Abstract
Flavones are plant secondary metabolites that have wide pharmaceutical and nutraceutical applications. We previously constructed a recombinant flavanone pathway by expressing in Saccharomyces cerevisiae a four-step recombinant pathway that consists of cinnamate-4 hydroxylase, 4-coumaroyl:coenzyme A ligase, chalcone synthase, and chalcone isomerase. In the present work, the biosynthesis of flavones by two distinct flavone synthases was evaluated by introducing a soluble flavone synthase I (FSI) and a membrane-bound flavone synthase II (FSII) into the flavanone-producing recombinant yeast strain. The resulting recombinant strains were able to convert various phenylpropanoid acid precursors into the flavone molecules chrysin, apigenin, and luteolin, and the intermediate flavanones pinocembrin, naringenin, and eriodictyol accumulated in the medium. Improvement of flavone biosynthesis was achieved by overexpressing the yeast P450 reductase CPR1 in the FSII-expressing recombinant strain and by using acetate rather than glucose or raffinose as the carbon source. Overall, the FSI-expressing recombinant strain produced 50% more apigenin and six times less naringenin than the FSII-expressing recombinant strain when p-coumaric acid was used as a precursor phenylpropanoid acid. Further experiments indicated that unlike luteolin, the 5,7,4′-trihydroxyflavone apigenin inhibits flavanone biosynthesis in vivo in a nonlinear, dose-dependent manner.
Flavones, such as chrysin, apigenin, and luteolin, belong to the large group of plant secondary metabolites known collectively as flavonoids. These compounds exhibit an array of pharmacological properties, including antianxiety effects (35-37), improvement of cardiac function after ischemia (17, 28, 31), and antiestrogenic effects in breast cancer cell cultures (24). Their basic chemical structure consists of two benzene rings linked through a heterocyclic pyrone or pyran ring in the middle (23). Flavones occur only in a relatively small food group that includes parsley, thyme, celery, and sweet red pepper (27). Thus, biosynthesis of these compounds from microorganisms, similar to the biosynthesis that has recently been demonstrated for other flavonoid molecules (12, 15, 38, 39), can be an attractive alternative to the current methods that employ plant extraction.
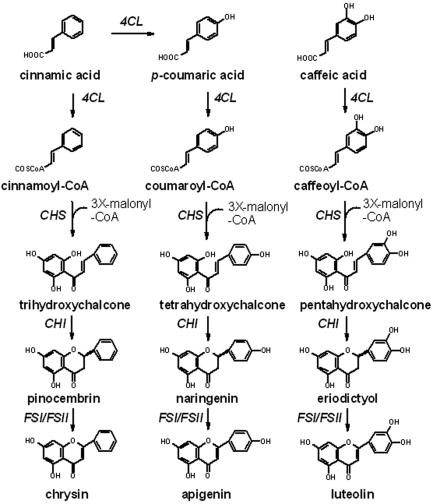
Flavone biosynthesis starts with the conversion of cinnamic acid to p-coumaric acid by a P450 monooxygenase, cinnamate 4-hydroxylase (C4H). p-Coumaric acid is then converted to 4-coumaroyl coenzyme A (4-coumaroyl-CoA) by 4-coumaroyl:CoA ligase (4CL). Next, chalcone synthase (CHS) catalyzes condensation of 4-coumaroyl-CoA with three molecules of malonyl-CoA, in which tetrahydroxychalcone is formed. Following this reaction, chalcone isomerase (CHI) performs stereospecific isomerization of tetrahydroxychalcone to (2S)-flavanone, which is the branch point precursor of many important downstream flavonoids, including flavones. In most cases, a membrane-bound cytochrome P450 monooxygenase, flavone synthase II (FSII), catalyzes the biosynthesis of flavones from (2S)-flavanones (1, 20). However, in certain species of Apiceae, this reaction is performed by soluble flavone synthase I (FSI) (Fig. 1) (19, 22).
FIG. 1.
Flavone biosynthetic pathway. 4CL, 4-coumaroyl:CoA ligase; CHI, chalcone isomerase.
In previous work, we demonstrated engineering of a recombinant, flavanone-producing Saccharomyces cerevisiae strain by simultaneous expression of four flavanone biosynthetic enzymes, C4H, 4CL, CHS, and CHI (39). Upon addition of various phenylpropanoid acids, the recombinant strain produced significant amounts of flavanones, such as pinocembrin, naringenin, and eriodictyol, which are the precursors of the vast majority of flavonoids. Because of the up-to-60-fold increase in the amount of flavanones produced by the recombinant yeast strain compared to the amount produced by an Escherichia coli recombinant described previously (39), we believe that yeast could be a better production platform for the general biosynthesis of plant-specific flavonoids. Additionally, S. cerevisiae, unlike E. coli, can functionally express P450 hydroxylases involved in flavonoid biosynthesis (26).
In an extension of our previous work, we describe here for the first time the development and optimization of flavone-producing recombinant yeast strains. The model microorganisms allowed evaluation of the two alternative ways that plants have evolved for flavone synthesis, namely, through the P450 monooxygenase FSII, which requires NADPH and is inhibited by Fe2+, and the soluble dioxygenase FSI, which requires 2-oxoglutarate, Fe2+, and ascorbate as cofactors (21). The overall flavonoid metabolism is different in the two strains, and the FSI-expressing recombinant strain exhibits better overall production. Overexpression of the yeast P450 reductase CPR1 and use of acetate as a carbon source led to improvement in flavone biosynthesis. Further in vivo experiments demonstrated that apigenin, but not luteolin, is a feedback inhibitor of the flavone biosynthetic circuit.
MATERIALS AND METHODS
Chemicals and media.
Cinnamic acid, p-coumaric acid, and caffeic acid were purchased from MP Biomedicals Inc. (Irvine, CA). Naringenin was purchased from Sigma-Aldrich (St. Louis, MO). Ferulic acid, eriodictyol, apigenin, and luteolin were purchased from Indofine (Hillsborough, NJ). Flavonoids were extracted from plants by using standard procedures (10).
Luria broth rich medium was purchased from Sigma-Aldrich. S. cerevisiae YPD rich medium and SC minimal selection medium with glucose or raffinose and galactose were prepared as previously described (9, 39). Minimal medium with acetate (MA) was prepared as described by Verduyn et al. without the antifoaming agent (34).
Strains, plasmids, and plant materials.
All strains and plasmids used are shown in Table 1. Plants were purchased from local nurseries and were exposed to the sun for a few hours before materials were collected. For mRNA extraction, leaflets of flat-leaved Petroselinum crispum (parsley) and flower petals of Antirrhinum majus cv. Montego Yellow (snapdragon) and Catharanthus roseus (Madagascar periwinkle) were used. Plant materials were quickly frozen with liquid nitrogen and stored at −80°C until they were used.
TABLE 1.
Strains and plasmids used in the present study
| Strain or plasmid | Genotype or properties | Source or reference |
|---|---|---|
| E. coli TOP 10F′ | F′[lacIq Tn10(Tetr)] mcrA (mrr-hsdRMS-mcrBC) 80lacZM15 lacX74 recA1 araD139 (ara-leu)7697 galU galK rpsL (Strr) endA1 n upG | Invitrogen |
| S. cerevisiae INVSc1 | MATa his3D1 leu2 trp1-289 ura3-52 | Invitrogen |
| pYES2.1/V5-His-TOPO | T/A cloning vector, URA3, Apr | Invitrogen |
| YEplac181 | Cloning vector, LEU2, Ampr | ATCC |
| YCplac22 | Cloning vector, TRP1, Ampr | ATCC |
| Ycc4c181 | Gal-inducible C4H Pc4CL-2 chs CHI-A | 39 |
| pYES-FSI | Gal-inducible PcFSI from parsley | This study |
| pYES-AFNS2 | Gal-inducible AFNS2 from snapdragon | This study |
| pYES-CPR | Gal-inducible cpr from C. roseus | This study |
| pYES-CPR1 | Gal-inducible CPR1 from S. cerevisiae | This study |
| YC-FSI | Gal-inducible PcFSI from parsley | This study |
| YC-AFNS2 | Gal-inducible AFNS2 from snapdragon | This study |
| YC-AFNS2 + CPR1 | Gal-inducible AFNS2 from snapdragon, CPR1 from S. cerevisiae | This study |
DNA manipulations.
All DNA manipulations were performed by using standard procedures (29). Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs and Promega. Reverse transcription-PCRs were performed using a Superscript One-step w/platinum Taq kit (Invitrogen, Carlsbad, CA). PCRs were performed using Roche's Expand High Fidelity PCR system. Flavone synthase I PcFSI cDNA from parsley, flavone synthase II AFNS2 cDNA from snapdragon, C. roseus P450 reductase cpr cDNA, and S. cerevisiae P450 reductase CPR1 cDNA were isolated in our lab based on DNA sequences available in the GenBank database (accession numbers AY230247, AB028151, X69791, and D13788, respectively). A QIAGEN RNeasy MiniKit was used for isolation of total RNA from plants or yeast. All amplified genes were initially cloned in the S. cerevisiae vector pYES2.1/V5-His-TOPO using T/A cloning according to the manufacturer's instructions (Invitrogen), which yielded plasmids pYES-FSI, pYES-AFNS2, pYES-CPR, and pYES-CPR1. Transformation of yeast was carried out using the lithium acetate method (7, 8). In all cases, the absence of undesired mutations introduced during PCR was verified by direct nucleotide sequencing.
Construction of plasmids.
The flavone biosynthetic genes were introduced into yeast using two coreplicable shuttle vectors, YEplac181 and YCplac22, which carry the LEU2 and TRP1 markers, respectively, to allow selection of transformants by growth in minimal medium lacking leucine and tryptophan.
Plasmid Ycc4c181, derived from plasmid YEplac181 and carrying cDNA for C4H from Arabidopsis thaliana, Pc4cL-2 from parsley, and CHI-A and chs from Petunia × hybrida, has been described previously (39).
Plasmid YC-FSI was constructed by amplifying the PcFSI cDNA together with the GAL1 promoter from plasmid pYES-FSI using a forward primer hybridizing to a vector DNA region that lies upstream of the GAL1 promoter and a reverse primer that hybridizes at the end of the cloned cDNA. The GAL1-PcFSI fusion was then inserted into vector YCplac22 between HindIII and KpnI sites, yielding plasmid YC-FSI. Plasmid YC-AFNS2 was constructed by a similar approach by inserting GAL1-AFNS2 into vector YCplac22 between BamHI and KpnI restriction sites. Insertion of GAL1-CPR1 between PstI and SalI restriction sites of vector YCplac22 and insertion of GAL1-AFNS2 between SacI and XbaI restriction sites yielded plasmid YC-AFNS2+CPR1.
In vitro AFNS2 assay.
Yeast recombinant strains harboring pYES-AFNS2, pYES-CPR1, and pYES-CPR were cultivated in liquid SC minimal medium lacking uracil (SC-Ura−) with glucose as the carbon source overnight at 30°C. For protein expression, the overnight culture was used to inoculate 250 ml SC-Ura− minimal medium with raffinose to obtain an initial absorbance at 600 nm (A600) of 0.2. When the culture reached an A600 of 0.8, sterile galactose was added into the culture to a final concentration of 2% to induce protein expression. After 24 h of incubation at 30°C, recombinant yeast cells were harvested and used for microsomal protein preparation as described by Schoehnbohm et al. (30). A BCA protein assay kit (Pierce Chemicals, Rockford, IL) was used for protein quantification. In vitro apigenin synthesis was performed in 100-μl reaction mixtures containing 0.1 μmol of naringenin substrate, NADPH at a final concentration of 1.5 mM, 20 μg of a microsomal preparation of AFNS2, and 50 μg of a microsomal preparation of the P450 reductase (either C. roseus CPR or yeast CPR1). The reaction was initiated by addition of a freshly prepared NADPH solution, and the mixture was incubated at 25°C for 2 h. The reaction products were extracted with an equal volume of ethyl acetate twice and evaporated to dryness under a vacuum. Acetonitrile-water (1:3, vol/vol) was then added to dissolve the organic compounds for high-performance liquid chromatography (HPLC) analysis.
Heterologous expression and fermentation.
In order to engineer flavone biosynthesis through two different flavone synthases, recombinant INVSc1 carrying plasmid Ycc4c181G was transformed with YC-FSI, YC-AFNS2, or YC-AFNS2+CPR1 separately, yielding recombinant strains INV-4G+FSI, INV-4G+AFNS2, and INV-4G+AFNS2+CPR1. Yeast colonies harboring both plasmids were selected by growth on SC agar plates lacking leucine and tryptophan (SC-Leu−Trp−). For flavone fermentation purposes, an individual recombinant yeast colony was grown overnight in 10 ml liquid SC-Leu−Trp− medium containing glucose at 30°C with shaking. The following day, a portion of this culture was used to seed the main culture in SC-Leu−Trp− medium containing galactose and raffinose or MA-Leu−Trp− containing galactose and acetate to obtain an A600 of 0.4. Then, 0.5 mM phenylpropanoid acid (cinnamic acid, p-coumaric acid, caffeic acid, or ferulic acid) was added to the main culture, and the mixture was incubated at 30°C with horizontal shaking for a maximum of 92 h. For fermentation of INV-4G+FSI, FeSO4, 2-oxoglutaric acid, and sodium ascorbate were added to a final concentration of 0.5 mM. Plasmid stability was checked by isolation from the recombinant strains using Zymoprep I (Zymo Research, Orange, CA) every 24 h, and the plasmids were subsequently used for restriction mapping and PCR analysis.
Analytical methods.
Flavonoids and phenylpropanoid acids were analyzed by HPLC, using an Agilent 1100 series instrument and a reverse-phase ZORBAX SB-C18 column (4.6 by 150 mm) that was maintained at 25°C. The compounds were separated by elution with an acetonitrile-water gradient at a flow rate of 1.0 ml/min. The HPLC conditions were as follows (profile 1): 20 to 27% acetonitrile (vol/vol) for 45 min and 27 to 95% acetonitrile (vol/vol) for 30 s. The retention times under these conditions for the standard authentic compounds were as follows: caffeic acid, 2.7 min; p-coumaric acid, 4.7 min; eriodictyol, 18.4 min; luteolin, 20.8 min; naringenin, 30.9 min; and apigenin, 33.2 min. Cinnamic acid, pinocembrin, and chrysin were separated by elution with an acetonitrile-water gradient at a flow rate of 1.0 ml/min under the following conditions (profile 2): 10 to 40% acetonitrile (vol/vol) for 10 min, 40 to 60% acetonitrile (vol/vol) for 5 min, and 60 to 10% acetonitrile (vol/vol) for 2 min. The retention times for cinnamic acid, chrysin, and pinocembrin were 12.1 min, 16.0 min, and 16.3 min, respectively. Flavanones were detected and quantified by monitoring the absorbance at 290 nm. Flavones were detected and quantified by monitoring the absorbance at 340 nm. Calibration curves were obtained with solutions containing authentic flavanones and flavones at various concentrations.
RESULTS
Optimization of AFNS2 activity.
AFNS2 is a class II P450 monooxygenase. Using molecular oxygen and two electrons, the P450 oxidizes its substrate and forms a water molecule. Catalysis requires a single redox partner, the P450-reductase that extracts two electrons from NADPH and channels them to the P450 iron center. On the other hand, FSI is an oxoglutarate-dependent dioxygenase, and its activity does not require electron transfer. Therefore, the enzyme reaction does not depend on the action of a P450-reductase. Even though it appears that the endogenous yeast cytochrome P450 reductase is capable of supporting plant P450 monooxygenase functions (39), it has been shown in the past that the activity of yeast recombinant P450 proteins improves in the presence of an abundant amount of the redox partner (26). For this reason, the electron transfer process is currently considered the rate-limiting step in the P450 monooxygenase reaction (4, 5).
In order to investigate and select the most effective P450 reductase for improving AFN2 activity in yeast, we tested C. roseus P450 reductase CPR and S. cerevisiae P450 reductase CPR1 for optimizing the AFNS2 reaction in vitro. We chose C. roseus CPR because in the past it has been used successfully for optimizing the activity of flavonoid P450 monooxygenases, such as cinnamate 4-hydroxylase and flavonoid 3′,5′-hydroxylase (11, 16). For this purpose, microsomal proteins of recombinant yeasts individually expressing AFNS2, CPR1, and CPR were prepared. The activity of AFNS2 was investigated by quantifying the conversion of naringenin to apigenin in the presence of NADPH and either CPR1 or CPR. After 2 h, approximately 40% and 90% increases in the amount of apigenin produced were observed in the presence of CPR and CPR1, respectively (Table 2). Based on these results, we concluded that overexpression of yeast CPR1 enhances the performance of AFNS2 more efficiently, and CPR1 was chosen for further investigation.
TABLE 2.
In vitro assay of AFNS2 from yeast microsomal preparation
| Incubation time (h) | Amt of apigenin produced (nmol)
|
% Increasea
|
|||
|---|---|---|---|---|---|
| AFNS2 | AFNS2+ CPR | AFNS2+ CPR1 | AFNS2+ CPR | AFNS2+ CPR1 | |
| 0.5 | 0.05 | 0.12 | 0.13 | 140 | 160 |
| 1 | 0.19 | 0.23 | 0.35 | 21 | 84 |
| 2 | 0.28 | 0.40 | 0.53 | 43 | 89 |
Values are percent increases compared with AFNS2.
Construction of flavone biosynthetic pathway in yeast.
Structural genes having heterologous plant origins were used to construct the recombinant flavonoid pathway in S. cerevisiae. The flavanone biosynthesis network was assembled as previously described (39). In this case, the yeast vector YEplac181 was used to carry the four-gene flavanone biosynthetic cluster, yielding plasmid Ycc4c181. This plasmid, with a 2 μm origin of replication and the LEU2 selection marker, can coreplicate with the vector YCplac22, which carries the CEN4 origin and the TRP1 selection marker and was utilized to clone the GAL1-PcFSI and GAL1-AFNS2 expression cassettes to create plasmids YC-FSI and YC-AFNS2, respectively. Another YCplac22-based plasmid, which harbors CPR1 in addition to AFNS2 cDNA (plasmid YC-AFNS2+CPR1), was also constructed.
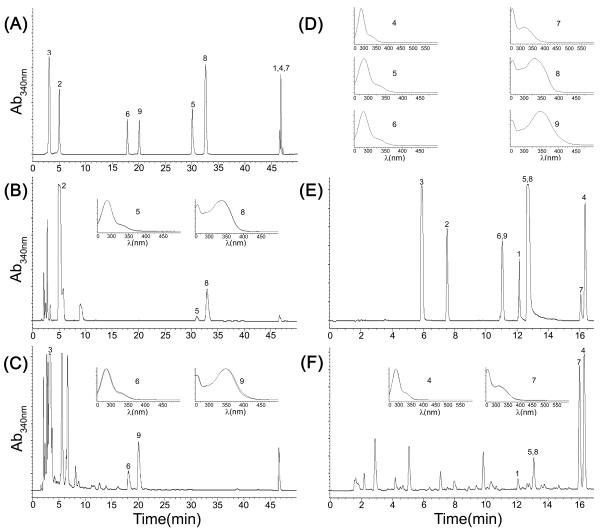
S. cerevisiae strain INVSc1 carrying Ycc4c181 was transformed with YC-FSI, YC-AFNS2, and YC-AFNS2+CPR1, generating recombinant strains INV-4G+FSI, INV-4G+AFNS2, and INV-4G+AFNS2+CPR1, respectively. We investigated the abilities of these three recombinant strains to produce flavones when phenylpropanoid acid precursors were added to the SC-Leu−Trp− culture medium. Expression of the recombinant proteins was induced with galactose, and glucose was replaced with raffinose as the carbon source because raffinose, unlike glucose, does not repress the GAL1 promoter (9). After 46 h, the fermentation products were extracted from the culture media and analyzed using HPLC. The identities of the flavonoid products were determined by cochromatography by matching the UV absorbance spectra (Fig. 2D) and retention times with those authentic standard compounds (Fig. 2A and E). In general, flavonoid accumulation occurred mostly in culture media, and intracellular flavonoid accumulation accounted for less than 10% of the overall flavonoid production. All recombinant strains produced the flavone apigenin (Fig. 2B) when nonhydroxylated cinnamic acid was utilized as the precursor phenylpropanoid acid. Additionally, all recombinant strains were able to metabolize caffeic acid to the corresponding flavone, luteolin, and the corresponding flavanone, eriodictyol (Fig. 2C). Chrysin, however, was detected only in INV-4G+FSI cultures (Fig. 2F). This result confirmed our later finding that snapdragon does not accumulate chrysin. However, it does not exclude the possibility that other FSII enzymes can accept pinocembrin (the flavanone precursor of chrysin) as a substrate. The flavanones pinocembrin (nonhydroxylated) and naringenin (monohydroxylated) were produced by all strains. Incorporation of CPR1 into the flavone biosynthetic pathway with AFNS2 (strain INV-4G+AFNS2+CPR1) resulted in increases in apigenin and luteolin production of 62% and 11%, respectively, compared to the production observed with INV-4G-AFNS2 (Table 3). Addition of ferulic acid did not result in flavanone or flavone production (results not shown).
FIG. 2.
HPLC analysis of recombinant strain INV-4G+FSI and INV-4G+FS2+CPR1 fermentation. (A) Standard compounds separated by using HPLC profile 1. 1, cinnamic acid; 2, p-coumaric acid; 3, caffeic acid; 4, pinocembrin; 5, naringenin; 6, eriodictyol; 7, chrysin; 8, apigenin; 9, luteolin. (B) Apigenin and naringenin produced by the recombinant strains fed p-coumaric acid. (C) Luteolin and eriodictyol produced by the recombinant strains fed caffeic acid. (D) UV absorbance spectra of authentic compounds. (E) Standard compounds separated by using HPLC profile 2. (F) Chrysin, apigenin, pinocembrin, and naringenin produced by recombinant strain INV-4G+FSI fed cinnamic acid. The insets show the UV-visible spectra of flavonoid substances produced by the recombinant strains superimposed with the spectra of the authentic compounds.
TABLE 3.
Comparison of flavonoid production by recombinant yeast strains grown in SC-Leu−Trp− minimal medium with raffinose as the carbon source or in MA-Leu−Trp− minimal medium with acetate as the carbon sourcea
| Compound | Raffinose
|
Acetate
|
% Increase in specific production | ||
|---|---|---|---|---|---|
| Specific production (μg liter−1A600−1) | A600 | Specific production (μg liter−1A600−1) | A600 | ||
| INV-4G+FSI | |||||
| Chrysin | 24 ± 2 | 11.1 ± 0.3 | 150 ± 30 | 6.0 ± 0.8 | 525 |
| Apigenin | 23 ± 5 | 11.1 ± 0.3 | 33 ± 3 | 6.0 ± 0.8 | 44 |
| Pinocembrin | 7 ± 0 | 11.1 ± 0.3 | 213 ± 20 | 6.0 ± 0.8 | 2,943 |
| Naringenin | 4 ± 2 | 11.1 ± 0.3 | 10 ± 3 | 6.0 ± 0.8 | 150 |
| Luteolin | 93 ± 45 | 13.6 ± 0.7 | 210 ± 60 | 7.5 ± 0.6 | 126 |
| Eriodictyol | 536 ± 100 | 13.6 ± 0.7 | 576 ± 30 | 7.5 ± 0.6 | 8 |
| INV-4G+AFNS2 | |||||
| Chrysin | 0 | 8.9 ± 0.5 | 0 | 6.3 ± 0.9 | |
| Apigenin | 29 ± 7 | 8.9 ± 0.5 | 48 ± 4 | 6.3 ± 0.9 | 66 |
| Pinocembrin | 70 ± 20 | 8.9 ± 0.5 | 220 ± 30 | 6.3 ± 0.9 | 214 |
| Naringenin | 4 ± 2 | 8.9 ± 0.5 | 11 ± 3 | 6.3 ± 0.9 | 175 |
| Luteolin | 148 ± 40 | 11.3 ± 0.5 | 160 ± 4 | 5.9 ± 1.3 | 8 |
| Eriodictyol | 500 ± 90 | 11.3 ± 0.5 | 752 ± 80 | 5.9 ± 1.3 | 50 |
| INV-4G+AFNS2+CPR1 | |||||
| Chrysin | 0 | 9.7 ± 1.8 | 0 | 7.2 ± 0.6 | |
| Apigenin | 46 ± 7 | 9.7 ± 1.8 | 57 ± 20 | 7.2 ± 0.6 | 24 |
| Pinocembrin | 87 ± 20 | 9.7 ± 1.8 | 194 ± 60 | 7.2 ± 0.6 | 123 |
| Naringenin | 8 ± 2 | 9.7 ± 1.8 | 9 ± 1 | 7.2 ± 0.6 | 13 |
| Luteolin | 165 ± 20 | 12.8 ± 0.2 | 190 ± 30 | 5.8 ± 0.9 | 15 |
| Eriodictyol | 358 ± 10 | 12.8 ± 0.2 | 423 ± 90 | 5.8 ± 0.9 | 18 |
Chrysin, apigenin, pinocembrin, and naringenin were derived from cinnamic acid. Luteolin and eriodictyol were derived from caffeic acid. The values are means ± standard deviations derived from three independent cultures.
Effect of carbon source on flavonoid production.
In an effort to further optimize flavonoid biosynthesis, we investigated the role of the carbon source (more specifically, acetate) in flavone production. The recombinant yeast strains were grown in MA-Leu−Trp− minimal medium with acetate as the sole carbon source. The fermentation resulted in a general increase in flavonoid production. More specifically, in the case of INV-4G+FSI, the specific production of flavones and flavanones increased 3-fold and 1.5-fold, respectively, when acetate was used as the carbon source instead of raffinose. In the case of INV-4G+AFNS2+CPR1, fermentation in MA medium increased flavone and flavanone specific production 1.2-fold and 1.4-fold, respectively, compared to that in SC minimal medium with raffinose. A summary of the specific productivities obtained with acetate and raffinose as the carbon sources is shown in Table 3.
Overall, these results demonstrated that acetate is a better carbon source for flavone biosynthesis and that flavone production by recombinant yeast can be a competitive alternative to the method currently used, plant extraction. To further prove the last point, we extracted apigenin and luteolin from parsley leaves (in which flavones are produced via the FSI route) and snapdragon petals (in which flavones are produced via the AFNS2 route). Approximately 3 mg of apigenin and 0.01 mg of luteolin per mg of leaves were extracted from parsley, and 0.2 μg of apigenin and 0.05 μg of luteolin per mg of petals were extracted from snapdragon. In both cases, no chrysin was identified.
Regulation of flavanone biosynthesis by flavones.
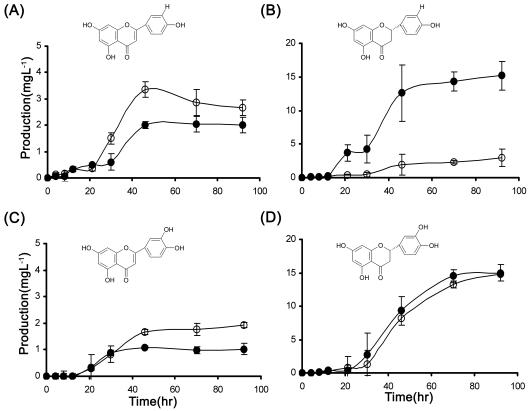
We tested the possibility that flavanone biosynthesis is feedback inhibited by apigenin because a 50% increase in production of apigenin in the FSI expression strain (compared with the AFNS2 analog) reduced naringenin accumulation by 480%. More specifically, as shown in Fig. 3A and B, in the stationary phase the levels of apigenin and naringenin production were 3 mg/liter and 2.4 mg/liter, respectively, for INV-4G+FSI and 2 mg/liter and 14 mg/liter, respectively, for INV-4G+AFNS2. Adding caffeic acid to both recombinant strains resulted in similar production of luteolin and accumulation of eriodictyol (Fig. 3C and D).
FIG. 3.
Flavone and flavanone biosynthesis by two yeast recombinant strains. (A) Biosynthesis of apigenin from p-coumaric acid; (B) biosynthesis of naringenin from p-coumaric acid; (C) biosynthesis of luteolin from caffeic acid; (D) biosynthesis of eriodictyol from caffeic acid. ○, INV-4G+FSI; •, INV-4G+AFNS2+CPR1. All fermentations were carried out in SC-Leu−Trp− minimal medium with raffinose as the carbon source.
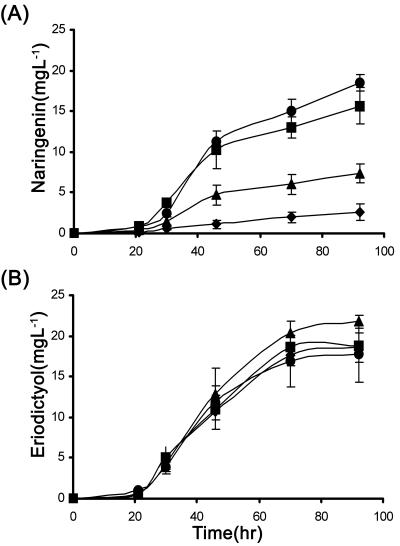
For this purpose, recombinant yeast strain INV-4G was created, which carried plasmid YP-4G (expressing the four flavanone biosynthetic enzymes) and empty plasmid YCplac22. INV-4G was then tested for its ability to produce naringenin from p-coumaric acid in the presence of different apigenin concentrations. As shown in Fig. 4A, in the presence of 1 mg/liter apigenin, the production of naringenin in the stationary phase was reduced by 14%. A more prominent effect was observed in the presence of 3 mg/liter and 10 mg/liter apigenin, which resulted in approximately 60% and 85% decreases in production. It is important to note that in all cases, the decrease in naringenin production was not due to growth reduction because all cultures displayed similar growth profiles. Overall, these data indicated that apigenin acts as a feedback inhibitor of flavanone biosynthesis in a dose-dependent manner.
FIG. 4.
Flavanone production by yeast recombinant strain INV-4G. (A) Naringenin produced from p-coumaric acid in the presence of various concentrations of apigenin; (B) eriodictyol produced from caffeic acid in the presence of various concentrations of luteolin. ▪, 1 mg/liter of flavone; ▴, 3 mg/liter of flavone; ⧫, 10 mg/liter of flavone; •, control (no flavone).
Similarly, we also tested the efficiency of eriodictyol production by INV-4G with caffeic acid as the precursor phenylpropanoid acid and in the presence of various luteolin concentrations. Unlike apigenin, luteolin did not regulate (feedback inhibit) flavanone biosynthesis (Fig. 4B).
DISCUSSION
We describe the construction and comparison of the biosynthetic circuits for three plant-specific flavone molecules, chrysin, apigenin, and luteolin, in S. cerevisiae. An attractive property of yeast is its ability to functionally express plant cytochrome P450 monooxygenases, many of which are involved in flavonoid hydroxylation reactions. Functional expression of eukaryotic membrane-bound enzymes in prokaryotes is not trivial, and it usually requires colocalization of the redox partner and the P450 (2, 3, 13, 25) or extensive protein engineering, such as fusions between the two proteins (11, 32). In the present work, we first tested the efficiency of two P450 reductases, the native S. cerevisiae CPR1 and the C. roseus-derived CPR, for enhancement of snapdragon AFNS2 activity. Despite the limited sequence similarity between yeast CPR1 and its plant equivalents, our in vitro assays demonstrated that AFNS2 converted the substrate naringenin into apigenin more efficiently in the presence of CPR1 than in the presence of CPR. It is therefore possible that expression of the yeast reductase is superior in its native system than expression of the plant-derived protein. Moreover, in support of the in vitro data, episomal overexpression of CPR1 led to increases as great as 60% in the total amounts of flavones produced by the yeast recombinant strain (Table 3). However, these increases in flavone production resulting from CPR1 overexpression were most likely due to increases in the enzyme activities of both P450 monooxygenases present in the recombinant pathway, namely, C4H and AFNS2.
The use of acetate as a sole carbon source led to overall increases in flavone-specific production that were as great as 515% in the case of chrysin produced by strain INV-4G+FSI (Table 3). We tested acetate because recently Daran-Lapujade et al. demonstrated that growth of yeast on acetate led to a 24-fold increase in the carbon flux through acetyl-CoA synthase, the enzyme that converts acetate to acetyl-CoA (6, 33). This was the greatest increase in acetyl-CoA synthase flux observed in that study for all of the different carbon sources tested. Since we have hypothesized that malonyl-CoA, a flavonoid precursor, is a limiting metabolite in overall flavonoid biosynthesis, we believe that the observed increase in flavone specific production was a direct consequence of an increase in the amount of the intracellular malonyl-CoA pool directly derived from acetyl-CoA.
The recombinant yeast strains also provided insight into the regulation of flavone biosynthesis. The INV-4G+FSI strain generated approximately 50% more apigenin and six times less naringenin than INV-4G+AFNS2+CPR1. To explain this phenomenon, we demonstrated that apigenin inhibited naringenin biosynthesis in a dose-dependent manner. Moreover, we demonstrated that there was a nonlinear relationship between the apigenin concentration and the total amount of naringenin produced by the recombinant flavanone biosynthetic pathway. On the other hand, eriodictyol synthesis did not decrease when luteolin was present. It is possible that apigenin, a stable analog of 2′,4,4′,6′-tetrahydroxychalcone, could act as a competitive inhibitor of 4-coumaroyl-CoA binding in the condensation reaction catalyzed by CHS.
Overall, the relatively low plant flavone contents (3 mg and 2 μg total flavones per mg of parsley and snapdragon tissue, respectively) could be a barrier for commercial large-scale production. In that respect, the results presented in this study demonstrate that recombinant yeast can be a competitive alternative to plant extraction for flavone production, without excluding the possibility of further optimization through further increases in the malonyl-CoA pool. One of our immediate goals is to test the effect of acetyl-CoA carboxylase (the enzyme that converts acetyl-CoA to malonyl-CoA) overexpression on flavone biosynthesis. Considering the nonlinear response of this enzyme activity to gene copy number and its complicated transcriptional regulation in E. coli (14, 18), such work may prove to be more challenging than initially expected.
.
Acknowledgments
This work was supported by a research grant from the U.S. National Science Foundation (BES-0331404) to M.A.G.K.
We thank Stefan Martens (Philipps-Universität Marburg, Germany) and Lixuan Huang (DuPont Central Research and Development, United States) for helpful discussions and suggestions. We also thank Amalia Koffas for providing her experience in plant cultivation.
REFERENCES
- 1.Akashi, T., M. Fukuchi-Mizutani, T. Aoki, Y. Ueyama, K. Yonekura-Sakakibara, Y. Tanaka, T. Kusumi, and S. Ayabe. 1999. Molecular cloning and biochemical characterization of a novel cytochrome P450, flavone synthase II, that catalyzes direct conversion of flavanones to flavones. Plant Cell Physiol. 40:1182-1186. [DOI] [PubMed] [Google Scholar]
- 2.Bell, S. G., R. J. Sowden, and L. L. Wong. 2001. Engineering the heme monooxygenase cytochrome P450(cam) for monoterpene oxidation. Chem. Commun. 7:635-636. [Google Scholar]
- 3.Blake, J. A. R., M. Pritchard, S. H. Ding, G. C. M. Smith, B. Burchell, C. R. Wolf, and T. Friedberg. 1996. Coexpression of a human P450 (CYP3A4) and P450 reductase generates a highly functional monooxygenase system in Escherichia coli. FEBS Lett. 397:210-214. [DOI] [PubMed] [Google Scholar]
- 4.Brewer, C. B., and J. A. Peterson. 1986. Single-turnover kinetics of oxy-cytochrome-P-450Cam with reduced putidaredoxin. Fed. Proc. 45:1506. [Google Scholar]
- 5.Brewer, C. B., and J. A. Peterson. 1986. Single turnover studies with oxy-cytochrome P-450 Cam. Arch. Biochem. Biophys. 249:515-521. [DOI] [PubMed] [Google Scholar]
- 6.Daran-Lapujade, P., M. L. Jansen, J. M. Daran, W. van Gulik, J. H. de Winde, and J. T. Pronk. 2004. Role of transcriptional regulation in controlling fluxes in central carbon metabolism of Saccharomyces cerevisiae. A chemostat culture study. J. Biol. Chem. 279:9125-9138. [DOI] [PubMed] [Google Scholar]
- 7.Gietz, R. D., and R. H. Schiestl. 1995. Transforming yeast with DNA. Methods Mol. Cell. Biol. 5:255-269. [Google Scholar]
- 8.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the Liac/S-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 9.Guthrie, C., G. R. Fink, M. I. Simon, and J. N. Abelson (ed.). 1991. Guide to yeast genetics and molecular biology. Methods Enzymol. 194:1-863. [PubMed] [Google Scholar]
- 10.Harbone, A. J. 1998. Phytochemical methods: a guide to modern techniques of plant analysis, 3rd ed. Springer, New York, N.Y.
- 11.Hotze, M., G. Schroder, and J. Schroder. 1995. Cinnamate 4-hydroxylase from Catharanthus roseus, and a strategy for the functional expression of plant cytochrome P450 proteins as translational fusions with P450 reductase in Escherichia coli. FEBS Lett. 374:345-350. [DOI] [PubMed] [Google Scholar]
- 12.Hwang, E. I., M. Kaneko, Y. Ohnishi, and S. Horinouchi. 2003. Production of plant-specific flavanones by Escherichia coli containing an artificial gene cluster. Appl. Environ. Microbiol. 69:2699-2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwata, H., K. Fujita, H. Kushida, A. Suzuki, Y. Konno, K. Nakamura, A. Fujino, and T. Kamataki. 1998. High catalytic activity of human cytochrome P450 co-expressed with human NADPH-cytochrome P450 reductase in Escherichia coli. Biochem. Pharmacol. 55:1315-1325. [DOI] [PubMed] [Google Scholar]
- 14.James, E. S., and J. E. Cronan. 2004. Expression of two Escherichia coli acetyl-CoA carboxylase subunits is autoregulated. J. Biol. Chem. 279:2520-2527. [DOI] [PubMed] [Google Scholar]
- 15.Jiang, H., K. V. Wood, and J. A. Morgan. 2005. Metabolic engineering of the phenylpropanoid pathway in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 71:2962-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaltenbach, M., G. Schroder, E. Schmelzer, V. Lutz, and J. Schroder. 1999. Flavonoid hydroxylase from Catharanthus roseus: cDNA, heterologous expression, enzyme properties and cell-type specific expression in plants. Plant J. 19:183-193. [DOI] [PubMed] [Google Scholar]
- 17.Lebeau, J., R. Neviere, and R. Cotelle. 2001. Beneficial effects of different flavonoids on functional recovery after ischemia and reperfusion in isolated rat heart. Bioorg. Med. Chem. Lett. 11:23-27. [DOI] [PubMed] [Google Scholar]
- 18.Li, S. J., and J. E. Cronan. 1993. Growth rate regulation of Escherichia coli acetyl coenzyme A carboxylase, which catalyzes the first committed step of lipid biosynthesis. J. Bacteriol. 175:332-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lukacin, R., U. Matern, K. T. Junghanns, M. L. Heskamp, L. Britsch, G. Forkmann, and S. Martens. 2001. Purification and antigenicity of flavone synthase I from irradiated parsley cells. Arch. Biochem. Biophys. 393:177-183. [DOI] [PubMed] [Google Scholar]
- 20.Martens, S., and G. Forkmann. 1999. Cloning and expression of flavone synthase II from Gerbera hybrids. Plant J. 20:611-618. [DOI] [PubMed] [Google Scholar]
- 21.Martens, S., and G. Forkmann. 1998. Genetic control of flavone synthase II activity in flowers of Gerbera hybrids. Phytochemistry 49:1953-1958. [Google Scholar]
- 22.Martens, S., G. Forkmann, U. Matern, and R. Lukacin. 2001. Cloning of parsley flavone synthase I. Phytochemistry 58:43-46. [DOI] [PubMed] [Google Scholar]
- 23.Middleton, E., Jr., C. Kandaswami, and T. C. Theoharides. 2000. The effects of plant flavonoids on mammalian cells: implications for inflammation, heart disease, and cancer. Pharmacol. Rev. 52:673-751. [PubMed] [Google Scholar]
- 24.Miksicek, R. J. 1995. Estrogenic flavonoids—structural requirements for biological activity. Proc. Soc. Exp. Biol. Med. 208:44-50. [DOI] [PubMed] [Google Scholar]
- 25.Parikh, A., E. M. J. Gillam, and F. P. Guengerich. 1997. Drug metabolism by Escherichia coli expressing human cytochromes P450. Nat. Biotechnol. 15:784-788. [DOI] [PubMed] [Google Scholar]
- 26.Pompon, D., B. Louerat, A. Bronine, and P. Urban. 1996. Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol. 272:51-64. [DOI] [PubMed] [Google Scholar]
- 27.Ross, J. A., and C. M. Kasum. 2002. Dietary flavonoids: bioavailability, metabolic effects, and safety. Annu. Rev. Nutr. 22:19-34. [DOI] [PubMed] [Google Scholar]
- 28.Rump, A. F. E., M. Schussler, D. Acar, A. Cordes, M. Theisohn, R. Rosen, W. Klaus, and U. Fricke. 1994. Functional and antiischemic effects of luteolin-7-glucoside in isolated rabbit hearts. Gen. Pharmacol. 25:1137-1142. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Schoenbohm, C., S. Martens, C. Eder, G. Forkmann, and B. Weisshaar. 2000. Identification of the Arabidopsis thaliana flavonoid 3′-hydroxylase gene and functional expression of the encoded P450 enzyme. Biol. Chem. 381:749-753. [DOI] [PubMed] [Google Scholar]
- 31.Schussler, M., J. Holzl, A. F. E. Rump, and U. Fricke. 1995. Functional and antiischaemic effects of monoacetyl-vitexinrhamnoside in different in vitro models. Gen. Pharmacol. 26:1565-1570. [DOI] [PubMed] [Google Scholar]
- 32.Sieber, V., C. A. Martinez, and F. H. Arnold. 2001. Libraries of hybrid proteins from distantly related sequences. Nat. Biotechnol. 19:456-460. [DOI] [PubMed] [Google Scholar]
- 33.Staral, V. J., and J. C. Escalante-Semerena. 2004. Acetyl-coenzyme A synthetase (AMP forming). Cell. Mol. Life Sci. 61:2020-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verduyn, C., E. Postma, W. A. Scheffers, and J. P. Vandijken. 1992. Effect of benzoic acid on metabolic fluxes in yeasts—a continuous culture study on the regulation of respiration and alcoholic fermentation. Yeast 8:501-517. [DOI] [PubMed] [Google Scholar]
- 35.Viola, H., C. Wasowski, M. L. Destein, C. Wolfman, R. Silveira, F. Dajas, J. H. Medina, and A. C. Paladini. 1995. Apigenin, a component of Matricaria recutita flowers, is a central benzodiazepine receptors ligand with anxiolytic effects. Planta Med. 61:213-216. [DOI] [PubMed] [Google Scholar]
- 36.Wolfman, C., H. Viola, A. Paladini, F. Dajas, and J. H. Medina. 1994. Possible anxiolytic effects of chrysin, a central benzodiazepine receptor ligand isolated from Passiflora coerulea. Pharmacol. Biochem. Behav. 47:1-4. [DOI] [PubMed] [Google Scholar]
- 37.Wolfman, C., H. Viola, C. Wasowski, M. L. Destein, R. Silveira, F. Dajas, J. H. Medina, and A. C. Paladini. 1995. Apigenin, a component of Matricaria recutita (L.) flowers, is a central benzodiazepine receptor ligand with anxiolytic effects. J. Neurochem. 65:S167-S167. [DOI] [PubMed] [Google Scholar]
- 38.Yan, Y., J. Chemler, L. Huang, S. Martens, and M. A. Koffas. 2005. Metabolic engineering of anthocyanin biosynthesis in Escherichia coli. Appl. Environ. Microbiol. 71:3617-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan, Y., A. Kohli, and M. A. Koffas. 2005. Biosynthesis of natural flavanones in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 71:5610-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]