Abstract
The biomass, phylogenetic composition, and photoautotrophic metabolism of green sulfur bacteria in the Black Sea was assessed in situ and in laboratory enrichments. In the center of the western basin, bacteriochlorophyll e (BChl e) was detected between depths of 90 and 120 m and reached maxima of 54 and 68 ng liter−1. High-pressure liquid chromatography analysis revealed a dominance of farnesyl esters and the presence of four unusual geranyl ester homologs of BChl e. Only traces of BChl e (8 ng liter−1) were found at the northwestern slope of the Black Sea basin, where the chemocline was positioned at a significantly greater depth of 140 m. Stable carbon isotope fractionation values of farnesol indicated an autotrophic growth mode of the green sulfur bacteria. For the first time, light intensities in the Black Sea chemocline were determined employing an integrating quantum meter, which yielded maximum values between 0.0022 and 0.00075 μmol quanta m−2 s−1 at the top of the green sulfur bacterial layer around solar noon in December. These values represent by far the lowest values reported for any habitat of photosynthetic organisms. Only one 16S rRNA gene sequence type was detected in the chemocline using PCR primers specific for green sulfur bacteria. This previously unknown phylotype groups with the marine cluster of the Chlorobiaceae and was successfully enriched in a mineral medium containing sulfide, dithionite, and freshly prepared yeast extract. Under precisely controlled laboratory conditions, the enriched green sulfur bacterium proved to be capable of exploiting light intensities as low as 0.015 μmol quanta m−2 s−1 for photosynthetic 14CO2 fixation. Calculated in situ doubling times of the green sulfur bacterium range between 3.1 and 26 years depending on the season, and anoxygenic photosynthesis contributes only 0.002 to 0.01% to total sulfide oxidation in the chemocline. The stable population of green sulfur bacteria in the Black Sea chemocline thus represents the most extremely low-light-adapted and slowest-growing type of phototroph known to date.
The Black Sea represents the largest extant anoxic water body worldwide. Its stratified water column comprises a ∼60-m-thick oxic top layer, a ∼40-m-thick suboxic intermediate zone devoid of sulfide and oxygen, and a ∼2,000-m-deep sulfidic bottom zone (32). Bottom water anoxia was initiated 7,000 to 8,000 years ago by the intrusion of saltwater from the Mediterranean via the Bosporus strait (44). Within the subsequent 3,000 years, the O2-H2S interface rose from the bottom at 2,200 m depth toward the surface (13). The presence of green sulfur bacterial photosynthetic pigments and their degradation products in subfossil sediments suggests that photic zone anoxia occurred already more than 6,000 years ago (41, 47).
Different bacteriochlorophyll e (Bchl e) homologs, as well as the carotenoids isorenieratene and β-isorenieratene, were detected in chemocline samples (43). These photosynthetic pigments are specific for brown-colored species of the green sulfur bacteria. Red autofluorescent cells became visible after fixation of chemocline samples (4). Since this autofluorescence is due to the formation of free bacteriopheophytins from bacteriochlorophylls (52), it was used to quantify green sulfur bacterial cell numbers, yielding a fraction of 10% of the total bacterial cell numbers (corresponding to ≤8 × 104 cells ml−1) (4). Theoretically, the green sulfur bacteria could be metabolically active in the chemocline and thus be relevant for the biogeochemical cycles in the Black Sea (43).
A brown-colored green sulfur bacterium, Chlorobium phaeobacteroides MN1, was previously enriched from chemocline water samples (34). First growth experiments indicated that this bacterium is adapted to low-light conditions and could divide at a light intensity of 0.25 μmol quanta m−2 s−1, but was inhibited at ≥200 μmol quanta m−2 s−1. No data are available on the physiology of this bacterium at lower light intensities. To date, the in situ light conditions in the Black Sea chemocline have not been determined. It is also unknown whether the enriched bacterium is representative for the natural assemblage of green sulfur bacteria present in the chemocline.
In order to quantify anoxygenic photosynthesis in the chemocline and to elucidate its role in the carbon and sulfur cycles of the Black Sea, the in situ light intensities and the photosynthetic metabolism of the dominant phototrophic organisms need to be elucidated under natural conditions. The Black Sea also serves as a model system for past oceanic anoxic events and green sulfur bacterial pigments have been used as indicators for photic zone anoxia (2, 28). A better understanding of the ecophysiology of the low-light-adapted green sulfur bacteria in the Black Sea therefore would also contribute to a more precise reconstruction of the biogeochemical processes in past sulfidic oceans.
MATERIALS AND METHODS
Study sites and sampling procedures.
Water samples were obtained during cruise number 51, leg 51-4, of the R/V Meteor between 12 December and 28 December 2001. Two of the sampling sites were located in the center of the western basin. Station GeoB number 7605 was positioned at a water depth of 2,162 m (42°30.7′N, 30°14.7′E) and station GeoB number 7620 (42°56.2′N, 30°01.9′E) at a depth of 2,006 m. A third station (Station GeoB number 7617; 43°38.9′N, 30°04.1′E) was located at the Bulgarian shelf edge at a water depth of 468 m.
Water samples were obtained by means of an integrated pumpcast system (Seabird Electronics, Bellevue, WA) mounted on a 12-bottle Hydrobios rosette water sampler (29), allowing us to sample the water column at 3-m intervals. Water samples for pigment and DNA analyses were collected in 20-liter polyethylene containers, those for physiological experiments in sterile 1-liter glass bottles. Bacterial cells for the extraction of genomic DNA were collected from 120 liters of water by a Pellicon 2 tangential flow device (Millipore, Bedford, MA) equipped with a VVPP-C-filter (0.1 μm pore size, Millipore), and further concentrated by subsequent filtration onto polycarbonate filters (diameter 47 mm, pore size 0.1 μm, Millipore) in a sterilized filtration unit (Sartorius).
Physicochemical parameters and total cell numbers.
Conductivity, temperature and molecular oxygen concentration were determined with sensors mounted to the Hydrobios water sampler. Underwater light intensities were measured with a LI-190SZ quantum sensor and a LI-200SZ pyranometer sensor, both connected to an LI-1400 data logger (LiCor Biosciences GmbH, Bad Homburg, Germany) and encased in a custom-built stainless steel pressure-proof chamber closed with a Perspex lid. Light meters were calibrated at the sea surface. The equipment was then lowered through the water column down to a depth of 130 m and the integrated light intensity was recorded in vertical intervals of 10 m above the chemocline, and in intervals of 3 m within the chemocline. At each depth, integration lasted for one minute. This method increased the sensitivity of the light measurements by three orders of magnitude, resulting in a detection limit of (3.9 ± 2.3) × 10−4 μmol quanta m−2 s−1.
Sulfide concentrations were determined in water samples of 20 to 50 ml. Samples were directly injected via syringe needles from the pumpcast-system into 100 ml serum bottles preloaded with 0.7 ml zinc acetate (20%, wt/vol) and 0.2 ml sodium hydroxide (4%, wt/vol), kept under a nitrogen atmosphere and sealed with 1-cm-thick butyl rubber stoppers to avoid oxidation. Precipitated sulfide was quantified with the methylene blue method (10).
For the determination of total bacterial cell numbers, samples were fixed in 2% glutaraldehyde and stored in autoclaved 22-ml screw cap glass tubes at 4°C. Total bacterial cell numbers were determined by epifluorescence microscopy on polycarbonate filters (Millipore) after staining with 4′,6-diamidino-2-phenylindol (DAPI) (22).
Analysis of photosynthetic pigments.
Seven liters from each sampling depth were filtered onto GF/F filters (Whatman) and the filters stored in the dark at −20°C. Although GF/F filters have a nominal pore size of 0.7 μm and theoretically may not retain all green sulfur bacterial cells (length, 1.8 ± 0.5 μm; width, 0.65 ± 0.1 μm; see Results section), losses were found to be insignificant due to rapid clogging of the filters by manganese oxides and colloidal organic matter present in the chemocline water. Filters were lyophilized for 6 h and subsequently extracted overnight with 4 ml of a methanol-acetone mixture (2:7, vol/vol) (18). The solvent was evaporated by a nitrogen flow. Total BChl e was quantified photometrically at a wavelength of 650 nm (Lambda 25 spectrophotometer, Perkin Elmer), after redissolving the pigments in acetone, employing the molar extinction coefficient of 48.9 mM−1 cm−1 (58.6 liters g−1 cm−1) (6).
For separation of different homologs, pigments were redissolved in 500 μl of acetone-methanol (1:5, vol/vol; for HPLC analysis); 150 μl of the sample were mixed with 15 μl of a 1 M ammonium acetate solution as ion pairing agent and injected into a Dionex HPLC system equipped with a P580 pump, an STH585 column oven, a PDA-100 photo diode array detector and an RF2000 online fluorescence detector (Dionex Softron). For separation, a Spherisorb ODS2 column (3 μm, 250 mm by 4.6 mm) in-line with a precolumn packed with the same material (CS Chromatographie Service, Langerwehe, Germany) was employed. The mobile phase consisted of a linear gradient of two different mixtures of acetonitrile, methanol, 0.01 M ammonium acetate and ethyl acetate at a flow rate of 0.7 ml min−1 (method B 1). Homologs were identified based on their retention time (18, 42). Fluorometric detection of pigments was carried out at an excitation wavelength of 476 nm and an emission at 676 nm. For standardization, extracts from Chlorobium phaeovibrioides DSMZ260T and Chlorobium phaeobacteroides Dagow III (18) were used.
Lipid analysis.
Particulate organic matter for lipid analyses was collected by in situ filtration of large volumes (∼1,000 liters) of water through 292 mm diameter, precombusted (at 450°C) GF/F filters using in situ pumps. The wet GF/F filters were extracted for 24 h with dichloromethane-methanol (7.5:1, vol/vol) in a Soxhlet extractor to obtain the total lipid extracts. Aliquots of the total extracts were saponified after addition of an internal standard (containing 1-nonadecanol, nonadecanoic acid, 5α-cholestane,and hexatriacontane) with aqueous 0.5 N KOH in methanol (3 h at 80°C). Nonsaponifiable (neutral lipids) and acid fractions were sequentially extracted with hexane at pH 14 and pH 2, respectively.
The neutral fractions were silylated with N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA) in pyridine and analyzed by gas chromatography-mass spectrometry (GC-MS) for identification and quantification of farnesol. Repeated concentration measurements were within ±10%. GC-isotope ratio monitoring MS was performed as described previously (46).
Isotope analysis of DIC.
Water samples for dissolved inorganic carbon (DIC) analysis were treated with 100 μM HgCl2 to inhibit biological activity. δ13C analysis of the total DIC in the Black Sea water column was performed by analysis of the headspace after the addition of 100 μl H3PO4 and an equilibration period of 2 h at 40°C. The headspace was analyzed six times using a Multiflow system connected to an Isoprime isotope ratio monitoring MS system (GV Instruments, Manchester, United Kingdom), with typical standard deviations of 0.1 to 0.2‰. Stable carbon isotope ratios were determined relative to lab standards calibrated on NBS-19 carbonate and CO-8 and reported in the Vienna Pee Dee Belemnite notation.
Enrichment of green sulfur bacteria.
Artificial seawater medium (12) was adjusted to the ionic strength of the Black Sea chemocline. The medium contained (per liter) 14.1 g NaCl, 2.9 g MgCl2 · 6 H2O, 0.6 g CaCl2 · 2 H2O, 0.4 g KCl, 2.3 g Na2SO4, 2.4 g HEPES, 0.2 ml selenite-tungstate solution (58), 100 mg KBr, 24.7 mg H3BO3, 23.8 mg SrCl2, 21.4 mg NH4Cl, 5.4 mg KH2PO4, and 2.9 mg NaF. After autoclaving, 1.4 mM NaHCO3, 1 mM Na2S · 9H2O, 1 ml of trace element solution SL10 (58), 0.1 ml of a solution of 10 vitamins (3), and 200 μM dithionite were added. The pH was adjusted to 7.2 and the medium was distributed into 50-ml screw cap bottles.
Different enrichment conditions were established by combining a variety of electron donors and carbon sources. As sulfur sources, sulfide (final concentration, 1 or 2.5 mM), polysulfides (1 mM S3.252−) (39), or thiosulfate (1 mM) was added. Acetate, pyruvate, glutamate, 2-oxoglutarate (each 1 mM), propionate, glycerol, and malate (each 0.5 mM) served as carbon sources. Yeast extract was freshly prepared (30) and employed at a dilution of 1% (vol/vol). The substrate combinations were also tested in standard saline SL10 medium (58) and in sterile filtered chemocline water.
For inoculation, 200 ml of freshly sampled chemocline water from 95 m depth at station 7620 was filtered through nitrocellulose filters (25 mm, pore size 0.2 μm, Sartorius) and one filter each was added to the different media. All sample manipulations were done in an anaerobic chamber (AtmosBag, Sigma-Aldrich) onboard the research vessel in order to maintain anoxic conditions throughout. Enrichments were incubated at 5 μmol quanta m−2 s−1 and at 15°C. Successful enrichments were used to inoculate deep agar dilution series (40) and liquid dilution series for purification of the cultures.
Absorption spectra of whole cells were recorded after resuspending cell pellets in 50 μl of medium which was then mixed with 500 μl of saturated sucrose solution. Absorption spectra were recorded between 400 and 1100 nm (Lambda 25, Perkin Elmer).
DNA extraction and PCR.
For amplification of 16S rRNA genes from laboratory cultures, cell pellets were resuspended in 10 mM Tris-HCl (pH 8.5) and 0.5 μl of the suspension was added to each PCR. Bacterial cells on filters were extracted according to Fuhrman et al. (14) and the extracts concentrated with Centricon YM-50 centrifugal filter devices (Millipore). DNA concentrations were determined using Pico Green (Molecular Probes).
The 16S rRNA gene fragments and 16S to 23S rRNA intergenic transcribed spacer regions (ITS) of green sulfur bacteria were amplified using primer combinations specific for green sulfur bacteria (8f/GSB822r, GC341f/GSB822r, and GSB822f/L1R) (36) (Tables 1 and 2 in the supplemental material). For the analysis of 16S rRNA gene sequences of chemocline bacteria, the primer combination GC341f/907r was employed (33). Standard conditions for PCR comprised 20 ng DNA or 0.5 μl of a cell suspension, 10 μM of each primer, 5 μl of GeneAMP 10x PCR buffer, 0.2 mM of each deoxynucleoside triphosphate, 3.5 mM MgCl2 and 2.5 U AmpliTaq Gold polymerase (Applied Biosystems) in a total volume of 50 μl. All reactions were run in a GeneAMP 9600 thermocycler PCR system (Applied Biosystems). Amplification products were analyzed by standard agarose gel electrophoresis.
Denaturing gradient gel electrophoresis.
PCR products were loaded onto 6% (wt/vol) polyacrylamide gels in 1x TAE (40 mM Tris-acetate, 1 mM EDTA, pH 7.4) containing a linear gradient of formamide and urea as the denaturant (33). Gels were stained with SybrGold (Molecular Probes) for 45 min, the gel images were captured with a digital camera (Spot RT color, Diagnostic Instruments Inc., MI) and processed with the Spot RT software (version 3.1). Individual DNA bands were excised from the gel and eluted in 40 μl of 10 mM Tris-HCl (pH 8.5) at 65°C for 45 min. For subsequent sequencing 1 μl of the eluate was reamplified. The reamplification products were purified with the QiaQuick PCR purification kit (QIAGEN).
Sequencing and phylogenetic analyses.
For sequencing of 16S rRNA genes, primers 8f, 517r, 341f, 515f, GSB532f, GSB822r, 907r, 926f, 1055r,1492r were used (in the supplemental material Table 1). The ITS region was sequenced with the primers 1525f and L1r (Table 1 in the supplemental material). Cycle sequencing was performed with the AmpliTaq FS BigDye Terminator cycle sequencing kit (Applied Biosystems) following the protocol supplied by the manufacturer. Samples were analyzed on a capillary sequencer (ABI Prism 377 DNA sequencer, Applied Biosystems).
The 16S rRNA gene sequences were analyzed using the ARB phylogeny software package (31). The Fast Aligner V1.03 tool was used for automatic alignment. The resulting alignments were then corrected based on the 16S rRNA secondary structure information for Chlorobium vibrioforme DSMZ 260T, as available through the Comparative RNA Web Site (www.rna.icmb.utexas.edu) (9). Phylogenetic trees were constructed including 16S rRNA gene sequences of available strains and environmental sequences. First, sequences longer than 1100 bp were used for the calculation, employing the maximum likelihood algorithm (Fast DNA_ML). The shorter environmental sequences were inserted afterwards without changing overall tree topology employing the parsimony interactive tool implemented in the ARB software package.
Light dependence of photosynthesis.
Green sulfur bacterial cells were grown to a titer of 108 to 109 cells ml−1 in artificial seawater medium supplemented with 200 μM dithionite and 1 mM malate at 0.1 or 3 μmol quanta m−2 s−1. The protein content of the cultures was determined (21) and the fraction of green sulfur bacteria was determined by fluorescence in situ hybridization, using the green sulfur bacteria-specific Cy3-labeled probe GSB-532 (MWG, Ebersberg, Germany) (52).
Photosynthetic rates were determined in a light cabinet consisting of seven compartments with light intensities adjusted between 0.006 and 11.3 μmol quanta m−2 s−1. The compartments were illuminated from below by two daylight fluorescent tubes (Osram daylight 5000 de luxe, 18 W). Aliquots of the enrichment culture were distributed into 22 ml glass scintillation vials and sealed with screw caps and Teflon-coated butyl rubber septa. Four parallels were incubated at each light intensity. Dark controls consisted of vials coated with two layers of aluminum foil. The average light intensity Ia in the culture vials was calculated from the light intensity at the bottom of the vial (I1) and at its top (I2, measured in a vial without screw cap), according to the equation Ia = (I1 − I2)/ln(I1/I2) (55).
Each vial was supplemented with 370 kBq of [14C]-sodium bicarbonate (Hartmann Analytics, Braunschweig, Germany) and incubated for 6.5 h at 15°C. After incubation, 2- to 4-ml aliquots were filtered onto nitrocellulose filters (pore size 0.22 μm, diameter 23 mm; Sartorius), the filters were transferred to scintillation tubes and inorganic carbonates were removed by acidification with hydrochloric acid. After evaporation of the acid, the filters were air dried before adding 10 ml of scintillation solution (Ultima Gold F; Packard). For the determination of total radioactivity, 500 μl aliquots from each vial were mixed with 10 ml scintillation liquid (Ultima Gold; Packard). Radioactivity was determined in a liquid scintillation counter (Packard Tri-Carb, GMI, Albertville, MN).
As a reference strain, the brown Chlorobium phaeovibrioides DSMZ 269T was used since this strain is the most low-light-adapted strain among all cultured green sulfur bacterial species (34). Cultures were grown at 3 μmol quanta m−2 s−1 in standard saline SL10 medium (34) supplemented with 1 mM acetate.
Nucleotide sequence accession numbers.
The 16S rRNA gene and ITS sequences obtained in this study are available at EMBL under accession numbers AJ972456 and AM039431, respectively.
RESULTS
Environmental conditions and total bacterial cell counts in the Black Sea water column.
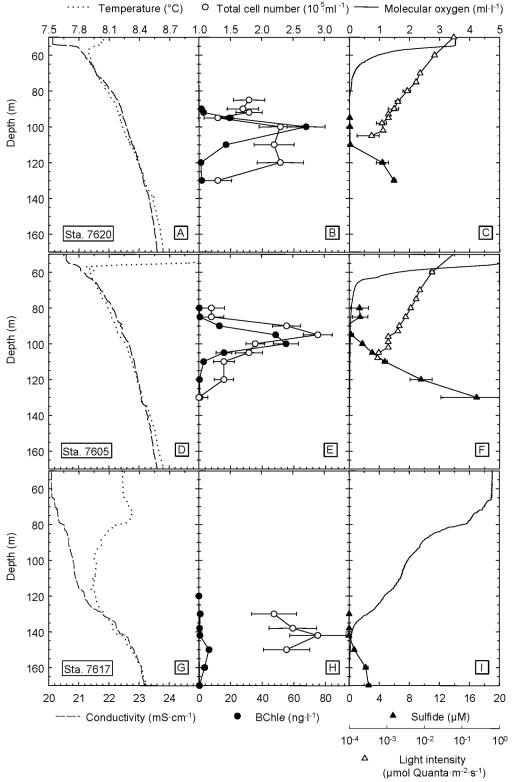
At all three stations, the typical stratification of the Black Sea water column was observed (Fig. 1). According to conductivity and temperature profiles the thermocline and halocline were positioned between 50 and 60 m depth at stations 7605 and 7620 (Fig. 1A and 1D). Anoxic conditions prevailed below 80 m depth and hydrogen sulfide was detected at 100 m and 120 m at stations 7605 and 7620, respectively (Fig. 1C and 1F). The oxic and sulfidic zones were thus separated by a suboxic zone 20 to 30 m thickness. Light intensities decreased exponentially at both stations (Fig. 1C and 1F). The lowest light intensities which were detectable with the integrating quantum meter amounted to 5.7 × 10−4 μmol quanta m−2 s−1 (station 7605) and 3.9 ×10−4 (station 7620) at a depth of 108 and 105 m, respectively.
FIG. 1.
Depth profiles of physicochemical parameters and bacterial biomass in the chemocline of the Black Sea at two stations in the western anoxic basin of the Black Sea (station 7620 and station 7605) and one station at the edge of the northwestern shelf (station 7617). Horizontal bars denote standard deviation.
The chemocline in the Black Sea is bowed, reaching the shallowest depths in the gyre centers. Towards the shelf areas, the discharge of the Danube, Dnjepr and Dnjestr rivers lead to an accumulation of freshwater, causing a deeper position of the chemocline and enhanced vertical mixing (49). Accordingly, the chemocline was located at significantly greater depth at the edge of the northwestern shelf (station 7617) than at the deep-water sites. Temperature and conductivity gradients were less pronounced, and the steepest increase in conductivity was observed at 130 m (Fig. 1G). The oxic and the sulfidic zones overlapped slightly at 140 m depth (Fig. 1I). In contrast to the other two stations, underwater light intensity in the chemocline was below the detection limit.
Maximum bacterial cell numbers were comparable between the three stations, ranging from 2.3 × 105 to 3 · 105 cells ml−1. At stations 7620 and 7605, maximum cell counts were detected at the top of the sulfidic zone at 100 to 120 m depth and at 95 m depth (Fig. 1B and E). At station 7617, comparable cell numbers were determined at the oxic/anoxic interface in 140 m depth (Fig. 1H).
Green sulfur bacteria in the chemocline.
In the center of the western basin, green sulfur bacteria reached their biomass maximum at 100 m depth, where maximum concentrations of BChl e were between 54 and 68 ng liter−1 (Fig. 1B and 1E). The integrated BChl e content of the Black Seawater column was 701 μg BChl e m−2 (station 7605) and 798 μg BChl e m−2 (station 7620). In pronounced contrast, a value of only 8 ng BChl e liter−1 and an integrated amount of BChl e of 113 μg BChl e m−2 were measured at 140 m depth at station 7617 (Fig. 1H).
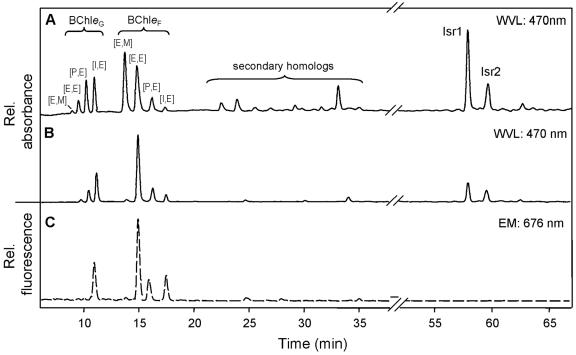
A series of BChl e homologs were detected by HPLC analysis of particulates from the chemocline (Fig. 2C). Dominant homologs were the ethyl/ethyl, propyl/ethyl and isobutyl/ethyl homologs of BChl e farnesyl esters ([E,E]-BChl eF, [P,E]-BChl eF, [I,E]-BChl eF), which together constituted 82.7% of the total BChl e. In addition, considerable amounts of the geranyl ester isobutyl/ethyl [I,E]-BChl eG with its typical very short retention time (42) were detected (Fig. 2C). Only traces of secondary BChl e homologs (mostly ethyl/ethyl-BChl eHen and isobutyl/ethyl-BChl eHen, esterified with hexadecenol) were present.
FIG. 2.
Comparison of high-pressure liquid chromatography traces of photosynthetic pigments extracted from BS-1 enrichment cultures and from a chemocline water sample. A. Pigment extract from a BS-1 culture grown at 3 μmol quanta m−2 s−1. Absorption detected at 470 nm. B. Pigment extract from a BS-1 culture grown at 0.1 μmol quanta m−2 s−1. Absorption detected at 470 nm. C. Pigment extract from a chemocline water sample from 100 m depth (station 7620). Due to the low concentrations in these samples, tetrapyrrol pigments could only be detected by fluorescence. Excitation wavelength was set to 476 nm, emission was recorded at 676 nm. The tentatively identified (see text) geraniol homologs ethyl/methyl [E,M]-BChl eG, ethyl/ethyl [E,E]-BChl eG, propyl/ethyl [P,E]-BChl eG and isobutyl/ethyl [I,E]-BChl eG eluted first. Homologs esterified with farnesol were ethyl/methyl [E,M]-BChl eF, ethyl/ethyl [E,E]-BChl eF, propyl/ethyl [P,E]-BChl eF, isobutyl/ethyl [I,E]-BChl eF. Trace amounts of the secondary homologs (mostly ethyl/ethyl-Bchl eHEN and isobutyl/ethyl-BChl eHEN esterified with hexadecenal) were also detected. Isr1, isorenieratene, Isr2, β-isorenieratene.
Farnesol concentrations in the chemocline reached maxima between 0.90 and 5.89 ng liter−1 in the center of the western basin. δ13C values for farnesol determined at the depth of the maximum farnesol concentrations were −12.1‰ and −11.4‰ (versus VPDB) at stations 7605 and 7620, respectively. The δ13C values of dissolved inorganic carbon (DIC) were −1.3 to −1.4‰ in the chemocline, corresponding to δ13C values of dissolved CO2 of −12.3 to −12.4‰ (45).
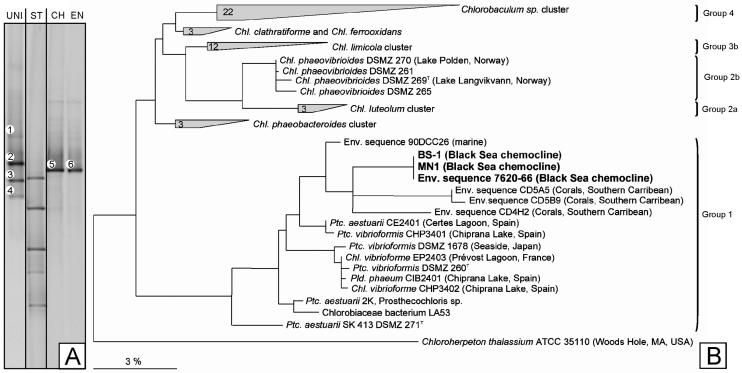
16S rRNA gene fingerprinting of green sulfur bacteria revealed the presence of only one single phylotype (melting type 5, Fig. 3A). The nucleotide sequence of this fragment grouped with the marine cluster of the Chlorobiaceae and was most closely related to environmental clones recovered from various marine habitats (Fig. 3B). Most notably, it was identical to the sequences of the phylotype BS-1 enriched in the present study (see below), and of Chlorobium phaeobacteroides MN1, which had been enriched 13 years earlier from the Black Sea chemocline (34) (Fig. 3B).
FIG. 3.
A. DGGE fingerprinting of the 16S rRNA gene fragment amplified with green sulfur bacterial primers from the chemocline (band 5, lane CH), compared to the fingerprint from BS-1 in the green sulfur bacterial enrichment culture (band 6, lane EN). Bacterial fingerprints amplified with primer pair GC341f/907r are depicted in lane UNI. Melting type 2 corresponded to the chemocline green sulfur bacterium. In addition, 16S rRNA gene sequences of two Cytophaga strains (bands 1 and 3, 97% similarity to GenBank accession number AJ011658, and 96% similarity to AJ296576, respectively) and a relative of Ilyobacter polytrophus DSM 2926T (94% similarity, band 4) were identified. ST DGGE-Standard. B. Phylogenetic position of the green sulfur bacteria BS-1 and MN1 enriched from the chemocline of the Black Sea and of the sequence obtained directly from the chemocline (7620-66, from band 5). Sequences retrieved in the present study given in bold face. For comparison, phylogenetic subgroups as proposed by Imhoff (23) are indicated. Bar denotes fixed substitutions per nucleotide.
Selective enrichment of green sulfur bacteria.
Using chemocline water samples collected at a depth of 100 m at station 7620, the green sulfur bacterium could be selectively enriched. A total of 29 combinations of electron donors and carbon substrates were tested in two different media and in sterile filtered chemocline water. Of all the combinations, only one single enrichment culture was successful, indicating a very low culturability of the cells in natural samples. This particular cultivation medium contained artificial seawater medium supplemented with 2.5 mM sulfide and 1% (vol/vol) of freshly prepared yeast extract solution. In contrast, no enrichment was observed in media containing 1 mM sulfide, 1 mM polysulfides or 1 mM thiosulfate, or a combination of 1 mM thiosulfate plus 1 mM acetate, 1 mM polysulfides plus 1 mM acetate, or in media containing 1 mM acetate, 0.5 mM propionate, 0.5 mM glycerol, 0.5 mM malate or a carbon source mixture (0.5 mM pyruvate, 0.5 mM glutamate, 0.5 mM 2-oxoglutarate). Similarly, fermented rumen extract (1%, vol/vol, of the extract) yielded no enrichment.
Unlike other green sulfur bacteria, the bacterium from the chemocline did never grow in deep agar dilution series. However, the enrichment culture could be partially purified in liquid dilution series. A total of 68 different carbon substrates were tested for stimulation of growth of the green sulfur bacterium. In contrast to the primary enrichment experiments, growth in culture could be stimulated by addition of either malate, acetate, l-aspartate or l-glutamate in the presence of sulfide as electron-donating substrate. Therefore, sulfide was employed as electron donor and 1 mM malate was routinely added as a carbon source for subsequent cultivation and physiological characterization.
DGGE analyses of the enrichment confirmed that the green sulfur bacterium present in the chemocline of the Black Sea had been selectively enriched. When the 16S rRNA gene fragment of the green sulfur bacterium was specifically amplified with primer pair GC341f/GSB822, its melting behavior was identical to that of the phylotype in the chemocline (Fig. 3A, compare bands 5 and 6). Green sulfur bacteria constituted ≥80% of the cells in the culture as determined by fluorescent in situ hybridization using the specific probe GSB-532. In order to identify the accompanying bacteria in the enrichment culture (compare Fig. 4B, arrows), a PCR/DGGE analysis with the eubacterial primer pair GC341f/907r was conducted (Fig. 3A, lane UNI). In addition to the fingerprint of the green sulfur bacterium (band 2) this analysis revealed the presence of three additional phylotypes. One phylotype (band 1) exhibited 97% sequence similarity to an unidentified Cytophaga sp. The second phylotype (band 3) showed 96% similarity to an environmental clone of the Cytophagales. The third additional phylotype (band 4) showed 94% similarity to Ilyobacter polytrophus DSM 2926T (Fusobacteriaceae), a fermentative bacterium specialized in the degradation of hydroaromatic compounds.
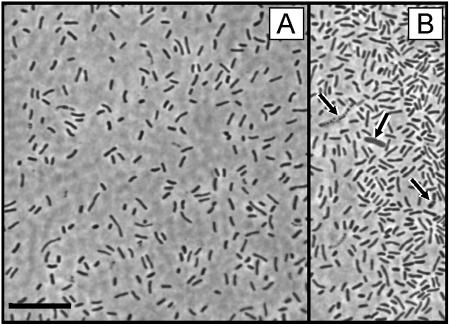
FIG. 4.
A. Phase contrast photomicrograph of the green sulfur bacterium BS-1 in the enrichment culture from the Black Sea chemocline. B. Microscopic field showing three cells of chemotrophic contaminants (arrows).
Employing group specific PCR primers, the almost complete 16S rRNA gene (1388 bp) and the ITS region (468 bp length) of the green sulfur bacterium BS-1 were selectively amplified and sequenced. The 16S rRNA gene and ITS sequences were both identical to the sequences determined in parallel for a conserved sample of Chlorobium phaeobacteroides MN1, which had been enriched from the Black Sea chemocline 13 years earlier (Fig. 3B) (34).
Characterization of the green sulfur bacterium BS-1.
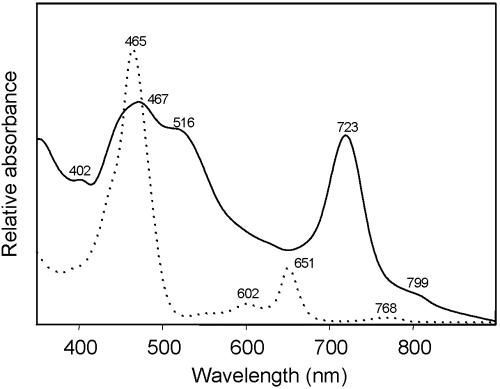
Cells of the enriched green sulfur bacterium were nonmotile short rods, 2.8 ± 1.2 μm long and 0.65 ± 0.1 μm wide when grown at 3 μmol quanta m−2 s−1 (Fig. 4A). After growth at 0.1 μmol quanta m−2 s−1, cells were significantly (P < 0.001) shorter (1.8 ± 0.5 μm). Absorption spectra of whole cells and crude pigment extracts (Fig. 5) were similar to those of other brown-colored green sulfur bacteria (6) and identical to the spectra reported for Chlorobium phaeobacteroides MN1 (34).
FIG. 5.
Absorption spectra of whole cells (—) and acetone pigment extracts (· · · · ·) of the enrichment culture of phylotype BS-1, grown at 0.1 μmol quanta m−2 s−1.
The pigment composition of the cells changed with incubation light intensity (Fig. 2A and B), whereby the BChl e homolog composition of cells grown at 0.1 μmol quanta m−2 s−1 approached that of samples from the chemocline (Fig. 2C). The BChl e farnesyl esters [E,E]-, [P,E]-, and [I,E]-BChl eF amounted to 52 to 64% of the total BChl e and thus represented the dominant homologs. A fourth farnesyl homolog, [E,M]-BChl eF could be detected in cells grown at 3 μmol quanta m−2 s−1 where it constituted 44% of all farnesyl homologs (Fig. 2A). In contrast, the relative amount of [E,M]-BChl eF was significantly decreased in cells grown at 0.1 μmol quanta m−2 s−1 (2.3% of all farnesyl homologs; Fig. 2B), as well as in the chemocline (1.1% of all farnesyl homologs; Fig. 2C).
Similar to the chemocline population, the geranyl ester isobutyl/ethyl [I,E]-BChl eG occurred also in the enrichment cultures. However, two other geranyl homologs were also detected and tentatively identified as ethyl/methyl-, ethyl/ethyl- and propyl/ethyl-BChl eG based on their retention times. Their relative amount was strongly influenced by the incubation light intensity. Cells grown at light saturation (i.e., 3 μmol quanta m−2 s−1) contained [E,E]-BChl eG, [P,E]-BChl eG and [I,E]-BChl eG at a ratio of 17:40:43 (Fig. 2A). This ratio changed towards a strong dominance of [I,E]-BChl eG (6:28:66) in low-light-adapted cells (Fig. 2B).
In order to determine the specific pigment content of the green sulfur bacterial cells, their cellular protein was estimated from the total protein content of the enrichment, and from the fraction of green sulfur bacterial among the cells as determined by FISH. This yielded a specific bacteriochlorophyll content in the enrichment culture of 97.4 ± 36 μg (mg protein)−1 for cells grown at 3 μmol quanta m−2 s−1 and increased values of 224 ± 95 μg (mg protein)−1 for cultures grown at 0.1 μmol quanta m−2 s−1.
Low light threshold of anoxygenic photosynthesis.
Due to the extremely low biomass density of green sulfur bacteria in the chemocline, photosynthetic CO2 fixation could not be determined directly in natural samples (data not shown), which is inconsistent with the results of another recent investigation (19). Our culture contained only one 16S rRNA gene sequence type of green sulfur bacteria and no other phototrophs. Furthermore, photosynthetic pigments other than those of green sulfur bacteria were never detected by HPLC. Therefore the enrichment was used to assess the light dependence of anoxygenic photosynthesis of the green sulfur bacteria by 14CO2 fixation.
The light intensities that were measured at the top of the green sulfur bacterial layer around solar noon were 0.0022 to 0.00075 μmol quanta m−2 s−1 at stations 7605 and 7620, respectively (Fig. 1C and 1F). These values correspond to 0.0009 to 0.0003% of the surface light intensities (236 to 239 μmol quanta m−2 s−1 on the sampling date). It was therefore mandatory to use a light cabinet which permitted the precise control of very low light intensities during the measurement of photosynthetic rates.
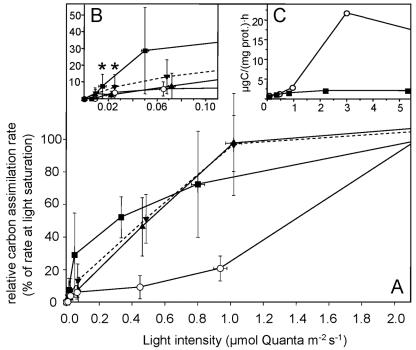
Cells precultured at 3 μmol quanta m−2 s−1 exhibited significant photosynthetic activity at light intensities as low as 0.022 μmol quanta m−2 s−1 (Fig. 6). Cultures grown at 0.1 μmol quanta m−2 s−1 exhibited a different light dependence of photosynthetic CO2-fixation and showed detectable photosynthetic activity at 0.015 μmol quanta m−2 s−1 (7.5% of the maximum rate) (Fig. 6B). However, the radiocarbon method was not suited to determine even lower rates of CO2- incorporation. The green sulfur bacterium from the Black Sea chemocline reached light saturation of photosynthesis at 1 μmol quanta m−2 s−1. In contrast, Chlorobium phaeovibrioides DSMZ 269T showed only marginal (4% of the maximum photosynthetic rate) photosynthetic CO2 incorporation at the low light intensities tested in the present study and reached only 20.7% of the maximum photosynthetic rate at 1 μmol quanta m−2 s−1. Photosynthesis of this strain was light saturated at 3 μmol quanta m−2 s−1 (Fig. 6C). The light-saturated specific photosynthetic rates of the Black Sea bacterium were markedly lower than those of the reference strain DSMZ 269T (Fig. 6C). At the same time, the fraction of growing cells in cultures of the Black Sea bacterium was unusually low (most probable numbers ∼0.1% of total cell counts; U. Henssge and J. Overmann unpublished) compared to cultures of other green sulfur bacteria. Therefore, the low values for light-saturated photosynthetic rates most likely are due to a large number of photosynthetically inactive cells in the culture.
FIG. 6.
Light dependence of [14C]bicarbonate assimilation in the culture of phylotype BS-1 and Chlorobium phaeovibrioides DSMZ 269T. A. Relative assimilation rates at light intensities between 0 and 2 μmol quanta m−2 s−1 given as percentages of the maximum photosynthetic rate at light saturation. Bars represent one standard deviation. B. Expanded view of curves measured below 0.1 μmol quanta m−2 s−1. Asterisks denote values which were significantly higher than dark controls (P = 0.1). C. Comparison of absolute values of biomass-specific photosynthetic rates. ○, Chlorobium phaeovibrioides DSMZ 269T, cells grown at 3 μmol quanta m−2 s−1; ▪, BS-1, grown at 0.1 μmol quanta m−2 s−1; ▾, BS-1, grown at 3 μmol quanta m−2 s−1 (experiment 1); ▴, BS-1, grown at 3 μmol quanta m−2 s−1 (experiment 2).
DISCUSSION
A single phylotype of green sulfur bacteria persists in the Black Sea chemocline.
The biomass of phototrophic sulfur bacteria in the Black Sea (≤0.8 mg BChl e m−2) is orders of magnitude lower than in any other environment studied so far (25 to 2,000 mg BChl e m−2) (54). In a previous study, 16S rRNA genes of green sulfur bacteria could not be detected in clone libraries established after PCR amplification with eubacterial primers (57). Obviously, the limited number of clones tested and the low frequency of green sulfur bacteria prevented their detection by nonspecific molecular methods. The group-specific PCR protocol employed in the present work permits the highly specific amplification of 16S rRNA genes from ≤200 target cells (17) thereby providing the required sensitivity. Only a single green sulfur bacterial phylotype was detected. Comparison of the 16S rRNA gene sequence with all known sequences in the databases revealed that the Black Sea bacterium so far is unique and has not been discovered in any other system.
Based on 16S rRNA gene and ITS sequence comparison, the green sulfur bacterium BS-1 is identical to a green sulfur bacterium enriched 13 years earlier from chemocline samples of the Black Sea (34). The same bacterium was detected again in chemocline samples obtained in May 2004 (R/V Marion Dufresne; A. K. Manske, unpublished). Also, no physiological differences were detected between strains MN1 and BS-1. These data indicate that a single phylotype of green sulfur bacteria continuously persisted in the chemocline of the Black Sea over a time period of at least 16 years. Both the monospecific assemblage and persistence indicate a particular adaptation of BS-1 to the extreme low-light conditions.
Low-light adaptation of BS-1.
The light intensity in the Black Sea chemocline corresponds to that available at a distance of 50 m from a little candle in otherwise total darkness. Compared to other environments (54), phylotype BS-1 thus experiences by far the most severe light limitation ever recorded for any phototrophic organism.
Theoretically, phylotype BS-1 could have acquired an alternative, chemoorganoheterotrophic mode of growth. However, growth of phylotype BS-1 was never observed in cultures incubated in the dark. All known green sulfur bacteria are obligate photolithoautotrophs which use the reverse citric acid cycle for CO2 fixation (48) resulting in green sulfur bacterial biomass which is depleted by 2.5 to 12‰ in 13C relative to CO2 (53). Balancing this depletion, membrane lipids and photosynthetic pigments are enriched in 13C by 1 to 15‰ relative to green sulfur bacterial biomass (16, 53). Based on our HPLC analyses, farnesol represents the dominant lipid biomarker of green sulfur bacteria in the Black Sea chemocline.
The δ13C values of farnesol and dissolved CO2 measured in the present study are nearly identical, suggesting that green sulfur bacteria in the Black Sea chemocline indeed fix CO2 via the reverse citric acid cycle. In conclusion, our data suggest that phylotype BS-1 grows photolithoautrophically in situ. Theoretically, farnesol could also originate from Archaea which have been detected based on their 16S rRNA gene sequences in the chemocline (57). No information on the abundance of archaeal cells in the chemocline is presently available. However, only few archaea and only species which are phylogenetically distant to those occurring in the chemocline of the Black Sea (57) contain very small traces of farnesol (51). It is therefore unlikely that archaeal farnesol contributed to the isotopic signal.
Previous studies indicated that low-light adaptation of the green sulfur bacterium from the Black Sea chemocline includes an increase in the specific BChl e content (15, 34). So far, only cells incubated at ≥0.25 μmol quanta m−2 s−1 had been studied and reached a specific pigment content of ∼220 μg BChl e (mg protein)−1. In the present study, similar values were determined for cells grown at lower light intensities. This specific pigment content of strain BS-1 surpasses that of other green sulfur bacteria (34). The cellular pigment content in our cultures grown at 0.1 μmol quanta m−2 s−1 was 3.1 × 10−5 ng BChl e cell−1. This value is comparable to in situ values of 1.7 to 3.5 × 10−5 ng cell−1 as derived from the pigment concentrations and the numbers of green sulfur bacteria determined by quantitative PCR in the Black Sea chemocline. Thus, the cellular pigment content is not increased further under the severe light limitation in the chemocline, and hence represents the maximum pigment content the cells can attain.
Compared to the results reported for the year 1988 (4, 43), the maximum concentrations of Bchl e determined by us 13 years later were decreased by a factor of 14 (68 ng liter−1 versus 940 ng liter−1). In 1988, green sulfur bacteria amounted to 10% of the total bacterial cell numbers (4), whereas a quantification of the cell number of phylotype BS-1 by real-time PCR indicated that this bacterium constituted only 0.5 to 1% of the total cell number in the chemocline in 2001 (E. Marschall and J. Overmann, unpublished). Both lines of evidence indicate a significant decrease in the population density of the green sulfur bacteria over the last decade, concomitant to the vertical displacement of their biomass maximum from 74 to 100 m. At the northwestern continental slope, the chemocline was located at a depth of 140 m and only traces of BChl e were detected. This pronounced dependence of green sulfur bacterial biomass on the vertical position of the chemocline implies that phylotype BS-1 reaches its lower limits for phototrophic growth between 100 and 140 m in the Black Sea.
Its ability to exploit minute light intensities renders phylotype BS-1 a valuable model system for the study of the molecular basis of low-light adaptation. Based on our data, this adaptation involves changes in pigment composition. Green sulfur bacteria use chlorosomes as light-harvesting structures. The latter contain rod-shaped aggregates of antenna bacteriochlorophylls. Similar to phylotype BS-1, a loss of [E,M]-Bchl eF with a concomitant increase in [I,E]-Bchl eF upon transfer to light limiting conditions has previously been documented for Chlorobium phaeobacteroides strain Dagow III (18) and may represent an adaptative trait to increase the photosynthetic efficiency. The alkyl side chain of porphyrin ring III is directly involved in the aggregation of BChl molecules (56). A high degree of alkylation therefore leads to a red shift of the QY absorption maximum by 7 to 11 nm (5), which has been hypothesized to facilitate the channeling of excitation energy towards the reaction center, thereby causing an increase in energy transfer efficiency of the chlorosomes (5).
A conspicuous feature of the low-light adaptation of phylotype BS-1 is the presence of geranyl homologs of BChl e which have never been described for any isolate of the green sulfur bacteria (5, 8, 18). The cellular concentrations of the geranyl ester homologs ([E,E]-BChl eG and [P,E]-BChl eG) were decreased in cells grown at 0.1 μmol quanta m−2 s−1 and absent in the severely light-limited chemocline population. Within the aggregates, bacteriochlorophylls seem to be arranged in a bilayer with the alcohol tails of bacteriochlorophylls in the inner layer oriented towards the central cavity of the rod (50). Because of spatial constraints, the structure of esterifying alcohols may therefore influence the stability of the rod structure and the function of the chlorosome as a whole (18).
Biogeochemical significance of anoxygenic photosynthesis in the Black Sea.
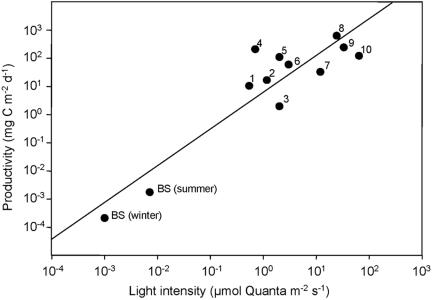
Based on our data, CO2 fixation rates of green sulfur bacteria in the chemocline can be estimated. For the calculation of the daily rate of anoxygenic photosynthesis, global irradiance and average day length in the Black Sea region at summer and winter solstice were taken from the Solar Irradiance Data Utility (http://sunbird.jrc.it/pvgis/sunraddayframe.php). In situ light intensities were then calculated using our light transmission data for the Black Sea water column. Experimentally, photosynthetic CO2 fixation rates could only be determined at ≥0.015 μmol quanta m−2 s−1. Therefore, rates of anoxygenic photosynthesis were calculated for each depth by linear interpolation of laboratory data to in situ light intensities, and considering the biomass of green sulfur bacteria present at each depth. This yielded an integrated anoxygenic photosynthetic rate of 211 ng C m−2 day−1 (corresponding doubling time = 26 years) for a clear December day, and a maximum value of 1,760 ng C m−2 day−1 (doubling time = 3.1 years) during summer solstice. These values are at least a thousandfold lower than the productivity of anoxygenic phototrophic bacteria in any other stratified environment investigated to date (Fig. 7).
FIG. 7.
Cross-system comparison of the integrated productivity of anoxygenic phototrophs in different chemocline environments. BS, data for Black Sea chemocline (this study) estimated for light intensities available in summer and winter; 1, Peter Lake (7, 37); 2, Paul Lake (7, 37); 3, Mary Lake (7, 37); 4, Big Soda Lake (11); 5, Knaack Lake (38); 6, Mirror Lake (7, 37); 7, Rose Lake (7, 37); 8, Lake Cisó (20); 9, Lake Vilar (20); and 10, Mahoney Lake (35). The line depicts a linear regression of all data.
By comparison, the phytoplankton primary production in the center of the Western basin amounts to 575 mg m−2 day−1 (25). Similar to laboratory enrichments, the cultivation success for green sulfur bacterial cells from natural samples was very low, indicating a low fraction of photosynthetically active cells in the chemocline. Even if this fraction was significantly higher in the chemocline than in the cultures used for the determination of photosynthetic activity, integrated anoxygenic photosynthesis would still contribute far less than 1% of total photosynthesis in the Black Sea.
It has been discussed previously (24) that the suboxic zone may be the result of sulfide oxidation by anoxygenic photosynthetic bacteria. Our data can be used to infer the significance of anoxygenic photosynthesis for sulfide turnover in the Black Sea. From the integrated anoxygenic photosynthetic rate, a sulfide oxidation rate of 8.8 to 293.3 nmol H2S m−2 day−1 can be deduced, depending on the oxidation product formed (sulfate or elemental sulfur). By comparison, the sulfide flux from below into the green sulfur bacterial layer is 370 to 530 μmol sulfide m−2 day−1 based on the vertical concentration profiles and the coefficient for turbulent diffusivity (27). Hence, green sulfur bacteria account for ≤0.1% of total sulfide oxidation in the Black Sea chemocline.
The results of modeling the upward fluxes of sulfide suggest that sulfide is actually consumed within the anoxic zone between the chemocline and 150 m depth, but not within the chemocline (27). Most likely molecular oxygen enters the anoxic zone by massive lateral injections of oxygen-enriched Mediterranean waters through the Bosporus plume, and leads to the formation and sedimentation of particulate MnO2 in the chemocline, which in turn serves as the oxidant of ∼70% of the sulfide diffusing upwards (26). Together with molecular diffusion of O2 from upper water layers (accounting for ∼10% of the sulfide oxidation) (27), the fluxes of molecular oxygen thus are sufficient to explain most of the sulfide oxidation in the Black Sea.
In conclusion, the monospecific assemblage of green sulfur bacteria in the Black Sea represents the most extremely low-light-adapted population of phototrophic organisms documented to date. Although not of significance for the carbon and sulfur cycles in the Black Sea, its specific adaptation renders phylotype BS-1 an indicator of marine low light habitats. Based on our results, the ITS sequence of BS-1 is the most promising biomarker which can be used to detect extreme light limitation during photic zone anoxia.
Supplementary Material
Acknowledgments
We thank the master and the crew of the R/V Meteor for their help during sampling. Particular thanks are due to the chief scientist of the cruise, Bo Barker Jørgensen, and our technical assistants, Martina Sterz and Gabi Klockgether. CTD data and sampling facilities were kindly provided by Falk Pollehne (Institut für Ostseeforschung, Warnemünde, Germany). Dissolved inorganic carbon measurements in cultures were performed by Bernhard Schnetger, ICBM Oldenburg. We thank Carsten Schubert (EAWAG, Switzerland) for help with δ13C measurements.
This work was supported by grants Ov 20/7-1, Ov 20/8-1, and Ov 20/8-2 from the Deutsche Forschungsgemeinschaft to J. Overmann.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Airs, R. L., J. E. Atkinson, and B. J. Keely. 2001. Development of a high resolution liquid chromatographic method for the analysis of complex pigment distributions. J. Chromatogr. A. 917:167-177. [DOI] [PubMed] [Google Scholar]
- 2.Anbar, A. D., and A. H. Knoll. 2002. Proterozoic ocean chemistry and evolution: a bioinorganic bridge? Science 297:1137-1142. [DOI] [PubMed] [Google Scholar]
- 3.Balch, W. E., G. E. Fox, L. J. Magrum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bird, D. F., and D. M. Karl. 1991. Microbial biomass and population diversity in the upper water column of the Black Sea. Deep-Sea Res. 38:1069-1082. [Google Scholar]
- 5.Borrego, C. M., and L. J. Garcia-Gil. 1995. Rearrangement of light harvesting bacteriochlorophyll homologs as a response of green sulfur bacteria to low light intensities. Photosynth. Res. 41:53-65. [DOI] [PubMed] [Google Scholar]
- 6.Borrego, C. M., J. B. Arellano, C. A. Abella, T. Gillbro, and L. J. Garcia-Gil. 1999. The molar extinction coefficient of bacteriochlorophyll e and the pigment stoichiometry in Chlorobium phaeobacteroides. Photosynth. Res. 60:257-264. [Google Scholar]
- 7.Borrego, C. M., J. Garcia-Gil, X. P. Cristina, X. Vila, and C. A. Abella. 1998. Occurrence of new bacteriochlorophyll d forms in natural populations of green photosynthetic sulfur bacteria. FEMS Microbiol. Ecol. 26:257-267. [Google Scholar]
- 8.Borrego, C. M., L. J. Garcia-Gil, X. Vila, X. P. Cristina, J. B. Figueras, and C. A. Abella. 1997. Distribution of bacteriochlorophyll homologs in natural populations of brown-colored phototrophic sulfur bacteria. FEMS Microb. Ecol. 24:301-309. [Google Scholar]
- 9.Cannone, J. J., S. Subramanian, M. N. Schnare, J. R. Collett, L. M. D'Souza, Y. Du, B. Feng, N. Lin, L. V. Madabusi, K. M. Muller, N. Pande, Z. Shang, N. Yu, and R. R. Gutell. 2002. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3:2. [Correction: BMC Bioinformatics 3: 15.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cline, J. D. 1969. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol. Oceanogr. 14:454-458. [Google Scholar]
- 11.Cloern, J. E., B. E. Cole, and R. S. Oremland. 1983. Autotrophic processes in meromictic Big Soda Lake, Nevada. Limnol. Oceanogr. 28:1049-1061. [Google Scholar]
- 12.Coolen, M. J. L., and J. Overmann. 2000. Functional exoenzymes as indicators of metabolically active bacteria in 124,000-year-old sapropel layers of the eastern Mediterranean Sea. Appl. Environ. Microbiol. 66:2589-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Degens, E. T., and P. Stoffers. 1976. Stratified waters as a key to the past. Nature 263:22-27. [Google Scholar]
- 14.Fuhrman, J. A., D. E. Comeau, Å. Hagström, and A. M. Chan. 1988. Extraction from natural planktonic microorganisms of DNA suitable for molecular biological studies. Appl. Environ. Microbiol. 54:1426-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuhrmann, S., J. Overmann, N. Pfennig, and U. Fischer. 1993. Influence of vitamin B12 and light on the formation of chlorosomes in green- and brown-colored Chlorobium species. Arch. Microbiol. 160:193-198. [Google Scholar]
- 16.Glaeser, J., and J. Overmann. 2003. Characterization and in situ carbon metabolism of phototrophic consortia. Appl. Environ. Microbiol. 69:3739-3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glaeser, J., and J. Overmann. 2004. Biogeography, evolution and diversity of the epibionts in phototrophic consortia. Appl. Environ. Microbiol. 70:4821-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glaeser, J., L. Bañeras, H. Rütters, and J. Overmann. 2002. Novel bacteriochlorophyll e structures and species-specific variability of pigment composition in green sulfur bacteria. Arch. Microbiol. 177:475-485. [DOI] [PubMed] [Google Scholar]
- 19.Gorlenko, V. M., P. V. Mikheev, I. I. Rusanov, N. V. Pimenov, and M. V. Ivanov. 2005. Ecophysiological properties of photosynthetic bacteria from the Black Sea chemocline zone. Microbiology 74:201-209. [PubMed] [Google Scholar]
- 20.Guerrero, R., E. Montesinos, C. Pedrós-Alió, I. Esteve, J. Mas, H. Van Gemerden, P. A. G. Hofman, and J. F. Bakker. 1985. Phototrophic sulfur bacteria in two Spanish lakes: vertical distribution and limiting factors. Limnol. Oceanogr. 30:919-931. [Google Scholar]
- 21.Hartree, E. F. 1972. Determination of protein: a modification of the LOWRY-method that gives a linear photometric response. Anal. Biochem. 48:422-427. [DOI] [PubMed] [Google Scholar]
- 22.Hobbie, J. E., R. J. Daley, and S. Jaspers. 1977. Use of nucleopore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imhoff, J. F. 2003. Phylogenetic taxonomy of the family Chlorobiaceae on the basis of 16S rRNA and fmo (Fenna-Matthews-Olson protein) gene sequences. Int. J. Syst. Evol. Microbiol. 53:941-951. [DOI] [PubMed] [Google Scholar]
- 24.Jørgensen, B. B., H. Fossing, C. O. Wirsen, and H. W. Jannasch. 1991. Sulfide oxidation in the anoxic Black Sea chemocline. Deep-Sea Res. 38:S1083-S1103. [Google Scholar]
- 25.Karl, D. M., and G. A. Knauer. 1991. Microbial production and particle flux in the upper 350 m of the Black Sea. Deep-Sea Res. 38(Suppl.):S921-S942. [Google Scholar]
- 26.Konovalov, S. K., G. W. Luther, G. E. Friederich, D. B. Nuzzio, B. M. Tebo, J. W. Murray, T. Oguz, B. Glazer, R. E. Trouwborst, B. Clement, K. J. Murray, and A. S. Romanov. 2003. Lateral injection of oxygen with the Bosporus plume -fingers of oxidizing potential in the Black Sea. Limnol. Oceanogr. 48:2369-2376. [Google Scholar]
- 27.Konovalov, S. K., L. I. Ivanov, and A. S. Samodurov. 2001. Fluxes and budgets of sulphide and ammonia in the Black Sea anoxic layer. J. Mar. Syst. 31:203-216. [Google Scholar]
- 28.Koopmans, M. P., J. Köster, H. M.E. van Kaam-Peters, F. Kenig, S. Schouten, W. A. Hartgers, J. W. de Leeuw, and J. S. Sinninghe Damsté. 1996. Diagenetic and catagenic products of isorenieratene: molecular indicators for photic zone anoxia. Geochim. Cosmochim. Acta 60:4467-4496. [Google Scholar]
- 29.Krüger, S., T. Leipe, F. Pollehne, and P. Wlost. 2001. First development of an integrated pumpcast-CTD system. In C. Hemleben, K. Hoernle, B. B. Jørgensen, and W. Roether (ed.), Ostatlantik-Mittelmeer-Schwarzes Meer 2001, Meteor-Berichte 03-1, Leitstelle Meteor, Universität Hamburg, Hamburg, Germany.
- 30.Leadbetter, J. R., T. M. Schmidt, J. R. Graber, and J. A. Breznak. 1999. Acetogenesis from H2 plus CO2 by spirochetes from termite guts. Science 283:686-689. [DOI] [PubMed] [Google Scholar]
- 31.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murray, J. W., H. W. Jannasch, S. Honjo, R. F. Anderson, W. S. Reeburgh, Z. Top, G. E. Friederich, L. A. Codispoti, and E. Izdar. 1989. Unexpected changes in the oxic/anoxic interface in the Black Sea. Nature 338:411-413. [Google Scholar]
- 33.Muyzer, G., S. Hottenträger, A. Teske, and C. Waver. 1995. Denaturing gradient gel electrophoresis of PCR-amplified 16S rDNA -a new molecular approach to analyse the genetic diversity of mixed microbial communities, p. 3.4.4.1-3.4.4.22. In A. D. L. Akkermans, J. D. Van Elsas, and F. J. De Bruijn (ed.) Molecular microbial ecology manual, 2nd ed. Kluwer, Dordrecht, The Netherlands.
- 34.Overmann, J., H. Cypionka, and N. Pfennig. 1992. An extremely low-light-adapted green sulfur bacterium from the Black Sea. Limnol. Oceanogr. 37:150-155. [Google Scholar]
- 35.Overmann, J., J. T. Beatty, K. J. Hall, N. Pfennig, and T. G. Northcote. 1991. Characterization of a dense, purple sulfur bacterial layer in a meromictic salt lake. Limnol. Oceanogr. 36:846-859. [Google Scholar]
- 36.Overmann, J., M. J. L. Coolen, and C. Tuschak. 1999. Specific detection of different phylogenetic groups of chemocline bacteria based on PCR and denaturing gradient gel electrophoresis of 16S rRNA gene fragments. Arch. Microbiol. 172:83-94. [DOI] [PubMed] [Google Scholar]
- 37.Parkin, T. B., and T. D. Brock. 1980. Photosynthetic bacterial production in lakes: the effects of light intensity. Limnol. Oceanogr. 25:711-718. [Google Scholar]
- 38.Parkin, T. B., and T. D. Brock. 1981. Photosynthetic bacterial production and carbon mineralization in a meromictic lake. Arch. Hydrobiol. 91:366-382. [Google Scholar]
- 39.Pfennig, N., and H. Biebl. 1976. Desulfomonas acetoxidans gen. nov. and sp. nov., a new anaerobic, sulfur-reducing, acetate-oxidizing bacterium. Arch. Microbiol. 110:2-12. [DOI] [PubMed] [Google Scholar]
- 40.Pfennig, N. 1978. Rhodocyclus purpureus gen. nov. sp. nov, a ring-shaped, vitamin B12-requiring member of the family Rhodospirillaceae. Int. J. Syst. Bacteriol. 28:283-288. [Google Scholar]
- 41.Repeta, D. J. 1993. A high resolution historical record of Holocene anoxygenic primary production in the Black Sea. Geochim. Cosmochim. Acta 57:4337-4342. [Google Scholar]
- 42.Repeta, D. J., and D. J. Simpson. 1991. The distribution and recycling of chlorophyll, bacteriochlorophyll and carotenoids in the Black Sea. Deep-Sea Res. 38:969-984. [Google Scholar]
- 43.Repeta, D. J., D. J. Simpson, B. B. Jørgensen, and H. W. Jannasch. 1989. Evidence for anoxygenic photosynthesis from the distribution of bacteriochlorophylls in the Black Sea. Nature 342:69-72. [DOI] [PubMed] [Google Scholar]
- 44.Ross, D. A., and E. T. Degens. 1970. Black Sea: recent sedimentary history. Science 170:163-165. [DOI] [PubMed] [Google Scholar]
- 45.Schouten, S., M. Strous, M. M. M. Kuypers, W. I. C. Rijpstra, M. Baas, C. J. Schubert, M. S. M. Jetten, and J. S. Sinninghe Damsté. 2004. Stable carbon isotopic fractionations associated with inorganic carbon fixation by anaerobic ammonium-oxidizing bacteria. Appl. Environ. Microbiol. 70:3785-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schouten, S., W. C. M. Klein-Breteler, P. Blokker, N. Schogt, K. Grice, W. I. C. Rijpstra, M. Baas, and J. S. Sinninghe Damsté. 1998. Biosynthetic effects on the stable isotopic compositions of algal lipids: implications for deciphering the carbon isotopic biomarker record. Geochim. Cosmochim. Acta 62:1397-1406. [Google Scholar]
- 47.Sinninghe Damsté, J. S., Wakeham, S. G., M. E. L. Kohnen, Hayes, J. M., and J. W. de Leeuw. 1993. A 6,000-year sedimentary molecular record of chemocline excursions in the Black Sea. Nature 362:827-829. [DOI] [PubMed] [Google Scholar]
- 48.Sirevåg, R. 1995. Carbon metabolism in green bacteria, p. 871-883. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Advances in photosynthesis, vol. II. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 49.Spencer, D. W., P. G. Brewer, and P. L. Sachs. 1972. Aspects of the distribution and trace element composition of suspended matter in the Black Sea. Geochim. Cosmochim. Acta 36:71-86. [Google Scholar]
- 50.Steensgaard, D. B., H. Wackerbarth, P. Hildebrandt, and A. R. Holzwarth. 2000. Diastereoselective control of bacteriochlorophyll e aggregation. 31-S-BChl e is essential for the formation of chlorosome-like aggregates. J. Phys. Chem. B. 104:10379-10386. [Google Scholar]
- 51.Tornabene, T. G., T. A. Langworthy, G. Holzer, and J. Oró. 1979. Squalenes, phytanes and other isoprenoids as major neutral lipids of methanogenic and thermoacidophilic “Archaebacteria”. J. Mol. Evol. 13:73-83. [DOI] [PubMed] [Google Scholar]
- 52.Tuschak, C., J. Glaeser, and J. Overmann. 1999. Specific detection of green sulfur bacteria by in situ hybridization with a fluorescently labeled oligonucleotide probe. Arch. Microbiol. 171:265-272. [DOI] [PubMed] [Google Scholar]
- 53.van der Meer, M. T. J., S. Schouten, and J. S. Sinninghe Damsté. 1998. The effect on the reversed tricarboxylic acid cycle on the 13C contents of bacterial lipids. Org. Geochem. 28:527-533. [Google Scholar]
- 54.van Gemerden, H., and J. Mas. 1995. Ecology of phototrophic sulfur bacteria, p. 49-85. In M. T. Madigan and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 55.van Liere, E., and A. E. Walsby. 1982. Interactions of cyanobacteria with light, p. 9-45. In N. G. Carr and B. A. Whitton (ed.), The biology of cyanobacteria. Blackwell Scientific Publications, Oxford, England.
- 56.van Rossum B-J., D. B. Steensgard, F. M. Mulder, G. J. Boender, K. Schaffner, A. R. Holzwarth, and H. J. M. de Groot. 2001. A refined model of the chlorosomal antennae of the green sulfur bacterium Chlorobium tepidum from proton chemical shift constraints obtained with high-field 2-D and 3-D MAS NMR dipolar correlation spectroscopy. Biochemistry 40:1587-1595. [DOI] [PubMed] [Google Scholar]
- 57.Vetriani, C., H. V. Tran, and L. J. Kerkhof. 2003. Fingerprinting microbial assemblages from the oxic/anoxic chemocline of the Black Sea. Appl. Environ. Microbiol. 69:6481-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Widdel, F., G. W. Kohring, and F. Mayer. 1983. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. III. Characterization of the filamentous gliding Desulfonema limicola gen. nov., sp. nov., and Desulfonema magnum sp. nov. Arch. Microbiol. 134:286-293. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.