Abstract
Rapid detection of infectious viruses is of central importance for public health risk assessment. By directly visualizing newly synthesized viral RNA with molecular beacons (MBs), we have developed a generalized method for the rapid and sensitive detection of infectious viruses from cell culture. An MB, CVB1, specifically targeting the 5′ noncoding region of the enterovirus genome was designed and synthesized. Introduction of MB CVB1 into permeabilized cells highly infected with coxsackievirus B6 resulted in brightly fluorescent cells that can be easily visualized with a fluorescence microscope. In contrast, no detectable signal was observed with noninfected cells or with nonspecific MBs. The number of fluorescent cells also increased in a dose-responsive manner, enabling the direct quantification of infectious viral dosages by direct counting of fluorescent foci. As little as 1 PFU of infectious coxsackievirus B6 was detected within 6 h postinfection. When combined with nuclease-resistant MBs, this method could be useful not only for the real-time detection of infectious viruses but is also useful to study the life cycle of viral processing in vivo.
The ability to detect infectious viruses is of critical importance in medical diagnostic and environmental/agricultural protection (18). Current methods to assess the presence of infectious viruses are based on mammalian cell culture and rely on the production of visible cytopathic effects (CPE) (15). Depending on the specific virus type and the concentration of viruses in the sample, it may take several days or weeks for CPE to appear. The speed of detection has been improved by the use of molecular techniques such as immunoassay or PCR (4). Direct antigen detection by immunofluorescence techniques or enzyme immunoassays has been reported but is often too insensitive for detecting low concentrations of viruses (17). PCR assays based on amplification of either viral DNA or RNA provide improved sensitivity and specificity over immunoassay, but these techniques unfortunately only indicate the presence or absence of viruses in a sample and do not provide any information on infectivity, which is directly related to health risk (6). This is a critical requirement, as many inactivated viruses that carry the required antigens or genomic sequence could persist over a relatively long period in the environment (13). Improved methods that can be used for rapid detection ofinfectious viruses are needed to enable rapid and quantitative determination of their presence for public health risk assessment.
The synthesis of viral RNA/mRNA is a key step in viral replication and could be used as a positive indication of infection (1). Although integrated cell culture-reverse transcription-PCR (RT-PCR) assay has been used to provide rapid detection of infectious viruses (6) by probing the presence of viral mRNA, this method requires additional mRNA extraction, RT-PCRs, and gel analysis, leading to the potential for contamination. Since a large portion of viral mRNA is synthesized early in the infectious cycle (1), in situ detection of viral mRNA directly from cell cultures may be used as a very sensitive and specific indicator for infectious viruses without amplification.
Recently, a new technique known as molecular beacon (MB) was reported for the construction of probes that are useful for real-time detection of nucleic acids (16). These probes are based on single-stranded nucleic acid molecules that possess a stem-and-loop structure. A fluorescent moiety is attached to the end of one arm, and a nonfluorescent quenching moiety is attached to the other end. When the probe encounters a single-strand target, it forms a hybrid with the target, undergoing a spontaneous conformational change that forces the arm sequences apart and causes fluorescence to occur. The interaction of MBs with their targets is extraordinarily specific. No increase in fluorescence is observed even in the presence of a target strand that contains only a single nucleotide mismatch (12). The spontaneous hybridization between molecular beacons and their target sequences has been exploited as a means to monitor intracellular mRNA expression under physiological conditions (8, 14). MBs have been introduced into living cells either by microinjection or as a liposome mixture. In the presence of complementary mRNA, over a 60-fold increase in fluorescence was detected within 15 min, and as few as 10 mRNA molecules could be detected (14). These studies suggest that real-time visualization and localization of mRNA is now possible. Because of the exquisite sensitivity and real-time capability, one possible extension of this technology is to enable real-time detection of virus replication. By targeting the appropriate viral mRNA/RNA, we envision that it is possible to monitor the progress of virus replication using MB.
In this work, we have developed a generalized method for direct visualization of newly synthesized viral RNA as an indication of viral infection. Enteroviruses, which are significant causes of morbidity in the United States (3), with nonpolio enteroviruses such as coxsackieviruses and echoviruses being the second most common cause of viral infections in humans (9), were chosen as the initial target. They belong to an important class of viruses that cause 10 to 15 million cases of symptomatic infection annually in the United States; 25,000 to50,000 of these result in hospitalization for aseptic meningitis alone (3). Coxsackievirus B6 (CV-B6), a member of the Enterovirus genus in the Picornaviridae family that causes aseptic meningitis (9), was used as a model enterovirus. We demonstrate that as little as 1 PFU could be detected within 6 h of infection.
MATERIALS AND METHODS
Cell culture and viruses.
Buffalo green monkey kidney (BGMK) cells were cultured in a growth medium (4.0% autoclavable Eagle's minimal essential medium modified Earles salts [Irvine Scientific, Santa Ana, CA], 8.0 mM HEPES, 0.0075% NaHCO3, 80.0 mM l-glutamine, 10.0 mM minimal essential medium nonessential amino acids [Gibco BBL, Grand Island, NY], 1,000.0 U/ml penicillin, 1,000.0 U/ml streptomycin, 2,000.0 U/ml nystatin, and 2.0 mg/ml kanamycin) containing 8.0% fetal bovine serum (HyClone, Logan, UT). Cells were cultured to confluence in six-well plates (Costar no. 3506; Corning Inc.) for plaque assay analysis and in 16-well chamber slides (LabTek no. 178599; Nalge Nunc International) for MB analysis. Tris-buffered saline solution (0.05 M Tris-HCl [Fisher Biotech, Fair Lawn, NJ], 0.01 M Tris [Fisher Biotech, Fair Lawn, NJ], 0.28 M NaCl, 0.98 mM Na2HPO4, 10.0 mM KCl, pH 7.4) was used for washing in both the plaque assay and MB analysis steps described below.
Coxsackievirus B6 Schmitt strain (ATCC VR-155) stocks were obtained by inoculation onto confluent BGMK cells and incubation for 48 h at 37°C in 4.0% CO2. Viruses were partially purified from infected lysates by chloroform extraction and stored at −80°C until used for experiments. For the infection studies, CV-B6 samples from 10-fold serial dilutions of virus stock in TBSS were distributed on confluent cells in 16-well chamber slides and incubated for 60 min at 37°C in 4.0% CO2. After incubation, the cells were washed twice with TBSS, and 100 ml of medium was applied to the cells in the chamber well. The slide was kept in the incubator at 37°C in 4.0% CO2 until fixation and permeabilization.
Plaque assay.
CV-B6 samples from 10-fold serial dilutions of virus stock in TBSS were distributed on confluent BGMK cells in six-well plates and incubated for 60 min at 37°C in 4.0% CO2. After incubation, the excess solution was removed and the cells were washed with Tris-buffered saline solution. A 1:1 plaque assay medium (4.0% autoclavable Eagle's minimal essential medium modified Earles salts [Irvine Scientific, Santa Ana, CA], 8.0 mM HEPES, 0.0075% NaHCO3, 80.0 mM l-glutamine, 10.0 mM minimal essential medium nonessential amino acids [Gibco BBL, Grand Island, NY], 1,000.0 U/ml penicillin, 1,000.0 U/ml streptomycin, 2,000.0 U/ml nystatin, and 2.0 mg/ml kanamycin)-1.0% agarose overlay was applied at 37°C in 4.0% CO2 for the 48-hour incubation period. Plaques were visualized after overlay removal, fixation of cells with ethanol, and staining with a solution containing 8.0% (wt/vol) crystal violet and 20.0% (vol/vol) 95% ethanol in deionized water.
Molecular beacon synthesis.
A molecular beacon (CVB1) (5′-FAM-GCCGCTCGCATTCAGGGGCCGGAGAGCGGC-DABCYL-3′) was designed to be perfectly complementary to an 18-bp region of the 5′ noncoding region of the enterovirus genome. MB CVB1 was synthesized by Midland Certified Reagent Company (Midland, TX). MBs were resuspended in 10 mM Tris-EDTA buffer, pH 8.0, to a final concentration of 16 mM, stored at −20°C, and used for subsequent studies.
Fixation, permeabilization, and transfection.
At the appropriate times after infection, the slide was removed from the 37°C incubator. After removal of the medium by aspiration, cells were washed three times with TBSS buffer. The cells were then fixed with 2% (wt/vol) paraformaldehyde in TBSS buffer for 30 min at room temperature, washed three times with TBSS buffer, and incubated with 0.1% Triton X-100 in TBSS buffer for 5 min at 4°C for permeabilization. Following three washes with TBSS buffer, cells were incubated with 76 μM of either MB CVB1 or MB CVB1 complexed with a complementary oligonucleotide for 1 h at room temperature in the dark. The slide was subsequently washed three times with TBSS buffer, dried, and observed under a fluorescence microscope.
Fluorescence microscopy and numeration of fluorescent foci.
A BX51TRF fluorescence microscope with UV burner BH2-RFL-T3 and a charge-coupled-device camera (Olympus Optical Co., Ltd., Japan) was used for visualization. Image acquisition and analysis were carried out using the Olympus MicroSuite-B3 software. For each slide, both fluorescent and phase-contrast pictures were taken. Typically, a magnification of ×100 (10× objective lens and 10× eyepiece) was used for enumeration of fluorescent foci, while a magnification of ×400 magnification (40× objective lens and 10× eyepiece) was used for observing fluorescent cells in detail. For each 2-cm-diameter well on the glass slide, approximately 400 different fields could be visualized at a magnification of ×100.
To calculate the percentage of infected cells, the total number of cells or the number of fluorescent cells was counted using phase-contrast or fluorescence microscopy for 20 different fields. The average values were used to calculate the percentages of infected cells. For direct counting of fluorescent foci, 20 fields within the well were randomly chosen and observed at a magnification of ×100. A picture of each area was acquired and recorded. The clusters of fluorescent cells (fluorescent foci) within that area were manually counted and recorded. The numbers for the 20 areas were totaled and averaged.
RESULTS
Design of MB targeting the accessible region of viral RNA.
Enteroviruses are positive-polarity, single-stranded RNA viruses that include important human pathogens such as polioviruses, coxsackieviruses, and echoviruses (9). Similar to the case for other positive single-stranded RNA viruses, the RNA genome is directly translated by the cellular protein synthesis machinery to produce specific viral proteins after entry (1). The viral genomic RNA is then transcribed into a complementary RNA (negative strand), which in turn is used as a template to amplify the genomic RNA (positive strand).
To design an MB suitable for targeting the viral RNAs of a wide range of enteroviruses, a target sequence was selected in a region that was predicted (by MFOLD) (7) to have the least secondary structure and to be the most conserved among 22 different enteroviruses. The unique replication mechanism of enteroviruses allows the flexibility to survey the entire viral RNA genome for potential targets. Based on these criteria, MB CVB1, targeting an 18-bp region of the 5′ noncoding region, was selected.
Visualization of viral RNA in infected BGMK cells.
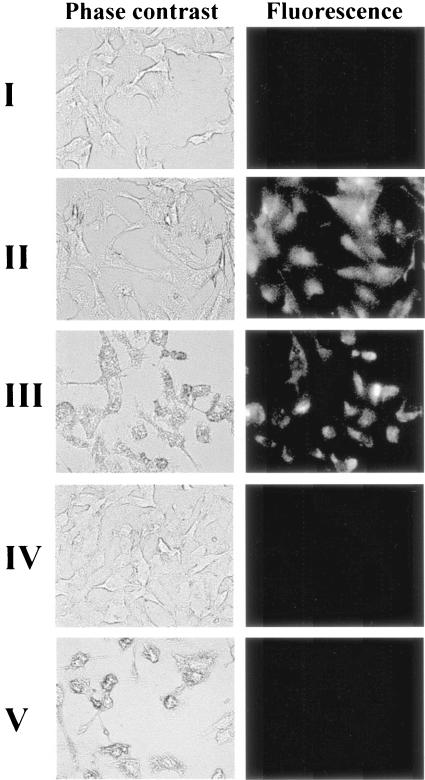
Unlike in other studies visualizing mRNA inside a living cell with MBs (11, 14), microinjection could not be used since it is impossible to predict a priori which particular cells are infected. To allow the introduction of a sufficient quantity of MB inside the entire population of cells, a simple fixation and permeabilization (with paraformaldehyde and Triton X-100) procedure typically used for in situ RT-PCR was adopted (5). The efficiency of the method was first tested by introducing fluorescent CVB1 MB/oligonucleotide hybrids into uninfected cells with or without permeabilization. As shown in Fig. 1, uninfected cells without the permeabilization procedure produced no detectable fluorescent signal (Fig. 1I). In contrast, 100% of permeabilized cells (Fig. 1II) were brightly fluorescent as visualized by phase-contrast and fluorescence microscopy, confirming the utility of the permeabilization procedure to successfully introduce sufficient quantities of MB to every targeted cell.
FIG. 1.
Visualization of infected or uninfected BGMK cells by use of MBs. Cells with or without permeabilization were incubated with 76μM of either MB CVB1/oligonucleotide hybrids (I and II) or MB CVB1 (III to V) for 1 h before being examined by phase-contrast and corresponding fluorescence microscopy. I, uninfected BGMK cells without permeabilization and with MB/oligonucleotide hybrids; II, uninfected BGMK cells with permeabilization and with MB/oligonucleotide hybrids; III, highly infected BGMK cells with permeabilization and with MBs; IV, uninfected BGMK cells with permeabilization and with MBs; V, highly infected BGMK cells with permeabilization and with nonspecific MBs.
To demonstrate the feasibility of visualizing the target viral RNA by MBs inside infected BGMK cells, cells were infected with a high dose (108 PFU) of CV-B6 for 24 h. The ability of the CVB1 MB to target viral RNA was tested by introducing the MB into either permeabilized infected or uninfected cells. High levels of cellular fluorescence were observed in infected cells (Fig. 1III), while no discernible fluorescence was detected in uninfected cells (Fig. 1IV), indicating that hybridization occurred only in the presence of the viral RNA targets and that no degradation of MB occurred during the incubation and detection procedure. Introduction of an unrelated MB containing no homology to the viral RNA genome again resulted in no discernible fluorescence signal from infected cells (Fig. 1V), confirming the specificity of the CVB1 MB.
Visualization of virus infection.
Although we were successful in detecting viral RNA in highly infected cells after 24 h, the challenge was to determine whether this procedure could be used to monitor the progress of virus infection, particularly early in the infection cycle. This is particularly important for the rapid detection of infectious viruses directly from cell culture. To investigate this feasibility, BGMK cells infected with 108 PFU of CV-B6 were visualized at different time points postinfection (p.i.). Since the typical infectious cycle of enteroviruses is around 6 to 7 h, with newly synthesized viral RNA accumulating exponentially during the initial 3 h of infection (10), visualization of viral RNA was conducted from 4 to 24 h p.i. As shown in Fig. 2, brightly fluorescent cells were clearly visible 4 h after infection. The percentage of fluorescent cells increased with increasing infection time, changing from 14% at 4 h to 90% at 24 h (Fig. 3). This was correlated with the progression of virus infection, providing the real-time tracking of virus infection in situ. More importantly, this result demonstrates the possibility of detecting the replication of viral RNA or infectious viruses within 4 h of infection, a time course consistent with the rapid replication cycle of CV-B6.
FIG. 2.
Progression of viral infection. Highly infected cells at various times p.i. were permeabilized and incubated with 76 μM of MB CVB1 before being examined by phase-contrast and corresponding fluorescence microscopy. The number of infected cells increased with infection time, as indicated by the increasing number of fluorescent cells.
FIG. 3.
Percentages of fluorescent (infected) cells at different times p.i. The total number of cells or the number of fluorescent cells was counted from phase-contrast or fluorescence microscopy for 20 different fields. The average values of four replicates were used to calculate the percentages. Error bars indicate standard deviations.
Detection and quantitative determination of infectious CV-B6 from cell cultures.
To test the validity of this method for rapid detection and quantification of infectious viruses, cultures infected with a calculated 1 PFU of CV-B6 were analyzed from 3 to 12 h p.i. in order to investigate the minimal time required to consistently detect fluorescent cells. The ability of the method to detect even a single infectious virus particle is critical, considering the low infectious doses of many enteroviruses. The actual infectious doses added to the cultures were independently confirmed using the plaque assay. Quantification of infection was carried out by direct counting of fluorescent cells with a fluorescence microscope at a magnification of ×100, and the average number of fluorescent foci per field was used as an indicator. As shown in Fig. 4, the number of fluorescent foci decreased rapidly with infection time, and fluorescent foci were barely detectable at 3 h p.i. Although the actual viral dosages used fluctuated from 0 to 4 PFU for the four different sets of experiment, our results consistently demonstrated that even 6 h p.i. was sufficient to detect a distinguishable fluorescent signal from the background, a time frame significantly shorter than the 48-h incubation period for the traditional plaque assay.
FIG. 4.
Visualization of cells infected with 1 PFU by phase-contrast and fluorescence microscopy at various times p.i. Cells infected with 1 PFU were permeabilized and incubated with 76 μM of MB CVB1 for 1 h before being examined by phase-contrast and corresponding fluorescence microscopy. Fluorescent cells were clearly visible within 6 h.
Since the traditional plaque assay can be used to accurately count up to 50 plaques per well, we were interested in whether this assay could provide a similar dynamic range of detection. Using the 6-h infection window, the efficacy of the assay to quantify the infectious dosage of CV-B6 was tested. Cultures were infected with 1 to 30 PFU and the average number of fluorescent foci was plotted as a function of PFU. A linear correlation was obtained over the entire range of interest (Fig. 5), suggesting that this assay may be useful not only as a detection tool but also for quantification of infectious dosages. To test this feasibility, samples infected with unknown quantities of CV-B6, and the average PFU calculated based on the calibration shown in Fig. 5 was compared with the results obtained using the traditional plaque assay. The infectious dosages obtained from the two assays were remarkably similar (Table 1), with a maximum of 25% difference, validating the utility of the assay for viral quantification.
FIG. 5.
Correlation between the average number of fluorescent foci per field and the corresponding infectious viral dosage (PFU) at 6 h p.i. Direct counting of fluorescent foci for 20 fields within the well was done at a magnification of ×100. The clusters of fluorescent cells (fluorescent foci) within that area were manually counted and recorded. The numbers for the 20 areas were totaled and averaged.
TABLE 1.
Comparison of PFU of infectious coxsackieviruses as determined by the traditional plaque assay and the fluorescent-focus assay
| Dosage (dilution) from viral stock | PFU (mean ± SD) from:
|
|
|---|---|---|
| Plaque assay | Fluorescent-focus assay | |
| 1:100 | 27.0 ± 5.0 | 31.0 ± 4.2 |
| 1:1,000 | 2.83 ± 1.50 | 3.57 ± 1.94 |
| 1:10,000 | 0.667 ± 0.150 | 0.616 ± 0.181 |
DISCUSSION
The goal of this study was to investigate the possibility of using MBs as a rapid and sensitive tool to quantify the presence of newly synthesized viral RNA as an indication of virus replication and infection. Introduction of MBs targeting the 5′ noncoding region of CV-B6 into highly infected cultures resulted in brightly fluorescent cells, while only background signals were obtained with noninfected cells or infected cells targeted with an unrelated MB. These results clearly demonstrate the viability of this method to target only viral RNA and that it may be used as a powerful approach to probe invasion by other important viruses.
The presence of brightly fluorescent cells allows simple quantification of infection by the direct counting of fluorescent foci. This was used to monitor the progress of infection, and even cells infected with 1 PFU were detected within 6 h p.i. This rapid detection may be attributed to the sensitivity of the MB method, which has been reported to detect as few as 10 RNA molecules, allowing even early viral replication to be visualized. The number of fluorescent cells also increased in a dose-responsive manner, enabling the direct quantification of infectious viral dosages. With the combination of the eightfold reduction in detection time and the quantitative capability, this direct counting method represents a powerful alternative for the rapid determination of infectious viruses for medical and environmental diagnostics.
Although the reported method does not allow real-time monitoring of viral infection, due to the short in vivo half-life of MBs, nuclease-resistant MBs that are suitable for prolonged probing of intracellular RNA have recently been synthesized (2). Introduction of nuclease-resistant MBs into living infected cells via liposome delivery not only may allow the real-time detection of infectious viruses but also will be useful for study of the life cycle of viral processing in vivo.
Acknowledgments
This work was funded by a grant (R828040) from the U.S. Environmental Protection Agency.
REFERENCES
- 1.Andino, R., N. Boddeker, and A. V. Gamarnik. 1999. Intracellular determinants of picornavirus replication. Trends Microbiol. 7:76-82. [DOI] [PubMed] [Google Scholar]
- 2.Bratu, D., M. M. B.-J. Cha, F. R. Mhlanga, F. R. Kramer, and S. Tyagi. 2003. Visualizing the distribution and transport of mRNAs in living cells. Proc. Natl. Acad. Sci. USA 100:13308-13313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2005. Non-polio enterovirus infections. [Online.] http://www.cdc.gov/ncidod/dvrd/revb/enterovirus/non-polio_entero.htm.
- 4.Elnifro, E. M., A. M. Ashshi, R. J. Cooper, and P. E. Klapper. 2000. Multiplex PCR: optimization and application in diagnostic virology. Clin. Microbiol. Rev. 13:559-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klinger, M. H., R. Kammerer, B. Hornei, and V. Gauss-Muller. 2001. Perinuclear accumulation of hepatitis A virus proteins, RNA, and particles and ultrastructural alterations in infected cells. Arch. Virol. 146:2291-2307. [DOI] [PubMed] [Google Scholar]
- 6.Ko, G., T. L. Cromeans, and M. D. Sobsey. 2003. Detection of infectious adenovirus in cell culture by mRNA reverse transcription-PCR. Appl. Environ. Microbiol. 69:7377-7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 8.Matsuo, T. 1998. In situ visualization of messenger RNA for basic fibroblast growth factor in living cells. Biochim. Biophys. Acta 1379:178-184. [DOI] [PubMed] [Google Scholar]
- 9.Melnick, J. L. 1996. Fields virology. Lippincott-Raven Publishers, Philadelphia, Pa.
- 10.Nugent, C. I., K. L. Johnson, P. Sarnow, and K. Kirkegaard. 1999. Functional coupling between replication and packaging of poliovirus replicon RNA. J. Virol. 73:427-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perlette, J., and W. Tan. 2001. Real-time monitoring of intracellular mRNA hybridization inside single living cells. Anal. Chem. 73:5544-5550. [DOI] [PubMed] [Google Scholar]
- 12.Piatek, A. S., S. Tyagi, A. C. Pol, A. Telenti, L. P. Miller, F. R. Kramer, and D. Alland. 1998. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat. Biotechnol. 16:359-363. [DOI] [PubMed] [Google Scholar]
- 13.Sobsey, M. D., D. A. Battigelli, G. A. Shin, and S. Newland. 1998. RT-PCR amplification detects inactivated viruses in water and wastewater. Water Sci. Technol. 38:91-94. [Google Scholar]
- 14.Sokol, D. L., X. Zhang, P. Lu, and A. M. Gewirz. 1998. Real time detection of DNA RNA hybridization in living cells. Proc. Natl. Acad. Sci. USA 95:11538-11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Storch, G. A. 2000. Essentials of diagnostic virology. Churchill Livingstone, New York, N.Y.
- 16.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303-308. [DOI] [PubMed] [Google Scholar]
- 17.Walpita, P., and S. Darougar. 1989. Double-label immunofluorescence method for simultaneous detection of adenovirus and herpes-simplex virus from the eye. J. Clin. Microbiol. 27:1623-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang, D., L. Coscoy, L., M. Zylberberg, P. C. Avila, H. A. Boushey, D. Ganem, and J. L. DeRisi. 2002. Microarray-based detection and genotyping of viral pathogens. Proc. Natl. Acad. Sci. USA 99:15687-15692. [DOI] [PMC free article] [PubMed] [Google Scholar]