Abstract
We developed a new cell surface display system in Pseudomonas putida KT2442 using OprF, an outer membrane protein of Pseudomonas aeruginosa, as an anchoring motif in a C-terminal deletion-fusion strategy. The Pseudomonas fluorescens SIK W1 lipase gene was fused to two different C-terminal truncated OprF genes, and the fusion genes were cloned into the broad-host-range plasmid pBBR1MCS2 to make pMO164PL and pMO188PL. Plasmid pMO188PL allowed better display of lipase and thus was chosen for further study. The display of lipase on the surface of P. putida KT2442 was confirmed by Western blot analysis, immunofluorescence microscopy, and measurement of whole-cell lipase activity. The whole-cell lipase activity of recombinant P. putida KT2442 harboring pMO188PL was more than fivefold higher than that of recombinant Escherichia coli displaying lipase in the same manner. Cell surface-displayed lipase exhibited the highest activity at 47°C and pH 9.0, and the whole-cell lipase activity was greater than 90% of the initial activity in organic solvents at 47°C for 1 week. In a biocatalytic application, enantioselective resolution of 1-phenyl ethanol was carried out in an organic solvent. (R)-Phenyl ethyl acetate was successfully produced with 41.9% conversion and an enantiomeric excess of more than 99% in a 36-h reaction. These results suggest that the OprF anchor can be used for efficient display of proteins in P. putida KT2442 and consequently for various biocatalytic applications.
Cell surface display is a technique for expressing heterologous peptides or proteins on the surface of gram-negative and gram-positive bacteria, fungi, or even mammalian cells by fusing the peptides or proteins to an appropriate fusion partner (6, 16, 28). This technique has a wide range of biotechnological and industrial applications, including the development of vaccines, peptide and antibody libraries, bioremediation, biocatalysis, and biosensing. Many different proteins have been employed as anchoring motifs, and these proteins include various outer membrane proteins, lipoproteins, autotransporters, subunits of surface appendages, and S-layer proteins (6, 7, 9, 16). Of these, outer membrane proteins have been widely used as anchoring motifs for the display of peptides and proteins because they have several advantages, such as efficient secretory signals, unique membrane-spanning structures that provide fusion sites, and strong anchoring structures. Several outer membrane proteins, such as OmpA, OprF, OmpS, FadL, LamB, PhoE, OmpC, and Lpp-OmpA, have been used successfully as anchoring motifs for displaying various peptides and proteins (2, 26, 29).
So far, many bacteria have been examined as host strains for cell surface display, including Escherichia coli, Salmonella, Caulobacter, Lactobacillus, Moraxella, Proteus, Pseudomonas, Streptococcus, Staphylococcus, and Bacillus (3, 26). Of these, Pseudomonas strains are well known for their tolerance of organic solvents (23). This characteristic of Pseudomonas strains has been used in various fields, including biocatalysis, bioremediation, and biosensing. However, there have been few studies of cell surface display in Pseudomonas strains.
In this paper, we describe a new cell surface display system in Pseudomonas putida in which Pseudomonas aeruginosa OprF is used as an anchoring motif. Pseudomonas fluorescens SIK W1 lipase was used as a model protein for display on the cell surface. Using P. putida KT2442 displaying lipase as a whole-cell biocatalyst, we observed successful chiral resolution of 1-phenyl ethanol in organic solvents.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All bacterial strains and plasmids used in this study are listed in Table 1. E. coli XL-1 Blue was used as a host strain for general cloning, and E. coli XL-10 Gold and P. putida KT2442 were used as host strains for cell surface display studies. For transformation of P. putida KT2442 with a plasmid, competent cells were prepared as follows. P. putida KT2442 cells were grown to an optical density at 600 nm (OD600) of 0.3, harvested by centrifugation at 3,500 × g for 5 min at 4°C, and resuspended in 10 ml of 0.1 M MgCl2 for 2 h. Then cells were harvested by centrifugation at 3,500 × g for 5 min at 4°C and resuspended in 0.1 M MgCl2 with 15% (wt/vol) glycerol. Competent cells were stored at −80°C until they were needed. All DNA manipulations, including restriction digestion, ligation, and agarose gel electrophoresis, were carried out using standard procedures (24).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| XL-10 Gold | Tetr Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac Hte [F′ proAB lacIqZΔM15 Tn10 (Tetr) Amy Camr] | Stratagenea |
| XL-1 Blue | supE44 hsdR17 recA1 endA1 gyrA+96 thi relA1 lac F′(proAB+lacIqlacZΔM15 Tn10(Tetr) | Stratagenea |
| Pseudomonas putida KT2442 | 4 | |
| Plasmids | ||
| pTacOprF164PL | P. fluorescens SIK W1 lipase gene | 14 |
| pTacOprF188PL | P. fluorescens SIK W1 lipase gene | 14 |
| pBBR1MCS2 | Broad-host-range vector | 13 |
| pMO164PL | pBBR1MCS2 derivative | This study |
| pMO188PL | pBBR1MCS2 derivative | This study |
Stratagene, La Jolla, CA.
Culture conditions.
Recombinant cells were cultivated in Luria-Bertani (LB) medium (10 g/liter Bacto tryptone, 5 g/liter Bacto yeast extract, 5 g/liter NaCl) supplemented with either 50 mg/ml of ampicillin or 34 mg/ml of kanamycin depending on the plasmid used. At an OD600 of 0.4, cells were induced with isopropyl-β-d-thiogalactopyranoside (IPTG) for production of recombinant proteins. Cells were cultured for 4 h after induction.
Western blotting.
Whole-cell lysates and the membrane fraction were analyzed by 10% (wt/vol) sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). To prepare outer membrane proteins, 3 ml of culture broth was centrifuged at 3,500 × g for 5 min at 4°C, and the cell pellet was washed with 1 ml of 10 mM Na2HPO4 buffer (pH 7.2); this was followed by centrifugation at 3,500 × g for 5 min at 4°C. The resulting cell pellet was resuspended in 0.5 ml of 10 mM Na2HPO4 buffer (pH 7.2). Crude extracts of recombinant E. coli cells were prepared by three cycles of sonication (20 s each at 15% of maximum output; high-intensity ultrasonic liquid processors; Sonics & Material Inc., Newtown, CT). Partially disrupted cells were removed by centrifugation of sonicated samples at 12,000 × g for 2 min at room temperature. Membrane proteins and the lipid layer were isolated by centrifugation at 12,000 × g for 30 min at 4°C, followed by resuspension in 0.5 ml of 10 mM Na2HPO4 buffer (pH 7.2) containing 0.5% (wt/vol) sarcosyl. After incubation at 37°C for 30 min, an insoluble pellet containing membrane proteins was obtained by centrifugation at 12,000 × g for 30 min at 4°C. Membrane proteins were obtained by washing the insoluble pellet with 10 mM Na2HPO4 buffer (pH 7.2), which was followed by resuspension in 50 μl of Tris-EDTA buffer (pH 8.0).
The Western blot analysis was performed by using standard protocols (24). For immunodetection of the fusion protein, rat antilipase antibodies (Peptron, Daejeon, Korea) and rabbit anti-rat immunoglobulin G (IgG)-horseradish peroxidase conjugate (Sigma Chemical Co., St. Louis, MO) were used. A light-emitting nonradioactive ECL kit (Amersham Life Sciences, Buckinghamshire, United Kingdom) was used for signal detection.
Immunofluorescence microscopy.
Cells were harvested by centrifugation at 3,500 × g for 5 min at 4°C, washed with phosphate-buffered saline (PBS), and resuspended in PBS supplemented with 3% (wt/vol) bovine serum albumin (BSA) (Sigma Chemical Co., St. Louis, MO). Cells were incubated with rat antilipase antibody (Peptron, Daejeon, Korea) diluted 1:1,000 in PBS containing 3% (wt/vol) BSA at 4°C for 4 h. After it was washed five times with PBS, the cell-antibody complex was incubated overnight at 4°C with rabbit anti-rat IgG conjugated with fluorescein isothiocyanate (FITC) (Sigma) at a dilution of 1:3,000. Prior to microscopic observation, cells were washed five times with PBS to remove unbound anti-rat IgG conjugated with FITC. For immunofluorescence microscopy, cells were mounted on poly-l-lysine-coated microscopic slide glasses and examined by confocal microscopy (Carl Zeiss, Jena, Germany). Photographs were taken with a Carl Zeiss LSM 410.
FACS analysis.
For the fluorescence-activated cell sorting (FACS) analysis, cells were harvested, and the cell pellet was washed three times with 1 ml of PBS; this was followed by resuspension in 1 ml of PBS (OD600, 0.01) containing 2% (wt/vol) BSA and rat antilipase antibody (1:1,000). The cells were incubated for 2 h at 4°C. After it was washed with PBS five times, the cell-antibody complex was incubated with FITC-conjugated anti-rat IgG antibody (1:100) on ice for 1 h. Prior to analysis, cells were washed five times with PBS to remove unbound rabbit anti-rat IgG conjugated with FITC. The FITC-labeled cells were examined using a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA).
Measurement of lipase activities.
Cells were cultivated in a 250-ml flask containing 100 ml LB medium at 30°C and 250 rpm. At an OD600 of 0.4, cells were induced with 0.1 or 1 mM IPTG and then cultured for 4 h. The cells were harvested by centrifugation for 5 min at 5,590 × g and 4°C, washed with distilled water, and lyophilized with a freeze dryer (TFD5505; Ilshin Lab, Gyeonggi-do, Korea) for 48 h.
Lipase activity was assayed by a spectrophotometric method using p-nitrophenyl decanoate as the substrate. The p-nitrophenyl decanoate was dissolved in acetonitrile at a concentration of 10 mM. Ethanol and 50 mM Tris-HCl (pH 8.0) were subsequently added to obtain a substrate solution having a ratio of 10 mM p-nitrophenyl decanoate in acetonitrile to ethanol to Tris-HCl of 1:4:95 (vol/vol/vol). Lyophilized cells (0.15 mg) or culture supernatant (500 μl) was added to 3 ml of the substrate solution for determination of lipase activity. After the reaction mixture was incubated at 37°C for 10 min, the activity was assayed by detecting the product, p-nitrophenol, spectrophotometrically at 405 nm. The reaction was terminated by adding 2 μl of 0.5 M EDTA.
Temperature-dependent lipase activities were examined by using the substrate solution described above at controlled temperatures ranging from 27 to 67°C. The optimal pH was determined at 37°C using substrate solutions having a ratio of 10 mM p-nitrophenyl decanoate in acetonitrile to ethanol to 50 mM potassium phosphate or 50 mM Tris-HCl of 1:4:95 (vol/vol/vol) at various pHs (range, pH 5 to 10). For examination of the stability at a high temperature in organic solvents, lyophilized cells (10 mg) were resuspended in 10 ml Tris-HCl (pH 8.0) or organic solvents, such as hexane and isopropanol, and incubated at 47°C for 1 week. Then 0.1-ml aliquots were removed at intervals, and the cells were harvested by centrifugation and resuspended in 1 ml substrate solution for measurement of the residual activity at 37°C for 10 min. For comparative studies, free P. fluorescens SIK W1 lipase (Genofocus, Daejeon, Korea) was used. Five microliters of free enzyme solution (20 g/liter of free lipase in 50 mM Tris-HCl [pH 8.0]) was added to 1 ml of substrate solution. The activity of the free enzyme was assayed in the same manner. For bioconversion studies, 300 mg of lipase was added to 30 ml of hexane. The reaction was controlled under the conditions described above.
Enantioselective resolution of 1-phenyl ethanol in hexane using P. putida KT2442 cells displaying lipase.
Lyophilized cells (300 mg) prepared by induction with 0.1 mM IPTG were resuspended in 30 ml hexane to which 300 mg racemic 1-phenyl ethanol, 212 mg vinyl acetate, and 3 g molecular sieve 4A (Aldrich, St. Louis, MO) were added. The reaction mixture was incubated at 47°C and 250 rpm. The progress of the reaction was monitored by high-performance liquid chromatography (HPLC) (1100 HPLC system; Agilent, Palo Alto, CA). Enantiomeric excess (ee) was determined as follows: 100 × (A − B)/(A + B), where A and B are the amounts of enantiomers. The conversion percentage was calculated by using ees/(ees + eep), where the subscripts s and p indicate the remaining alcohol and product, respectively.
Analytical methods.
Cell growth was monitored by measuring the optical density at 600 nm using a spectrophotometer (BECKMAN DU650). The yield and optical purity of chemicals were determined with an HPLC equipped with a Chiralcel OB-H column (Daicel Chemical Industries, Tokyo, Japan). A mixture of hexane and isopropanol (90:10, vol/vol) was used as the mobile phase at a flow rate of 0.3 ml/min. The reaction products and substrates were detected by measuring the absorbance at 210 nm using a diode array detector (1100 series DAD; Agilent).
RESULTS
Construction of cell surface display system.
For display of the P. fluorescens SIK W1 lipase (49.9 kDa) (1) on the surface of P. putida KT2442, we first searched for possible fusion sites in the anchoring motif OprF. Since Lys164 and Val188 of P. aeruginosa PAO1 OprF served as successful fusion sites for display of lipase in an active form on the surface of E. coli (14), these sites were selected as the fusion sites in this study.
For production of lipase in P. putida KT2442, the broad-host-range plasmid pBBR1MCS2 (13) and the tac promoter were used. Truncated oprF (oprFt) genes encoding 164 and 188 amino acids from the N terminus fused to the lipase gene were obtained from pTacOprF164PL and pTacOprF188PL, respectively. These plasmids were digested with SspI, and the fragment containing the tac promoter and the oprFt (oprFt164 and oprFt188)-lipase fusion genes were cloned into the EcoRV site of pBBR1MCS2 to obtain pMO164PL and pMO188PL, respectively. Recombinant P. putida KT2442 cells harboring pMO164PL and pMO188PL were cultivated at 30°C and induced with 0.1 and 1 mM IPTG. Growth defects were not observed for recombinant cells.
Confirmation of lipase display on the surface of P. putida KT2442.
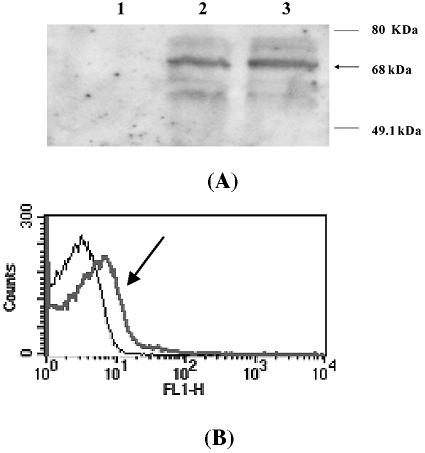
For confirmation of lipase display on the cell surface, we first analyzed the whole-cell lysates and outer membrane proteins of P. putida KT2442(pMO188PL) by SDS-PAGE and Western blotting. The Western blot analysis was carried out using the rat antilipase antibody, followed by detection with horseradish peroxidase-conjugated rabbit anti-rat IgG. As shown in Fig. 1A, approximately 68 kDa of fusion protein was detected in whole-cell lysates (lane 2) and the outer membrane fraction (lane 3). However, a signal was not detected in whole-cell lysates of the negative control, P. putida KT2442 (lane 1).
FIG. 1.
Confirmation of lipase display on the surface of P. putida KT2442. (A) Confirmation by immunoblotting of P. putida expressing OprFt and OprFt-lipase fusion protein. Lane 1, whole-cell lysate of P. putida; lane 2, whole-cell lysate of P. putida harboring pMO188PL; lane 3, outer membrane fraction of P. putida harboring pMO188PL. (B) Confirmation by flow cytometry analysis of P. putida and P. putida harboring pMO188PL. The arrow indicates the profile of recombinant P. putida(pMO188PL). The number of cells analyzed in each experiments was 50,000.
The surface display of lipase was further confirmed by FACS analysis. Fluorescence was monitored for P. putida KT2442 and the recombinant P. putida KT2442(pMO188PL) incubated with antilipase antibody, followed by FITC-conjugated secondary antibody. As shown in Fig. 1B, the mean fluorescence values of P. putida KT2442 and recombinant P. putida KT2442(pMO188PL) were 5.8 and 18.7, respectively.
To examine the display of lipase on the surface of P. putida KT2442 more directly, recombinant cells expressing the OprFt188-lipase fusion protein were observed by immunofluorescence microscopy. P. putida KT2442(pMO188PL) producing the fusion protein became fluorescent due to binding of the antilipase antibody, followed by binding of the FITC-conjugated secondary antibody, indicating that lipase was expressed on the surface of P. putida (data not shown). On the other hand, P. putida KT2442 cells were not fluorescent (data not shown). These results suggest that the lipases were successfully displayed on the cell surface of P. putida KT2442.
After confirming that lipase was displayed on the cell surface of P. putida, we examined whether lipase was active. The whole-cell lipase activities of recombinant P. putida KT2442 harboring pMO164PL and pMO188PL after incubation with 0.1 and 1 mM IPTG were measured and compared with the activities displayed on E. coli. When the whole-cell lipase activity of P. putida(pMO188PL) obtained with 0.1 mM IPTG induction was defined as 100, the relative lipase activities obtained for P. putida(pMO188PL) with 1 mM IPTG, for P. putida(pMO166PL) with 0.1 mM IPTG, for P. putida(pMO166PL) with 1 mM IPTG, for E. coli XL-10 Gold(pTacOprF164PL) with 0.1 mM IPTG, and for E. coli XL-10 Gold(pTacOprF188PL) with 0.1 mM IPTG were 58.3, 72.3, 29.9, 17.2, and 12.4, respectively. The whole-cell activities of P. putida were generally much higher than those obtained with E. coli. The highest activity was observed for P. putida harboring pMO188PL induced with 0.1 mM of IPTG; this activity was more than fivefold higher than the highest activity obtained with E. coli displaying the same lipase. Lipase activity was not detected in the supernatants. These results suggest that lipases were successfully displayed in an active form using OprFt as an anchoring motif on the surface of P. putida KT2442 without significant cell lysis.
Characterization of cell surface-displayed lipase.
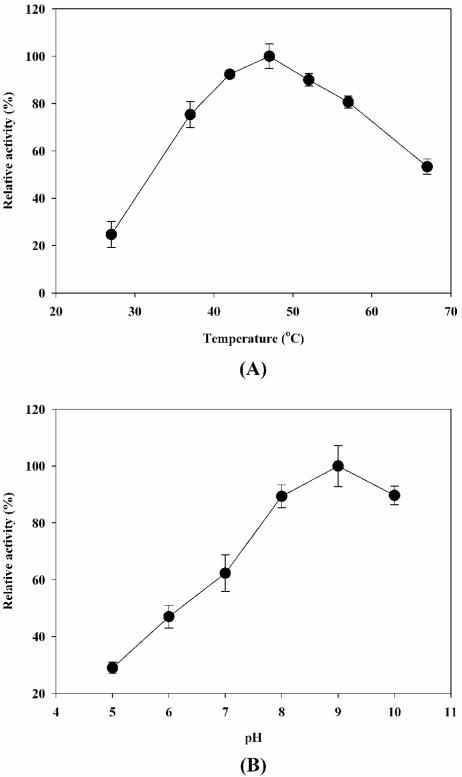
To investigate the optimal conditions for the cell surface-displayed lipase, whole-cell lipase activities of P. putida KT2442 harboring pMO188PL were measured at different temperatures (27 to 67°C) and at pH 5 to 10. The results are shown in Fig. 2. The maximum activity of cell surface-displayed lipase was observed at 47°C and pH 9.0. In the temperature range from 42 to 52°C and in the pH range from 8 to 10, the whole-cell activities were more than 90% of the maximum activity. These characteristics are similar to those of free enzyme but different from those of E. coli displayed lipase (14, 17, 18).
FIG. 2.
Effect of temperature (A) and pH (B) on the activity of surface-displayed lipase. To examine the temperature-dependent activity profiles, enzyme activities were determined at pH 8.0; the relative activity was calculated by assuming that the activity observed at 47°C was 100%. For the pH-dependent activity profiles, enzyme activities were measured at 37°C; the relative activity was calculated by assuming that the activity observed at pH 9.0 was 100%.
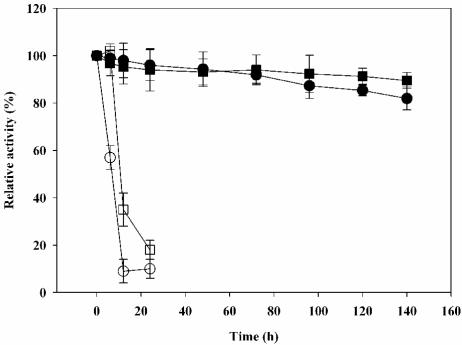
After optimization of the reaction conditions, the enzyme stability in the presence of heat and organic solvents was examined, as stability under these conditions is important for industrial applications. Cell surface-displayed lipase was incubated at 47°C in hexane and isopropanol for 1 week, during which the whole-cell lipase activity was measured periodically. As shown in Fig. 3, the recombinant P. putida cells displaying lipase were quite stable and retained 80% of the initial activity in nonaqueous solvents during the entire reaction time. On the other hand, the whole-cell lipase activity of E. coli XL-10 Gold harboring pTacO164PL rapidly decreased within 30 h. The lipase activity in the supernatant was negligible throughout the entire reaction period, suggesting that cell lysis was not a problem even in an organic solvent.
FIG. 3.
Stability of lipase displayed on the cell surface of E. coli XL-10 Gold harboring pTacO164PL and P. putida KT2442 harboring pMO188PL during prolonged incubation in organic solvents at 47°C. The enzyme activities were determined at 37°C and pH 8.0 using p-nitrophenyl decanoate as the substrate. The relative activity was calculated by assuming that the initial activity was 100%. ○, recombinant E. coli in hexane; (□, recombinant E. coli in isopropanol; •, recombinant P. putida in hexane; ▪, recombinant P. putida in isopropanol.
Biocatalytic applications of cell surface-displayed lipase.
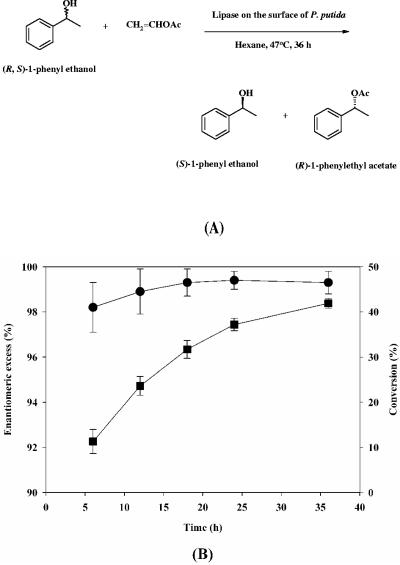
For possible application of the organism as a biocatalyst, we investigated the enantioselective resolution of 1-phenyl ethanol in an organic solvent. This reaction is shown in Fig. 4A. Time profiles for the reaction during enantioselective resolution are shown in Fig. 4B. The enantiomeric excess of the product (R)-phenyl ethyl acetate and the level of conversion obtained in 36 h were 99.5% and 41.9%, respectively. These results suggest that P. putida displaying lipase using the OprFt anchor can be successfully used for enantioselective resolution of chemicals in organic solvents.
FIG. 4.
Reaction (A) and time profiles (B) of the enantioselective resolution of 1-phenyl ethanol in hexane using lipase displayed on the surface of P. putida KT2442. The time profiles for conversion (▪) and enantiomeric excess (•) of reaction products are shown. OAc, acetate.
DISCUSSION
Enzyme reactions in organic solvents have been receiving much attention for the production of enantiomerically pure compounds from racemic substrates due to the increasing demand for chiral intermediates and drugs (5, 11, 21). However, the practical use of enzymes in organic solvents has been limited due to the lower catalytic activity and lower stability compared with the activity and stability of enzymes in an aqueous phase. Numerous immobilization techniques, including adsorption, entrapment, emulsion, and covalent attachment, have been proposed as alternatives to enhance activity and stability (10, 20, 22). However, many of these processes are expensive and have various limitations for actual application to industrial production. As another way to overcome this problem, we examined a genetic immobilization technique, namely, cell surface display of an enzyme in Pseudomonas.
Surprisingly, cell surface display in Pseudomonas has not been studied extensively in spite of the high biocatalytic potential of Pseudomonas displaying enzymes. Shimazu et al. (27) reported successful display of organophosphorus hydrolase on the surface of P. putida using ice nucleation protein as an anchoring motif, and the whole-cell activity of recombinant P. putida was 10-fold greater than that of E. coli. Here, we describe a new cell surface display system for P. putida KT2442 in which the P. aeruginosa outer membrane protein OprF is used as an anchoring motif via a C-terminal deletion-fusion strategy. P. putida cells displaying lipase (harboring pMO188PL) exhibited 5- to 10-fold-higher catalytic activity than E. coli cells displaying lipase. It seems that the efficiencies of membrane translocation and synthesis of the OprFt-lipase fusion protein are higher in P. putida than in E. coli because both OprF and lipase originated from Pseudomonas strains. The number of lipase molecules displayed per cell was estimated as reported previously (14). It was estimated that 1,534 lipase molecules were displayed per cell of P. putida harboring pMO188PL. This value is quite high compared with the value for E. coli cells displaying lipase using OmpC or OprF as an anchoring motif (14, 15). It was also found that induction with a low IPTG concentration resulted in better display of lipase. The fusion protein band was visible on an SDS-PAGE gel when P. putida(pMO188PL) was induced with 0.1 mM IPTG but not when it was induced with 1 mM IPTG, which suggested that the gene expression was better with a low IPTG concentration. The amount of fusion protein displayed on the surface of P. putida(pMO188PL) induced with 0.1 mM IPTG was estimated to be 3.1% of the total membrane proteins.
Most importantly, lipase displayed on the surface of P. putida showed much higher stability in organic solvents than lipase displayed on the surface of E. coli. Lyophilized E. coli and P. putida cells displaying lipase were stable in organic solvents at 37°C. However, the Pseudmonas strain exhibited much higher stability at a high temperature (47°C). This property should be useful for enantioselective biocatalysis in an organic solvent. Lipases have been used as target proteins for several cell surface display systems (8, 12, 14, 15, 19, 25). Of these, only yeast and E. coli have been employed for biocatalysis in an organic solvent at a moderate temperature. Lipase displayed on the surface of P. putida exhibited a conversion value of 41.9% with an enantiomeric excess of more than 99% in 36 h. These results are much better than those obtained with the E. coli display system (14). This performance was possible because the reaction could be carried out at 47°C without inactivation or instability of the whole-cell enzyme.
We also compared the performance of free lipase with the performance of the cell surface-displayed lipase with respect to heat stability and activity in an organic solvent. The activity of the free enzyme was less than 20% of the initial activity after 1 h of incubation at 37°C and further decreased to zero after 4 h of incubation. The performance of the free enzyme in hexane was also investigated by measuring the conversion of 1-phenyl ethanol to (R)-phenyl ethyl acetate. The conversion rate was less than 5% even after 48 h of incubation, which suggests that cell surface-displayed lipase outperforms the free enzyme in terms of heat stability as well as in terms of reacting in an organic solvent.
In conclusion, we developed a new cell surface display system for P. putida using P. aeruginosa OprF as an anchoring motif. Using this system, Pseudomonas sp. lipase was successfully displayed on the surface of P. putida, which exhibited much higher lipase activity and stability in an organic solvent than the enzyme displayed on E. coli. These improved characteristics are expected to result in the use of the Pseudomonas cell surface display system in a wide range of biocatalytic applications.
Acknowledgments
This work was supported by MOCIE grants from the Intelligence Bioinformatics and Application Center (TGW10011093) at the KRIBBand the Center for Ultramicrochemical Process Systems sponsored by KOSEF and by the BK21 project. Further support provided by an LG Chem Chair Professorship and the IBM-SUR program is appreciated.
REFERENCES
- 1.Ahn, J. H., J. G. Pan, and J. S. Rhee. 1999. Identification of the tliDEF ABC transporter specific for lipase in Pseudomonas fluorescens SIK W1. J. Bacteriol. 181:1847-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benhar, I. 2001. Biotechnological applications of phage and cell display. Biotechnol. Adv. 19:1-33. [DOI] [PubMed] [Google Scholar]
- 3.Chen, W., and G. Georgiou. 2002. Cell-surface display of heterologous proteins: from high-throughput screening to environmental applications. Biotechnol. Bioeng. 79:496-503. [DOI] [PubMed] [Google Scholar]
- 4.Christensen, B. B., C. Sternberg, J. B. Andersen, L. Eberl, S. Moller, M. Givskov, and S. Molin. 1998. Establishment of new genetic traits in a microbial biofilm community. Appl. Environ. Microbiol. 64:2247-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drauz, K., and H. Waldmann. 2002. Enzyme catalysis in organic synthesis: a comprehensive handbook, 2nd ed. Wiley-VCH Verlag GmbH, Weinheim, Germany.
- 6.Georgiou, G., C. Stathopoulos, P. S. Daugherty, A. R. Nayak, B. L. Iverson, and R. I. Curtiss. 1997. Display of heterologous proteins on the surface of microorganisms: from the screening of combinatorial libraries to live recombinant vaccines. Nat. Biotechnol. 15:29-34. [DOI] [PubMed] [Google Scholar]
- 7.Jose, J., R. Bernhardt, and F. Hannemann. 2002. Cellular surface display of dimeric Adx and whole cell P450-mediated steroid synthesis on E. coli. J. Biotechnol. 95:257-268. [DOI] [PubMed] [Google Scholar]
- 8.Jung, H. C., S. Ko, S. J. Ju, E. J. Kim, M. K. Kim, and J. G. Pan. 2003. Bacterial cell surface display of lipase and its randomly mutated library facilitates high-throughput screening of mutants showing higher specific activities. J. Mol. Catal. B Enzym. 26:177-184. [Google Scholar]
- 9.Jung. H. C., J. M. Lebeault, and J. G. Pan. 1998. Surface display of Zymomonas mobilis levansucrase by using the ice-nucleation protein of Pseudomonas syringae. Nat. Biotechnol. 16:576-580. [DOI] [PubMed] [Google Scholar]
- 10.Kamori, M., T. Hori, Y. Yamashita, Y. Hirose, and Y. Naoshima. 2000. Immobilization of lipase on a new inorganic ceramics support, toyonite, and the reactivity and enantioselectivity of the immobilized lipase. J. Mol. Catal. B Enzym. 9:269-274. [Google Scholar]
- 11.Klibanov, A. M. 2001. Improving enzymes by using them in organic solvents. Nature 409:241-246. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi, G., K. Fujii, M. Serizawa, H. Yamamoto, and J. Sekiguchi. 2002. Simultaneous display of bacterial and fungal lipases on the cell surface of Bacillus subtilis. J. Biosci. Bioeng. 93:15-19. [DOI] [PubMed] [Google Scholar]
- 13.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 14.Lee, S. H., J. H. Choi, S. H. Park, J. Choi, and S. Y. Lee. 2004. Enantioselective resolution of racemic compounds by cell surface displayed lipase. Enzyme Microb. Technol. 35:429-436. [Google Scholar]
- 15.Lee, S. H., J. Choi, M.-J. Han, J. H. Choi, and S. Y. Lee. 2005. Display of lipase on the cell surface of Escherichia coli using OprF as an anchor and its application to enantioselective resolution in organic solvent. Biotechnol. Bioeng. 90:223-230. [DOI] [PubMed] [Google Scholar]
- 16.Lee, S. Y., J. H. Choi, and Z. Xu. 2003. Microbial cell surface display. Trends Biotechnol. 21:45-52. [DOI] [PubMed] [Google Scholar]
- 17.Lee, Y. P., G. H. Chung, and J. S. Rhee. 1993. Purification and characterization of Pseudomonas fluorescens SIK W1 lipase expressed in Escherichia coli. Biochim. Biophys. Acta 1169:156-164. [DOI] [PubMed] [Google Scholar]
- 18.Lee. Y. P., and J. S. Rhee. 1996. Protein aggregation and adsorption upon in vitro refolding of recombinant Pseudomonas lipase. J. Microbiol. Biotechnol. 6:456-460. [Google Scholar]
- 19.Matsumoto, T., M. Ito, H. Fukuda, and A. Kondo. 2004. Enantioselective transesterification using lipase-displaying yeast whole-cell biocatalyst. Appl. Microbiol. Biotechnol. 64:481-485. [DOI] [PubMed] [Google Scholar]
- 20.Palomo, J. M., G. Fernandez-Lorente, C. Mateo, C. Ortiz, R. Fernandez-Lafuente, and J. M. Guisan. 2002. Modulation of the enantioselectivity of lipases via controlled immobilization and medium engineering: hydrolytic resolution of mandelic acid esters. Enzyme Microb. Technol. 31:775-783. [Google Scholar]
- 21.Patel, R. N. 2000. Stereoselective biocatalysis, Marcel Dekker, New York, N.Y.
- 22.Persson, M., I. Mladenoska, E. Wehtje, and P. Adlerceutz. 2002. Preparation of lipases for use in organic solvents. Enzyme Microb. Technol. 31:833-841. [Google Scholar]
- 23.Ramos, J. L., E. Duque, M.-T. Gallegos, P. Godoy, M. I. Ramos-Gonzalez, A. Rojas, W. Teran, A. Segura. 2002. Mechanisms of solvent tolerance in gram-negative bacteria. Annu. Rev. Microbiol. 56:743-768. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Samuelson, P., F. Cano, A. Robert, and S. Ståhl. 1999. Engineering of a Staphylococcus carnosus surface display system by substitution or deletion of a Staphylococcus hyicus lipase propeptide. FEMS Lett. 179:131-139. [DOI] [PubMed] [Google Scholar]
- 26.Samuelson, P., E. Gunneriusson, P. A. Nygren, and S. Ståhl. 2002. Display of proteins on bacteria. J. Biotechnol. 96:129-154. [DOI] [PubMed] [Google Scholar]
- 27.Shimazu, M., A. Nuyen, A. Mulchandani, and W. Chen. 2003. Cell surface display of organophosphorus hydrolase in Pseudomonas putida using ice nucleation protein anchor. Biotechnol. Prog. 19:1612-1614. [DOI] [PubMed] [Google Scholar]
- 28.Wittrup, K. D. 2001. Protein engineering by cell-surface display. Curr. Opin. Biotechnol. 12:395-399. [DOI] [PubMed] [Google Scholar]
- 29.Xu, Z., and S. Y. Lee. 1999. Display of polyhistidine peptides on the Escherichia coli cell surface by using outer membrane protein C as an anchoring motif. Appl. Environ. Microbiol. 65:5142-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]