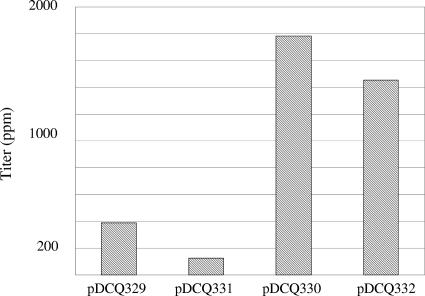
Idi catalyzes the isomerization reaction between IPP and DMAPP, which are needed in equal amounts as substrates for the first step of chain elongation (Fig. 2). This reaction was identified as one of the rate-limiting steps for isoprenoid synthesis. An increase in the enzymatic activity of this reaction by overexpression of idi could relieve this bottleneck. It has been reported that engineering the E. coli native type I idi (1, 19) or expression of an exogenous type I idi (8) could enhance production of carotenoids in E. coli. We compared the four different gene clusters to assess the effect of the type II idi gene on carotenoid production in E. coli (Fig. 4). Four plasmids were constructed from the clusters (Table 3) by expression of all of the genes transcribed in the same orientation. These plasmids, which lacked the crtZ gene transcribed in the opposite orientation, produced β-carotene almost exclusively in E. coli, which facilitated measurement of simple carotenoid titers by condensation of multiple peaks of zeaxanthin and glucosides into a single β-carotene peak. Plasmids pDCQ330 and pDCQ332 expressing the idi-containing gene clusters in E. coli showed high carotenoid titers, approximately 1,500 to 1,800 ppm. Plasmids pDCQ329 and pDCQ331 expressing the gene clusters lacking idi in E. coli showed low carotenoid titers, approximately 150 to 400 ppm. The pigmentation of the colonies also agreed with the titer measurement. The E. coli strains containing pDCQ330 or pDCQ332 were bright yellow, whereas the E. coli strains containing pDCQ329 or pDCQ331 were pale yellow. However, it is interesting that the color of the native host did not correlate with the presence of idi in the gene cluster. For example, DC404 was a lighter yellow than most of the other isolates, although the idi-containing plasmid pDCQ330 derived from DC404 gave the strongest yellow color in E. coli. Differences in host background and carotenoid regulation most likely accounted for the difference in color of the native strains.
FIG. 4.
Comparison of β-carotene titers in E. coli strains containing the four different β-carotene synthesis plasmids. The β-carotene titer (expressed in ppm) indicates the amount of β-carotene (μg per g[dry weight] of cells).
TABLE 3.
Plasmids used in this study
| Plasmid | Genes | Source of genes |
|---|---|---|
| pDCQ329 | crtE crtX crtY crtI crtB | DC260 |
| pDCQ330 | crtE idi crtY crtI crtB | DC404 |
| pDCQ331 | crtE crtX crtY crtI crtB | DC416 |
| pDCQ332 | crtE idi crtX crtY crtI crtB | DC413 |
| pDCQ350 | crtE crtY crtI crtB | DC413 |
| pDCQ380 | crtE idi crtY crtI crtB | DC413 |
To rule out the possibility that the higher-titer E. coli strains with the idi-containing clusters were due to higher activities of the associated crtEXYIB genes, we compared the same crtEYIB genes with and without idi. Plasmids pDCQ350 and pDCQ380 were constructed from the crtEYIB genes from DC413. An E. coli strain with plasmid pDCQ350 lacking idi yielded β-carotene at a level of 536 ± 69 ppm, and an E. coli strain with the idi-containing plasmid pDCQ380 yielded β-carotene at a level of 2,655 ± 20 ppm. This confirmed that the type II idi associated with the crtEXYIB gene cluster could increase the carotenoid titer approximately fivefold in E. coli. The higher titer obtained with pDCQ380 than with pDCQ332 might have been due to more efficient expression of the gene cluster after removal of the crtX gene.
Acknowledgments
We thank the DuPont Macromolecule Analysis Lab for DNA sequencing.
REFERENCES
- 1.Albrecht, M., N. Misawa, and G. Sandmann. 1999. Metabolic engineering of the terpenoid biosynthetic pathway of Escherichia coli for production of the carotenoids β-carotene and zeaxanthin. Biotechnol. Lett. 21:791-795. [Google Scholar]
- 2.Anderson, M. S., M. Muehlbacher, I. P. Street, J. Proffitt, and C. D. Poulter. 1989. Isopentenyl diphosphate:dimethylallyl diphosphate isomerase. An improved purification of the enzyme and isolation of the gene from Saccharomyces cerevisiae. J. Biol. Chem. 264:19169-19175. [PubMed] [Google Scholar]
- 3.Campbell, M., F. M. Hahn, C. D. Poulter, and T. Leustek. 1998. Analysis of the isopentenyl diphosphate isomerase gene family from Arabidopsis thaliana. Plant Mol. Biol. 36:323-328. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, Q., S. M. Thomas, K. Kostichka, J. R. Valentine, and V. Nagarajan. 2000. Genetic analysis of a gene cluster for cyclohexanol oxidation in Acinetobacter sp. strain SE19 by in vitro transposition. J. Bacteriol. 182:4744-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerster, H. 1993. Anticarcinogenic effect of common carotenoids. Int. J. Vitam. Nutr. Res. 63:93-121. [PubMed] [Google Scholar]
- 6.Granado, F., B. Olmedilla, and I. Blanco. 2003. Nutritional and clinical relevance of lutein in human health. Br. J. Nutr. 90:487-502. [DOI] [PubMed] [Google Scholar]
- 7.Hahn, F. M., A. P. Hurlburt, and C. D. Poulter. 1999. Escherichia coli open reading frame 696 is idi, a nonessential gene encoding isopentenyl diphosphate isomerase. J. Bacteriol. 181:4499-4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kajiwara, S., P. D. Fraser, K. Kondo, and N. Misawa. 1997. Expression of an exogenous isopentenyl diphosphate isomerase gene enhances isoprenoid biosynthesis in Escherichia coli. Biochem. J. 324:421-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaneda, K., T. Kuzuyama, M. Takagi, Y. Hayakawa, and H. Seto. 2001. An unusual isopentenyl diphosphate isomerase found in the mevalonate pathway gene cluster from Streptomyces sp. strain CL190. Proc. Natl. Acad. Sci. USA 98:932-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwon, S. W., S. J. Go, H. W. Kang, J. C. Ryu, and J. K. Jo. 1997. Phylogenetic analysis of Erwinia species based on 16S rRNA gene sequences. Int. J. Syst. Bacteriol. 47:1061-1067. [DOI] [PubMed] [Google Scholar]
- 11.Olmedilla, B., F. Granado, I. Blanco, and M. Vaquero. 2003. Lutein, but not alpha-tocopherol, supplementation improves visual function in patients with age-related cataracts: a 2-y double-blind, placebo-controlled pilot study. Nutrition 19:21-24. [DOI] [PubMed] [Google Scholar]
- 12.Page, R. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 13.Putra, S. R., A. Disch, J. M. Bravo, and M. Rohmer. 1998. Distribution of mevalonate and glyceraldehyde 3-phosphate/pyruvate routes for isoprenoid biosynthesis in some gram-negative bacteria and mycobacteria. FEMS Microbiol. Lett. 164:169-175. [DOI] [PubMed] [Google Scholar]
- 14.Rohmer, M., M. Seemann, S. Horbach, S. Bringer-Meyer, and H. Sarm. 1996. Glyceraldehyde-3-phosphate and pyruvate as precursors of isoprenic units in an alternative non-mevalonate pathway for terpenoid biosynthesis. J. Am. Chem. Soc. 118:2564-2566. [Google Scholar]
- 15.Sandmann, G. 1994. Carotenoid biosynthesis in microorganisms and plants. Eur. J. Biochem. 223:7-24. [DOI] [PubMed] [Google Scholar]
- 16.Takagi, M., K. Kaneda, T. Shimizu, Y. Hayakawa, H. Seto, and T. Kuzuyama. 2004. Bacillus subtilis ypgA gene is fni, a nonessential gene encoding type 2 isopentenyl diphosphate isomerase. Biosci. Biotechnol. Biochem. 68:132-137. [DOI] [PubMed] [Google Scholar]
- 17.Tao, L., A. Schenzle, J. M. Odom, and Q. Cheng. 2005. Novel carotenoid oxidase involved in biosynthesis of 4,4′-diapolycopene dialdehyde. Appl. Environ. Microbiol. 71:3294-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang, C. W., M. K. Oh, and J. C. Liao. 1999. Engineered isoprenoid pathway enhances astaxanthin production in Escherichia coli. Biotechnol. Bioeng. 62:235-241. [DOI] [PubMed] [Google Scholar]
- 20.Watzl, B., A. Bub, K. Briviba, and G. Rechkemmer. 2003. Supplementation of a low-carotenoid diet with tomato or carrot juice modulates immune functions in healthy men. Ann. Nutr. Metab. 47:255-261. [DOI] [PubMed] [Google Scholar]
- 21.Wilding, E. I., J. R. Brown, A. P. Bryant, A. F. Chalker, D. J. Holmes, K. A. Ingraham, S. Iordanescu, C. Y. So, M. Rosenberg, and M. N. Gwynn. 2000. Identification, evolution, and essentiality of the mevalonate pathway for isopentenyl diphosphate biosynthesis in gram-positive cocci. J. Bacteriol. 182:4319-4327. [DOI] [PMC free article] [PubMed] [Google Scholar]