Abstract
The origins and the evolutionary history of tetrahydromethanopterin-linked C1 transfer reactions that are part of two environmentally important biotransformations, methylotrophy and methanogenesis, are still not well understood. In previous studies, we have expanded the known phylogenetic diversity of these reactions by identifying genes highly diverging from the ones associated with cultivated Proteobacteria, Planctomycetes, or Archaea (M. G. Kalyuzhnaya, M. E. Lidstrom, and L. Chistoserdova, Microb. Ecol. 48:463-472, 2004; M. G. Kalyuzhnaya, O. Nercessian, M. E. Lidstrom, and L. Chistoserdova, Environ. Microbiol. 7:1269-1274, 2005). Here we used a metagenomic approach to demonstrate that these divergent genes are present with high abundance in the microbial community inhabiting Lake Washington sediment. We also gained preliminary insights into the genomic composition of the organisms possessing these genes by sequencing genomic fragments from three uncultured microbes possessing the genes of interest. Phylogenetic analyses suggested that, although distantly related to each other, these organisms deeply diverge from known Bacteria and Archaea, with more relation to the former, suggesting their affiliation with a new bacterial phylum. We also demonstrate, via specific mRNA detection, that these divergent genes are expressed in the environment, pointing toward their potential role in local carbon cycling.
The top layer of Lake Washington sediment is a habitat in which steep gradients of methane and oxygen occur (1, 14). This habitat has served as a model for studying microbial populations involved in bacterially mediated methane oxidation in pristine freshwater environments, using both culture-based and culture-independent approaches (1-3, 5, 6). Recently, we have used a novel set of environmental detection tools in this site, targeting reactions linked to tetrahydromethanopterin (H4MPT; encoded by fae, mtdB, mch, and fhcD), thus targeting the C1 transfer capacity of natural populations in a broader sense (11, 13). Using these tools, we have uncovered not only sequences belonging to known methanotrophs, but also a variety of sequences with phylogenetic positions similar to the ones of proteobacterial methylotrophs not capable of methane oxidation, as well as to the ones of Proteobacteria and Planctomycetes not thus far associated with methylotrophy (11, 13). In addition, sequences of fae, mtdB, and fhcD have been recovered that were deeply divergent from the ones associated with any known organisms, pointing toward the existence of phylogenetically divergent organisms capable of H4MPT-linked C1 transfer in the site. These sequences potentially representing a new group of microbes using H4MPT as a C1 transfer cofactor have been of great interest, since these may provide both a better understanding of the nature of microbes involved in environmental C1 cycling and a means for improved phylogeny of genes and proteins involved in H4MPT-linked reactions. The microbes possessing these divergent genes remain unknown and uncultured. However, we observed enrichment for those divergent sequences in microcosms exposed to formaldehyde, suggesting that the microbes possessing these sequences may be involved in formaldehyde metabolism (13, 16). We have also overexpressed two of the divergent homologs of mtdA/mtdB genes in Escherichia coli and demonstrated that these genes, entitled mtdC, encode novel methylene-H4MPT dehydrogenases whose substrate range and specificity differ from those of either MtdA or MtdB (20). Thus far, the diversity and the representation of these divergent sequences in Lake Washington sediment has been characterized by analyzing PCR-amplified gene libraries (11, 13, 16). In these libraries, representation and diversity of the novel genes differed between fae, mtdA/B, and fhcD, possibly due to primer and/or cloning biases, whereas no deeply diverging sequences have been detected in the mch library. It also remained unclear whether the divergent fae, mtdA/B, and fhcD belonged to the genomes of the same species. We used a metagenomic approach to obtain further insights into the occurrence, frequency, and diversity of the novel H4MPT-linked C1 transfer genes in Lake Washington and to provide the first glimpses into the genomes of the organisms possessing these genes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Vector pCR2.1 (Invitrogen) was used for cloning PCR fragments, and these were propagated in E. coli Top10 (Invitrogen), as directed by the manufacturer. A fosmid vector pCC1FOS (Epicenter) was used for large DNA insert library construction, and these were propagated in E. coli EPI300-1 (Epicenter) as directed by the manufacturer.
Sediment sample collection.
Sediment samples were collected on 24 July 2003 and on 11 January 2005 from a 63-m deep station in Lake Washington, Seattle, Wash. (47°38.075′N, 122°15.993′W), as described previously (11). The former sample was used for constructing fosmid libraries, and the latter was used for RNA extraction.
Metagenome library construction.
DNA isolation from the sediment was carried out as described before (11, 13). The DNA was electrophoresed in 1% low-melting-point agarose (Invitrogen), and the fraction representing fragments of approximately 40 kb was excised from the gel and recovered using agarase, as directed by the manufacturer (Fermentas). The resulting DNA fragments were cloned into the CopyControl pCC1FOS vector (Epicenter) as instructed by the manufacturer. The ligated DNA was packaged into MaxPlax lambda (Epicenter) and transfected into the EPI300-T1 plating cells, as instructed by the manufacturer. Fosmid-containing E. coli colonies were selected on Luria-Bertani (LB) solid medium supplemented with chloramphenicol (15 μg/ml). The packaging extract was titered and appropriately diluted to yield 1,000 E. coli colonies per plate. Library 1 was constructed by pooling approximately 36,000 colonies resulting from multiple transfections, as described below. To construct library 2, a total of 36 separate transfections were performed, resulting in a metagenomic library consisting of 36,000 clones distributed between 36 separate plates (1,000 colonies per plate). Colonies from each of the 36 plates were washed off with LB medium as pools, and cells were precipitated and resuspended in a minimal medium (9). Each pool was divided into two aliquots, one of which was frozen after adding 10% (vol/vol) dimethyl sulfoxide and stored at −80°C, while the second was used for DNA extraction using the QIAGEN Miniprep kit, as instructed by the manufacturer.
DGGE analysis.
Fragments of the small subunit rRNA gene of approximately 195 and 585 bp, respectively, were PCR amplified from each of the 36 pools using the 341fGC/536r and 341fGC/926r primer pairs (21). PCR amplifications were carried out under the following conditions: 95°C for 3 min, followed by 25 cycles of 95°C, 55°C for 40 s, and 72°C for 1.5 min, with a final extension for 10 min. Denaturing gradient gel electrophoresis (DGGE) was performed using the DGene system (Bio-Rad). Aliquots (20 μl) of the PCR products were loaded onto 10% acrylamide gel (37.5:1; Bio-Rad) containing a linear gradient of formamide-urea from 30 to 60%. Three reference samples of 16S rRNA gene fragments amplified from E. coli EPI300 were included per each gel. Gels were run for 15 h at 60 V in 0.5× TAE electrophoresis buffer (18). After electrophoresis, gels were soaked in ethidium bromide solution for 30 min, illuminated in UV, and manually analyzed.
Analysis of fae and fhcD genes in the metagenomic library.
DNA preparations isolated from each of the 36 pools of library 2 were used as templates to PCR amplify fae and fhcD genes. Details of the amplification protocols have been described previously (11, 13). The resulting PCR products were cloned into the pCR2.1 vector, and three to five randomly selected clones from each cloning were sequenced by using the M13F primer and the sequencing kit BigDye3.1 (Applied Biosystems) according to the manufacturer's instructions. Sequence analysis was carried out by the Department of Biochemistry DNA sequencing facility at the University of Washington, using an ABI 3700 high-throughput capillary DNA analyzer.
DNA-DNA hybridization.
Library 1 and pool 10 of library 2 were appropriately diluted and plated onto solid media to produce single colonies. Clones were manually arrayed on nylon filters, and the filters were treated as previously described (18). DNA probes were labeled with dCT32P by using the Random Primed DNA Labeling Kit (Roche). Hybridizations were carried out overnight at 45°C in 30 ml of hybridization buffer (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 5× Denhardt solution, 20% formamide, 0.1% sodium dodecyl sulfate) containing 50 μl of the labeled probe. Filters were then washed in 0.5× SSC-0.1% sodium dodecyl sulfate buffer three times for 15 min at 50°C and then dried and exposed to X-ray film (Kodak). Clones identified as positive were inoculated into 100 ml of liquid LB medium, grown to early exponential phase (A600 = 0.4), and induced by 1 ml of the CopyControl induction solution (Epicenter) for 5 h. Fosmid DNA was extracted by alkaline lysis and ethanol precipitation, as previously described (18). DNA was resuspended in 0.3 ml of H2O and further purified by using the QIAGEN PCR purification kit according to the manufacturer's instructions.
RNA extraction and RT-PCR amplification.
RNA from the sediment sample was extracted as described previously (16), with the following modification. An additional purification step was carried out after the DNase I digestion step, using the RNeasy columns (QIAGEN). Reverse transcription-PCR (RT-PCR) amplifications were carried out by using the One-Step RT-PCR kit (QIAGEN) and 0.2 μg of RNA. Reaction mixtures were incubated at 60°C (fhcD and fae) or 55°C (mch) for 2.5 h, followed by 15 min of denaturation at 96°C and then 45 cycles of 95°C for 40 s, 60° or 55°C for 40 s, and 72°C for 1.5 min, with a final extension at 72°C for 10 min. The resulting PCR products were cloned into the pCR2.1 vector, and 20 randomly selected clones from each cloning were sequenced by using the M13F primer and the BigDye3.1 kit. To increase specificity of RT-PCR amplifications targeting divergent fhcD, mch, and fae genes, new primer sets were designed as follows. All available sequences of divergent fhcD, mch, and fae were aligned by using the AlignX program of the VectorNTI package (Invitrogen). Regions of high conservation were identified and the following primers were designed for RT-PCR amplification: fae, fae-NGf (5′-CACACATCGACCTGATCATSGG-3′), and fae-NGr (5′-GGATGAAVACGCCGACCAGGA-3′); fhcD, fhcD-NG58f (GAGGCYTTCGACATGCGSGCGG-3′), and fhcD-NG895r (5′-GGAAGTGGTGCTTSCCGAG-3′); and mch, mch-NG422f (5′-GGCCTCSCAGTACGCCGGCTGGG), and mch-NG1000r (5′-GGGATCGAYCTTRTAGAAGTC-3′). The new primer sets were tested on fosmid DNA pools, as well as on total DNA isolated from the Lake Washington sediment, with positive results (data not shown). As negative controls, DNA samples isolated from cultured proteobacteria were used, with negative results (data not shown).
Fosmid insert sequencing and sequence annotation.
Three chosen fosmid inserts were entirely sequenced by primer walking using the BigDye3.1 kit. Sequence assembly and editing were performed by using the VectorNTI software package. Open reading frame (ORF) identification and gene annotation were performed by using the BLAST programs (National Center for Biotechnology Information). Translated protein sequences were also compared to the sequences in the Gemmata obscuriglobus genome (http://tigrblast.tigr.org/ufmg/) by using TBLASTN analysis.
Phylogenetic analysis.
Polypeptide sequences were aligned by using the CLUSTAL W program (17) and manually curated. Phylogenetic analyses were carried out by using the PHYLIP program package (8). Distance, parsimony, and maximum-likelihood analyses were performed, with 100 bootstrap analyses for each.
Nucleotide sequence accession numbers.
Sequences of the fosmid inserts have been deposited with GenBank under the accession numbers DQ084247, DQ084248, and DQ084250. Partial sequences of fhcD, fae, and mch genes identified in the present study have been deposited under accession numbers DQ173653 to DQ173667 and DQ176037, DQ173643 to DQ173652, and DQ176322 to DQ176324, respectively.
RESULTS
General characteristics of Lake Washington sediment metagenomic libraries.
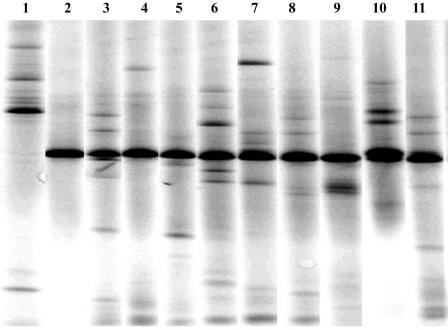
Two large DNA insert libraries were constructed in the CopyControl pCC1FOS vector as described in Materials and Methods. Library 1 consists of approximately 36,000 clones pooled together. Library 2 consists of approximately 36,000 clones divided into 36 pools containing approximately 1,000 clones in each. To analyze the average size of the inserts in the libraries, 25 randomly selected clones were digested with PstI and HindIII, and the resulting restriction mixtures were analyzed by electrophoresis in 0.8% agarose gels (data not shown). The average insert size was determined to be 36.8 ± 3.5 kb. Using this average size, each library was calculated to contain approximately 1,325 Mb of DNA. Assuming an average microbial genome to be 4 Mb in size, each library covers an equivalent of about 330 genomes. Library 2 was further analyzed for the abundance of bacterial 16S rRNA genes. Fragments of 16S rRNA genes were PCR amplified from each of the 36 pools, and the PCR products were analyzed by DGGE (Fig. 1). One to nine bands of different mobility were detected in each clone pool for both the smaller and the larger fragments amplified (see Materials and Methods) totaling 159 fragments for the entire library (Fig. 2), or one bacterial 16S rRNA gene per approximately 8.33 Mb of the metagenome. This is the minimal estimate of bacterial 16S rRNA gene representation in the library, since bands with different electrophoretic mobilities may have contained nonidentical gene fragments.
FIG. 1.
Sample of 16S rRNA gene DGGE fingerprint patterns obtained with the primer set 341fGC/536r. Lanes: 1, DGGE patterns of 16S rRNA gene PCR products obtained from sediment DNA; 2, DGGE pattern for E. coli EPI100; 3 to 11, DGGE patterns of PCR products obtained from metagenomic library 2 clone pools 3 to 11.
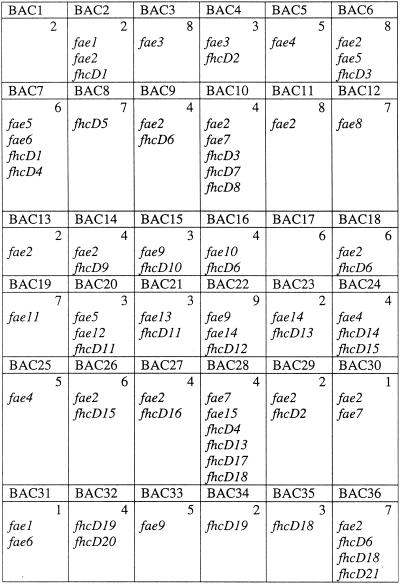
FIG. 2.
Distribution of 16S rRNA, fae, and fhcD genes in clone pools of library 2. Numbers in the top right corners of each box in the grid indicate the number of bands detected by DGGE analysis. Phylotypes were designated as sequences revealing <95% identity to each other at the amino acid level.
Abundance of fhcD and fae in the metagenome.
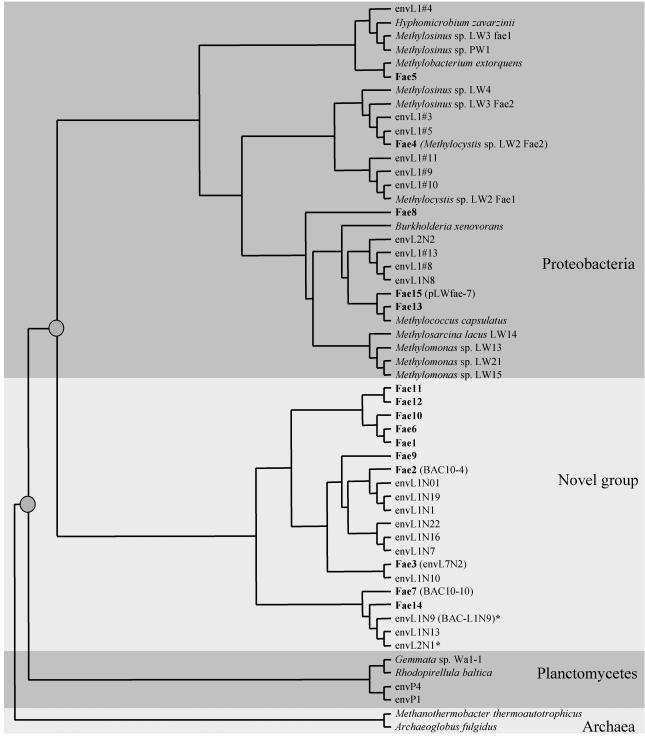
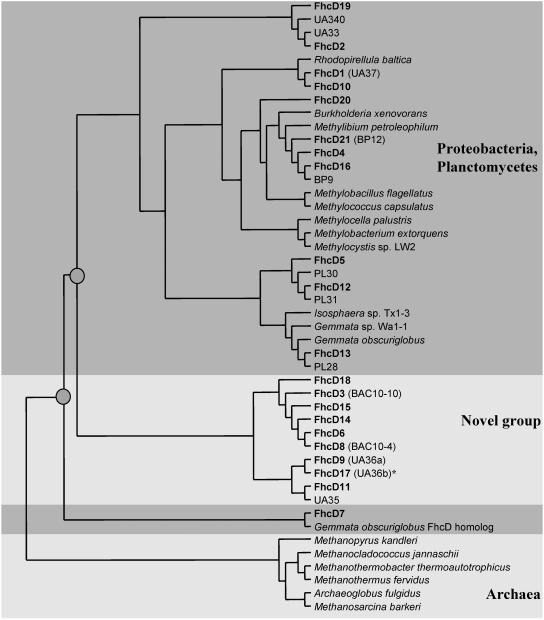
To evaluate the abundance of H4MPT-linked C1 transfer genes in the library, fhcD and fae were PCR-amplified from each of the 36 clone pools of library 2. Using fhcD-specific primers (13), PCR products of expected size were amplified from 24 of the 36 pools. Using fae-specific primers (11), PCR products of expected size were amplified from 30 of the pools (Fig. 2). The resulting PCR products were cloned into the pCR2.1 vector, and three to five clones from each cloning were analyzed by sequencing, revealing that 6 of the pools contained more than one and up to four fhcD genes and that 8 of the pools contained at least two different fae genes each. The sequences were categorized into phylotypes, based on a 95% cutoff at the amino acid level (16). Using this cutoff value, the fae and the fhcD sequences deduced from the metagenome were represented by 15 and 21 phylotypes, respectively. Each phylotype was then compared to the sequences deposited with GenBank. From these comparisons, only four of the newly sequenced fae phylotypes and four of the newly sequenced fhcD phylotypes overlapped with the phylotypes previously detected in PCR-amplified libraries originating from the Lake Washington sediment (11, 13). Phylogenetically, 2 of the 15 unique fae phylotypes clustered with alphaproteobacteria, 1 was deeply branching within beta/gammaproteobacteria, and 2 clustered with gammaproteobacteria, while the remaining 10 clustered with the sequences previously described as a novel, nonaffiliated group (Fig. 3). Of the 21 fhcD phylotypes, 4 clustered with betaproteobacterial sequences, 6 clustered with planctomycete sequences, 2 clustered with unaffiliated bacterial sequences, and 9 clustered with the deeply diverging group of sequences previously identified in Lake Washington (13) (Fig. 4).
FIG. 3.
Consensus phylogenetic tree showing relations of the Fae phylotypes uncovered in the present study (in boldface) to previously known Fae sequences. In parentheses, alternative names are shown (11, 16; the present study). Sequences that were also detected via RT-PCR are marked by asterisks. The nodes that separate the novel sequences (denoted by gray circles) are supported by bootstrap values of at least 94% in at least two out of three analyses performed (see Materials and Methods).
FIG. 4.
Consensus phylogenetic tree showing relations of the FhcD phylotypes uncovered in the present study (in boldface) to previously known FhcD sequences. In parentheses, alternative names are shown (13; the present study). The sequence that was also detected via RT-PCR is marked by an asterisk. The nodes that separate the novel sequences (denoted by gray circles) are supported by bootstrap values of at least 75% in at least two out of three analyses performed (see Materials and Methods).
Sequencing of fosmid inserts containing divergent fae and fhcD.
To obtain further information on the organisms containing the divergent formaldehyde handling genes, three fosmid clone inserts bearing divergent fae, fhcD, and/or mtdA/B genes were chosen to be sequenced. Clone LWBAC-L1N9 was identified by screening a total of 4,000 clones of library 1 by hybridization with the probe consisting of a mixture of fae fragments amplified from the previously described divergent clones L1N9, L1N13, and L1N19 (11). Clones LWBAC10-4 and LWBAC10-10 were identified by screening a total of 1,000 clones of pool 10 of library 2 by hybridization with the fhcD probe consisting of a mixture of three divergent fhcD fragments that had been PCR amplified from this pool as described in Materials and Methods (FhcD3, FhcD7, and FhcD8). The same set of filters was hybridized with another probe, a divergent mtdA/B gene homolog identified previously (clone env97 [20]), and clone LWBAC10-4 was found to be positive for this probe.
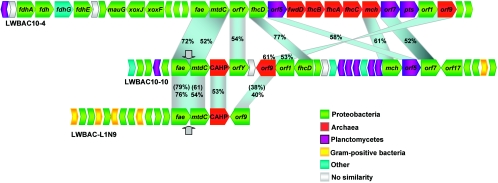
Clone LWBAC-L1N9 was shown to contain an insert of approximately 25.3 kb with an average G+C DNA content of 68%. The fae gene identified within this fragment corresponded to the L1N9 sequence. The sequence of a part of the fosmid insert estimated to be approximately 50 bp remained unresolved, apparently due to a strong hairpin structure (indicated by an arrow in Fig. 5). A total of 14 potential ORFs were identified within the insert (Fig. 5), including three of the H4MPT-linked C1 transfer gene homologs: the fae, the previously described mtdC (20), and an orf9 homolog. The predicted functions of the remaining 11 ORFs and their best hits in the databases are listed in Table S1 in the supplemental material. None of these ORFs showed high similarity to any known sequences. Identities with top hit sequences ranged between 23 and 50%, and top hit sequences belonged to Proteobacteria, gram-positive bacteria, and Archaea.
FIG. 5.
Gene content and organization of fosmid clones analyzed in this work. Genes conserved between the genomic fragments are connected by shaded areas. Other bacterial genera include Cytophaga, Flavobacterium, Cyanobacteria, Aquificae, Chloroflexi, Bacteroides, etc. Arrows show location of gaps in sequence. Numbers show percent amino acid identities between genes in BAC10-4 and BAC10-10; numbers in parentheses show the percent identity between genes in BAC10-4 and BAC-L1N9.
Clone LWBAC10-4 was shown to contain an insert of 39.9 kb, with an average G+C DNA content of 66%. The fhcD gene identified in this insert was the FhcD8 phylotype. A total of 31 potential ORFs were identified within this insert (Table S1 in the supplemental material and Fig. 5). Twelve of these were identified as the H4MPT-linked C1 transfer genes. Five of these showed highest similarity to proteobacterial counterparts (30 to 68% identity at the amino acid level), two showed highest similarity to planctomycete counterparts (48 to 49%), and five were most similar to archaeal counterparts (40 to 48%). In addition, a gene homologous to the fwdD gene encoding the D subunit of the tungsten formylmethanofuran dehydrogenase in Archaea was present in the cluster. Homologs of this gene have not been identified in C1 clusters previously characterized in Proteobacteria or Planctomycetes. Moreover, like in Archaea belonging to Methanosarcinales and Archaeoglobus, the fwdD homolog in clone LWBAC10-4 is part of the fwdD-fhcB(fwdB)-fhcA(fwdA)-fhcC(fwdC) gene cluster. Other genes of interest located in the insert are homologs of the xoxFJ genes and the adjacent gene potentially encoding a cytochrome c peroxidase (Fig. 5). The xox genes are predicted to encode a PQQ-linked dehydrogenase similar to methanol dehydrogenase (4, 10). Although the function of this enzyme and its substrate specificity remain unknown, xox genes are often found adjacent to the H4MPT-linked C1 transfer gene clusters (4, 7, 10) and thus possibly encode an enzyme producing formaldehyde. Next to the xox gene cluster, a cluster of genes is found predicted to encode an anaerobic formate dehydrogenase. The remaining potential ORFs were not closely related to known genes. Their top hits ranged between 22 and 55% and were distributed between Proteobacteria, gram-positive bacteria, Planctomycetes, Archaea, and bacteria of deeply branching lineages (Table S1 in the supplemental material). Some of the translated ORFs had no significant hits in the nonredundant database.
Clone LWBAC10-10 was shown to contain an insert of approximately 33.6 kb, with an average G+C DNA content of 67%. The fhcD sequence identified in this insert corresponded to the FhcD3 phylotype. The sequence of a part of the insert estimated to be approximately 50 base pairs remained unresolved, apparently due to a strong hairpin structure (indicated by an arrow in Fig. 4). Sequence analysis revealed the presence of 30 potential ORFs. Of these, 10 were homologous to the H4MPT-linked C1 transfer genes, with identities ranging from 26 to 59%, distributed between Proteobacteria, Planctomycetes, and Archaea. Immediately upstream of orf17, two genes are located, predicted to encode selenocysteine biosynthesis enzymes. The remaining ORFs did not reveal significant similarity to known sequences, with top hit identity levels ranging from 24 to 46%, distributed between Proteobacteria, gram-positive bacteria, Planctomycetes, Archaea, and bacteria of deeply diverging divisions (Table S1 in the supplemental material).
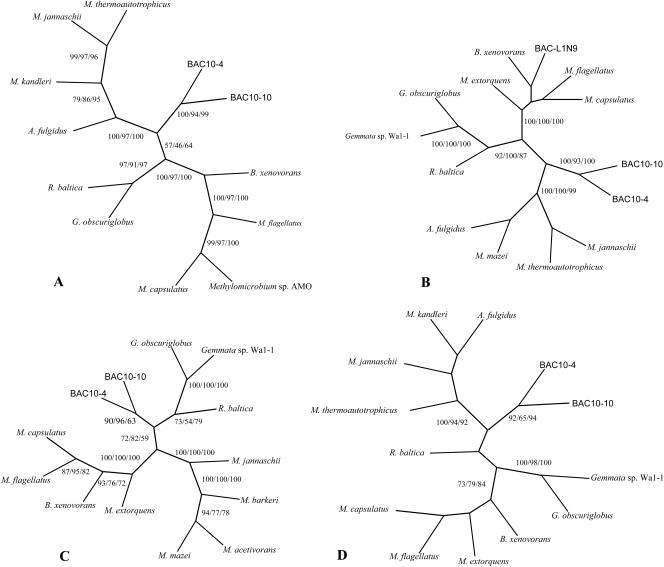
No strong conservation in gene order or content was found between the three analyzed genomic fragments, with the exception that the fae-mtdC pair conservation is common to Planctomycetes (12, 20). Only one hypothetical protein-coding ORF (located downstream of mtdC and encoding a conserved archaeal hypothetical protein) was conserved between the inserts of LWBAC-L1N9 and LWBAC10-10. Although nine H4MPT-linked C1 transfer genes were shared between LWBAC10-10 and LWBAC10-4 inserts, little conservation in their order was observed, with the exception that the orf1-orf9 pair is typical of Proteobacteria (12). The stretch of genes in the LWBAC10-10 insert, mch-orf5-orf7-orf17 has been found highly conserved in Proteobacteria (12). The polypeptide sequences translated from the C1 transfer genes present in the inserts were compared to each other and to the sequences in public databases, using BLAST analyses. The newly identified sequences were found more related to each other than to the previously known sequences (with an exception of orf9 in the BAC-L1N9 insert that was more related to proteobacterial orf9 sequences), with the levels of sequence identity ranging from 52 to 79% (Fig. 4). Phylogenetic analyses of these sequences further confirmed that they all diverged significantly from the sequences known for cultivated microbes belonging to Proteobacteria, Planctomycetes, or Archaea, forming deep branches on phylogenetic trees (Fig. 6). In many cases, however, the branching pattern differed for different proteins and different analyses and in general, nodes for the novel sequences and the planctomycete sequences were poorly resolved, apparently resulting from high protein divergence within both the novel group and the planctomycete sequences. Based on the low sequence similarity, the low degree of gene clustering conservation with known microbial groups and the phylogenetic tree patterns, it is likely that the fosmid insert sequences described above belong to microbes of a as-yet-undescribed, uncultivated phylum within Bacteria. Possibly, these sequences represent yet unknown, deeply branching members of Planctomycetes.
FIG. 6.
Consensus phylogenetic trees of Mch (A), Orf9 (B), Orf5 (C), and Orf7 (D) polypeptides. Bootstrap values are shown for distance, parsimony, and maximum-likelihood analyses. Nodes without bootstrap values are not strongly supported. M. extorquens, B. xenovorans, M. capsulatus, and M. flagellatus are Proteobacteria; G. obscuriglobus, Gemmata sp. strain Wa1-1, and R. baltica are Planctomycetes; M. thermoautotrophicus, M. kandleri, M. mazei, M. barkeri, M. acetivorans, A. fulgidus, and M. jannaschii are Archaea.
Expression of divergent C1 transfer genes in Lake Washington.
Expression of the divergent C1 transfer genes in Lake Washington was tested by using the RT-PCR technique. To specifically target the divergent genes of interest, new sets of primer pairs were designed for PCR amplification, as described in Materials and Methods section, targeting fae, fhcD, and mch. These were used in RT-PCRs with total RNA isolated from the top layer of Lake Washington sediment, and the resulting PCR products were cloned into the pCR2.1 vector. 20 randomly selected clones from each cloning were analyzed by sequencing. As a result, two unique fae phylotypes, two unique fhcD phylotypes, and four unique mch phylotypes were uncovered, all of these falling within the deep branches represented by the prototype sequences in the respective phylogenetic trees. Both fae sequences, one of the two fhcD sequences and one of the four mch sequences (not shown), were highly similar (98 to 99%) to the previously detected sequences, respectively, phylotype envL1N9, phylotype envL2N1 (fae genes), phylotype FhcD17, and the mch gene within the LWBAC10-4 insert.
DISCUSSION
Elemental cycling in natural habitats such as freshwater sediments is mainly maintained by prokaryotes (15, 19). We are using Lake Washington, a pristine freshwater lake, as a model to understand and monitor the microbial populations participating in environmental processes in such environments. Our focus thus far has been on one specific functional group, the bacteria involved in cycling of C1 (containing no carbon-carbon bonds) compounds. In our previous work, we applied a variety of approaches to characterize the main players in C1 cycling in the site, such as enrichments and pure culture isolations, PCR surveys of phylogenetic and functional genes, RT-PCR-based rRNA and mRNA analyses, and stable isotope probing (11, 13, 16). A picture emerging from these experiments is the one of a complex, multitiered microbial community involved in environmental C1 cycling. Comparisons between culture-based and culture-independent surveys, however, indicated that many members of this community remain uncultured and phylogenetically unaffiliated (11, 13, 16). Of special interest to us is a group of microbes represented by divergent sequences of the genes involved in the formaldehyde oxidation pathway linked to H4MPT (11, 13), for the following reasons. (i) If, as suggested by the present study, the microbes possessing these sequences are abundant in the environment, they are likely to play a significant ecological role, and this role needs to be uncovered. (ii) If the divergent genes are reflective of the deeply divergent nature of these microbes, their analysis may uncover existence of a new phylum and thus add a missing link to the universal tree of life. In the present study, we used a metagenomic approach to assess the abundance of the novel, divergent genes for the H4MPT-linked C1 transfer pathway in the Lake Washington sediment community and to obtain initial glimpses into the genomic structure of these microbes.
The metagenome analysis confirmed the relative abundance of the genes involved in H4MTP-linked C1 transfers in the microbial population inhabiting Lake Washington sediment. A comparison of the total number of fae and fhcD genes recovered from the metagenome to the number of 16S rRNA genes suggests that approximately 21 to 25% of the microbes represented in the metagenome possess these genes. Likely, this number is an underestimation for the total population since no gammaproteobacterial sequences related to the Methylomonas/Methylobacter group were uncovered, apparently due to cloning biases noted previously (11), and it is known that methanotrophs of this group are abundant in the sediment (1, 5, 6). Our analyses demonstrate that a large fraction of the H4MPT-linked C1 transfer genes is represented by the divergent genes (38% in the fhcD sequence subset and 77% in the fae sequence subset), a finding consistent with the previous PCR-based surveys using total environmental DNA (11, 13).
Three fosmid inserts carrying divergent fae and fhcD genes were sequenced in order to obtain data supporting (or rejecting) the deeply branching nature of the organisms in question. Indeed, we demonstrated that genes representative of the divergent groups previously detected in three independent PCR-surveys (fae, fhcD, and mtdA/B) cluster together within the genomic fragments sequenced. Other C1 transfer genes identified within the fosmid inserts also diverged deeply from the genes previously identified in cultured Proteobacteria, Planctomycetes, or Archaea. The genes outside of the C1 transfer gene clusters provided little useful phylogenetic signal, since they shared extremely low levels of similarity with known genes, and their top hits were distributed between various representatives of Bacteria or Archaea (Table S1 in the supplemental material). The three sequenced genomic fragments, while more similar to each other than to known organisms, still showed a high level of divergence in gene sequence for the homologous ORFs, in gene order conservation and in gene content, suggesting that the organisms from which these genomic fragments originated are not closely related to each other. These data provide additional evidence that these organisms may comprise a deeply branching clade. This new clade would most likely fall within the bacterial kingdom of life, based on higher hits with bacterial sequences for most genes and based on phylogenetic analyses. Possibly, the new sequences represent novel, deeply branching Planctomycetes. Our data indicate that this novel clade represents a significant fraction of the total microbial population in Lake Washington sediment, thus implying a potential ecological significance. The data on mRNA detection presented here demonstrate that the novel C1 transfer genes are expressed under in situ conditions, suggesting a function in C1 cycling. The proposed function in C1 cycling is also supported by previous enrichment data (13, 16). However, the exact role in C1 cycling and the nature of primary substrates for these organisms in the environment remain unknown. These will be addressed in our future studies, which will include expanded metagenome sequencing and expression analyses.
Supplementary Material
Acknowledgments
We acknowledge support from the Microbial Observatories program funded by the National Science Foundation (MCB-0131957).
Mark Dodobara is acknowledged for technical assistance, and Sergey Stolyar and David Stahl are acknowledged for providing access to the DGGE apparatus. The Institute for Genomic Research is acknowledged for early release of the G. obscuriglobus genomic sequence (funded by the U.S. Department of Energy).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Auman, A. J., S. Stolyar, A. M. Costello, and M. E. Lidstrom. 2000. Molecular characterization of methanotrophic isolates from freshwater lake sediment. Appl. Environ. Microbiol. 66:5259-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auman, A. J., and M. E. Lidstrom. 2002. Analysis of sMMO-containing type I methanotrophs in Lake Washington sediment. Environ. Microbiol. 4:517-524. [DOI] [PubMed] [Google Scholar]
- 3.Auman, A. J., C. C. Speake, and M. E. Lidstrom. 2001. nifH sequences and nitrogen fixation in type I and type II methanotrophs. Appl. Environ. Microbiol. 67:4009-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chistoserdova, L., and M. E. Lidstrom. 1997. Molecular and mutational analysis of a DNA region separating two methylotrophy gene clusters in Methylobacterium extorquens AM1. Microbiol. 143:1729-1736. [DOI] [PubMed] [Google Scholar]
- 5.Costello, A. M., A. J. Auman, J. L. Macalady, K. M. Scow, and M. E. Lidstrom. 2002. Estimation of methanotroph abundance in a freshwater lake sediment. Environ. Microbiol. 4:443-450. [DOI] [PubMed] [Google Scholar]
- 6.Costello, A. M., and M. E. Lidstrom. 1999. Molecular characterization of functional and phylogenetic genes from natural populations of methanotrophs in lake sediments. Appl. Environ. Microbiol. 65:5066-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denef, V. J., J. Park, T. V. Tsoi, J. M. Rouillard, H. Zhang, J. A. Wibbenmeyer, W. Verstraete, E. Gulari, S. A. Hashsham, and J. M. Tiedje. 2004. Biphenyl and benzoate metabolism in a genomic context: outlining genome-wide metabolic networks in Burkholderia xenovorans LB400. Appl. Environ. Microbiol. 70:4961-4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felsenstein, J. 2003. Inferring phylogenies. Sinauer Associates, Inc., Sunderland, Mass.
- 9.Harder, W., M. Attwood, and J. R. Quayle. 1973. Methanol assimilation by Hyphomicrobium spp. J. Gen. Microbiol. 78:155-163. [Google Scholar]
- 10.Harms, N., J. Ras, S. Koning, W. N. M. Reijnders, A. H. Stouthammer, and R. J. M. van Spanning. 1996. Genetics of C1 metabolism regulation in Paracoccus denitrificans, p. 126-132. In M. E. Lidstrom and F. R. Tabita (ed.), Microbial growth on C1 compounds. Kluwer Academic Press, Dordrecht, The Netherlands.
- 11.Kalyuzhnaya, M. G., M. E. Lidstrom, and L. Chistoserdova. 2004. Utility of environmental primers targeting ancient enzymes: methylotroph detection in Lake Washington. Microb. Ecol. 48:463-472. [DOI] [PubMed] [Google Scholar]
- 12.Kalyuzhnaya, M. G., N. Korotkova, G. Crowther, C. J. Marx, M. E. Lidstrom, and L. Chistoserdova. 2005. Analysis of gene islands involved in methanopterin-linked C1 transfer reactions reveals new function and provides evolutionary insights. J. Bacteriol. 187:4607-4614. [DOI] [PMC free article] [PubMed]
- 13.Kalyuzhnaya, M. G, O. Nercessian, M. E. Lidstrom, and L. Chistoserdova. 2005. Development and application of polymerase chain reaction primers based on fhcD for environmental detection of methanopterin-linked C1-metabolism in bacteria. Environ. Microbiol. 7:1269-1274. [DOI] [PubMed] [Google Scholar]
- 14.Lidstrom, M. E., and L. Somers. 1984. Seasonal study of methane oxidation in Lake Washington. Appl. Environ. Microbiol. 47:1255-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nealson, K. H. 1997. Sediment bacteria: who's there, what are they doing, and what's new? Annu. Rev. Earth Planet Sci. 25:403-434. [DOI] [PubMed] [Google Scholar]
- 16.Nercessian, O., E. Noyes, M. G. Kalyuzhnaya, M. E. Lidstrom, and L. Chistoserdova. 2005. Bacterial population active in metabolism of C1 compounds in the sediment of lake Washington, a freshwater lake. Appl. Environ. Microbiol. 71:6885-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 19.Spring, S., R. Schlze, J. Overmann, and K. H. Schleifer. 2000. Identification and characterization of ecologically significant prokaryotes in the sediment of freshwater lakes: molecular and cultivation studies. FEMS Microbiol. Rev. 24:573-590. [DOI] [PubMed] [Google Scholar]
- 20.Vorholt, J. A., M. G. Kalyuzhnaya, C. H. Hagemeier, M. E. Lidstrom, and L. Chistoserdova. 2005. MtdC, a novel class of methylene tetrahydromethanopterin dehydrogenases. J. Bacteriol. 187:6069-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe, K., Y. Kodama, and S. Harayama. 2001. Design and evaluation of PCR primers to amplify bacterial 16S ribosomal DNA fragments used for community fingerprinting. J. Microbiol. Methods 44:253-262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.