Abstract
The first inducible Arthrobacter overexpression system, based on the promoter/operator and the repressor of the 6-d-hydroxynicotine oxidase gene of Arthrobacter nicotinovorans, is described here. Nicotine-dependent overproduction and affinity purification of recombinant proteins are presented. The system will allow the production of complex enzymes and genetic complementation studies in Arthrobacter species.
The genus Arthrobacter of gram-positive soil bacteria plays an important role in the mineralization of natural and man-made organic matter in the environment. In spite of the enormous ecological and potential industrial importance of these bacteria, recombinant DNA work with these bacteria has been seriously hampered because until now there were no suitable plasmid vectors available for gene cloning and expression purposes for Arthrobacter species.
Most of the Arthrobacter genes characterized so far have been cloned and expressed in Escherichia coli (10, 15-17) and other bacterial hosts (16, 18, 19), or alternatively, the proteins have been purified directly from crude lysates of Arthrobacter. Heterologous expression, however, can lead to inactive proteins due to misfolding, a lack of appropriate enzyme cofactors and proper insertion of these cofactors into apoenzymes, or posttranslational modifications. The development of an Arthrobacter expression system would be beneficial for the production of active Arthrobacter enzymes, for use in genetic complementation assays, or for the engineering of Arthrobacter strains with new, biotechnologically significant catabolic features.
Our laboratory has investigated protein biochemistry and nicotine degradation in Arthrobacter nicotinovorans for over 2 decades. A large catabolic plasmid, pAO1, confers on Arthrobacter nicotinovorans the ability to degrade nicotine (3). Various enzymes of the nicotine degradation pathway in A. nicotinovorans (1, 3, 4) have been identified and characterized, and recently the sequencing of the 165-kb pAO1 plasmid was completed (8). We have previously cloned and biochemically characterized the 6-d-hydroxynicotine oxidase gene (hdnO) from pAO1 (12, 13) and characterized its promoter region (hdnOp) (2). Recently, we also cloned the hdnO repressor gene (hnoR), biochemically characterized its gene product, and identified the inducer molecule for hdnO expression (20).
It has been reported previously that shuttle vectors, developed from cryptic plasmids of phylogenetically related species such as Corynebacterium glutamicum (22) and Brevibacterium lactofermentum (9), can also replicate in some Arthrobacter species, and some reports have previously described the cloning of genes in Arthrobacter species (10, 14, 21) using such plasmids.
Shuttle plasmids for inducible gene expression in Arthrobacter species.
We have created two vectors, pART2 and pART3, for the expression of genes in A. nicotinovorans as a host. The plasmids are made of DNA modules derived from A. nicotinovorans, C. glutamicum, and E. coli and required a number of cloning steps, which are described in Table 1.
TABLE 1.
Construction of pART2 and pART3 vectors and their derivatives
| Plasmid | Description and cloning strategy | Source |
|---|---|---|
| pAO1 | PCR template for amplification of the 6-d-hydroxynicotine oxidase gene (hdnO) promoter, the 2,6-dihydroxypyridine 3-hydroxylase (dhpH) gene, and the hdnO repressor gene (hnoR); purified pAO1 plasmid was used | 8 |
| p25435 | Carries the 1.9-kb DNA fragment of the cryptic plasmid pCG100 from C. glutamicum ATCC 13058, which allows independent replication in Arthrobacter species, and the Kanr gene from Tn903 used for the construction of pART vectors | 22 |
| pQEtriSystem-PhdnO | The 171-bp hdnO promoter/operator (hdnOp) was amplified by PCR using the primers 5′-TATATACCATGGTCTTGACAAGG-3′ and 5′-TATATAGGATCCATTTCCAACTCC-3′, restricted with NdeI/BamHI, and ligated into the corresponding sites of pQEtriSystem (Qiagen) | This work |
| pQEtriSystem-PhdnO-bla | The 861-bp bla gene was amplified by PCR from pET21a(+) (Novagen) using the primers 5′-ATATATGGATCCGAGTATTCAACATTTCC-3′ and 5′-ATATATCTCGAGCCAATGCTTAATCAG-3′, followed by BamHI/XhoI restriction and ligation into the BamHI/XhoI sites of pQEtriSystem-PhdnO | This work |
| pLigI | 1.97-kb E. coli vector encoding hdnOp-driven Ampr gene; 1.29-kb hdnOp-bla-His8 DNA fragment was excised with NdeI/NcoI from pQEtriSytem-PhdnO-bla and ligated to the 693-bp ColE1 ori restricted with NcoI/NdeI; ColE1 ori was amplified by PCR with the primers 5′-TATATACCATGGAGCGTCAGACC-3′ and 5′-ATATATCATATGTGAGCAAAAGGCC-3′ and with pThioA (Invitrogen) as a template | This work |
| pART1 | 5.44-kb E. coli-A. nicotinovorans shuttle vector; the NcoI-restricted pLigI plasmid was ligated to the 3.47-kb pCG100 Kanr DNA fragment, excised with NcoI from p25435 | This work |
| pART1A | The same as pART1, except that the XhoI (1041) site was changed to XbaI by site-directed mutagenesis with the primers 5′-GATTAAGCATTGGTCTAGACACCACCATC-3′ and 5′-GATGGTGGTGTCTAGACCAATGCTTAATC-3′ | This work |
| pART2 | 4.63-kb E. coli-A. nicotinovorans shuttle plasmid for hdnOp-driven constitutive expression; a BamHI-AatII-SalI-DraI-AvrII-SpeI-KpnI-PstI-XbaI fragment derived from annealing of two complementary, overhanging 5′-phosphorylated oligonucleotides (5′-GATCCGACGTCGTCGACTTTAAACCTAGGACTAGTGGTACCCTGCAGT-3′ and 5′-CTAGACTGCAGGGTACCACTAGTCCTAGGTTTAAAGTCGACGACGTCG-3′) was ligated into the BamHI/XbaI-restricted pART1A vector | This work |
| pART2-gfp | The 720-bp gfp gene was amplified by PCR from pEGFP-N1 (Clontech), using the primers 5′-CATGGATCCCAAGGGCGAGG-3′ and 5′-CGCGGCCTCTAGACTTGTACAGC-3′, restricted with BamHI/XbaI, and ligated into the corresponding sites of the pART2 vector | This work |
| pART2-dhpH | The 1.2-kb dhpH gene of A. nicotinovorans was amplified by PCR with the primers 5′-CACTAAGGAAGATCTCATGAGTCCCAC-3′ and 5′-GTTTTTCCTTATCTAGAATTAGTGAC-3′, restricted with BglII/XbaI, and ligated into compatible sites (BamHI/XbaI) of the pART2 vector | This work |
| pART3 | 5.4-kb E. coli-A. nicotinovorans shuttle plasmid for nicotine-inducible, hdnOp-driven gene expression; the hdnO repressor gene (hnoR) was amplified by PCR with the primers 5′-CTTTGTGGATCCGCCACCTGGGATGCC-3′ and 5′-AAAGGCGGATCCCCATAAGGAGCAAGG-3′, restricted with BamHI, and ligated into a compatible BglII site of the pART2 plasmid | This work |
| pART3-gfp | Same cloning strategy and primers as for pART2-gfp | This work |
| pART3-dhpH | Same cloning strategy and primers as for pART2-dhpH | This work |
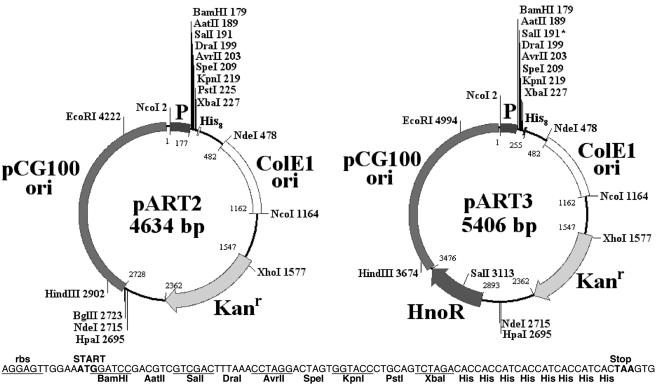
pART plasmids contain a 1.9-kb fragment from the cryptic plasmid pCG100 from C. glutamicum ATCC 13058 and can replicate autonomously in A. nicotinovorans and Arthrobacter globiformis. pART plasmid DNA can be reisolated from these species, and DNA restriction gives a pattern of digestion identical to the one expected for pART vectors (data not shown). These plasmids contain the ColE1 ori from E. coli and also the Kanr gene from Tn903, which confers resistance to kanamycin in all tested species. The expression of genes is driven by the promoter/operator of the 6-d-hydroxynicotine oxidase gene from A. nicotinovorans plasmid pAO1. pART2 and pART3 maps as well as details about the start codon, the multiple cloning sites (MCS), and the polyhistidine stretch sequence are presented in Fig. 1.
FIG. 1.
DNA maps of plasmids pART2 and pART3. The following plasmid features are presented: pCG100 and ColE1 origins of replication; Kanr, kanamycin resistance gene; P, promoter/operator of the hdnO gene (hdnOp); MCS; His8, eight-histidine tag coding sequence; and plasmid size in bp. Relevant restriction sites, including unique restriction sites, and bp positioning are represented. pART3 additionally carries the hdnO repressor gene (hnoR). The bottom sequence shows the following features: rbs, inferred ribosomal binding site; ATG, the translation start codon; MCS with nine unique restriction sites; coding sequence for a His8 tag; and TAA, translation stop codon. Note that the SalI* site is present twice in the pART3 vector. DNA analysis and map displays were prepared with BioEdit (7).
pART2 is a vector for constitutive gene expression, whereas pART3 also carries the hnoR gene and therefore is a vector for controlled gene expression in A. nicotinovorans. The HnoR protein binds to the hdnO operators IR1 and IR2 and blocks the expression of genes from this promoter (20). pART3 allows nicotine- and 6-hydroxynicotine-dependent gene expression in A. nicotinovorans and A. globiformis.
Overproduction of GFP in Arthrobacter sp.
A simple experiment to test the functionality of the pART plasmids in A. nicotinovorans is to express the green fluorescent protein (GFP) gene (gfp) from these plasmids (Table 1) and to monitor the fluorescence of bacteria by fluorescence microscopy.
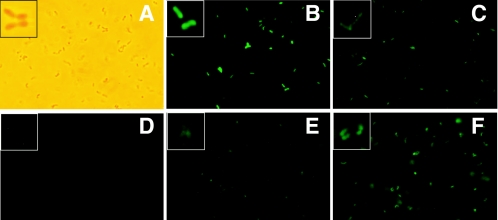
As shown in Fig. 2, A. nicotinovorans produces bright fluorescence when transformed with pART2-gfp (panel B).A. nicotinovorans containing plasmid pAO1 (A. nicotinovorans/pAO1) transformed with pART2-gfp produces slightly reduced fluorescence (panel C). This is explained by the presence of HnoR, produced from the pAO1 plasmid, which leads to the repression of hdnOp-driven gfp expression. The pART2 vector is designed for constitutive gene expression in Arthrobacter species, independent of the growth conditions. However, the expression of nicotine uptake and nicotine catabolic genes in A. nicotinovorans/pAO1 is poor in rich medium. Therefore, nicotine-inducible gene expression from the pART3 plasmid was carried out on citrate minimal medium. When A. nicotinovorans/pAO1 was transformed with pART3-gfp, no fluorescent bacteria were detectable under the fluorescence microscope (panel D). The presence of HnoR, produced from pART3 and additionally from pAO1, leads to efficient repression of hdnOp-driven gfp expression. The addition of nicotine to the cultures resulted in the induction of gfp expression, as demonstrated by the appearance in the cultures of fluorescent bacteria following 60 min of growth in the presence of nicotine (panel E) and an increased fluorescence of bacteria in cultures grown with nicotine overnight (panel F). The expression of gfp from the pART3 vector was also tested in A. globiformis, and we observed the appearance of nicotine- and 6-hydroxy-nicotine-dependent GFP fluorescence (data not shown). Cells induced with 6-hydroxynicotine expressed brighter fluorescence than those induced with nicotine, which is in agreement with the different induction efficiencies of the two compounds. These experiments demonstrate the functionality of pART vectors in A. nicotinovorans and other Arthrobacter spp. Nevertheless, not every species can be used as a host for nicotine-inducible expression from pART3, unless it has a nicotine uptake system. A. globiformis was described previously to degrade nicotine, and thereby it should have a nicotine uptake system (11).
FIG. 2.
Fluorescent A. nicotinovorans cells upon gfp expression. (A) Light microscopy image of A. nicotinovorans bacteria. Inset, magnified individual cell field. (B) Fluorescence microscopy image of A. nicotinovorans transformed with pART2-gfp; (C) A. nicotinovorans/pAO1 transformed with pART2-gfp; (D to F) A. nicotinovorans/pAO1 transformed with pART3-gfp and grown in the absence of nicotine (D), in the presence of nicotine for 60 min (E), and in the presence of nicotine overnight (F). Bacteria shown in panels A to C were grown in LB medium, and bacteria shown in panels D to F were grown in citrate medium supplemented with 0.5% yeast extract, mineral salts (5), and, as required, 0.05% l-nicotine. Pictures of A. nicotinovorans bacteria were taken using an Zeiss Axioskop 50 epifluorescence microscope equipped with a Plan-Nofluar 100× (1.3-numerical-aperture) objective and a triple-pass filter set. Digital images were recorded with a Nikon D100 camera.
Affinity purification of overproduced proteins in A. nicotinovorans.
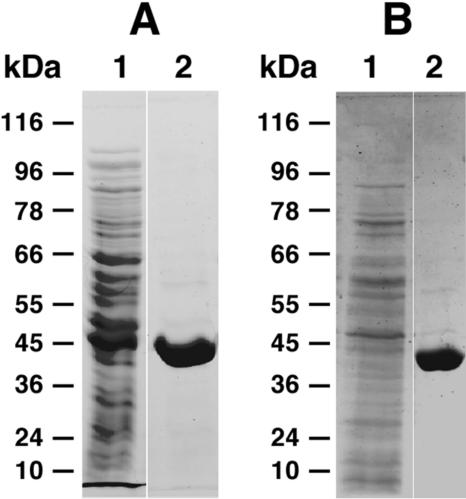
To assess whether the pART vectors can be used for protein overproduction and purification in A. nicotinovorans, we have chosen as a protein model the enzyme 2,6-dihydroxypyridine 3-hydroxylase (DhpH; see Table 1 for dhpH cloning details). DhpH-His8 is a yellow protein, a characteristic given by the flavin adenine dinucleotide cofactor, which makes it easy to monitor binding and elution of the protein from the column. Additionally, the hydroxylation of 2,6-dihydroxypyridine (2,6-DHP) to 2,3,6-trihydroxypyridine (2,3,6-THP) is accompanied by the formation of a blue pigment in the enzyme assay (1). In Fig. 3A, a gel is presented for a typical purification of DhpH-His8 from extracts of A. nicotinovorans transformed with pART2-dhpH. The yield of DhpH-His8 produced in A. nicotinovorans transformed with pART2 and grown in LB medium was up to 5% of the total protein present in the bacterial lysates. The specific activity of DhpH-His8 was ≈18 U per mg protein, which corresponds to the previously reported specific activity of the enzyme (1). DhpH-His8 was also produced from A. nicotinovorans/pAO1 transformed with pART3-dhpH, grown in citrate minimal medium, and induced with nicotine (Fig. 3B). The yield of DhpH-His8 from A. nicotinovorans/pAO1 lysates was approximately 3%.
FIG. 3.
Purification of DhpH-His8 from A. nicotinovorans extracts. (A) Ni2+-chelating Sepharose affinity chromatography purification of DhpH-His8 from crude extracts of A. nicotinovorans transformed with pART2-dhpH and grown in LB medium without nicotine induction. Lane 1, crude extract; lane 2, purified protein fraction, eluted with 200 mM imidazole. (B) Ni2+-affinity purification of DhpH-His8 from extracts of A. nicotinovorans/pAO1 transformed with pART3-dhpH and grown in citrate minimal medium in the presence of 0.05% l-nicotine. Lane 1, crude extract; lane 2, purified protein fraction, eluted with 200 mM imidazole. The transformation of A. nicotinovorans with pART plasmids was achieved by electroporation with competent cells as described previously (6), with selection on kanamycin (140 μg ml−1). E. coli was selected with 20 μg ml−1 kanamycin.
Since pART2 and pART3 are shuttle vectors, they may also be used in E. coli for the production and purification of proteins. Genes expressed from pART2 lead to constitutive expression from hdnOp, whereas expression from pART3 is reduced due to the presence of the hnoR gene.
We have proven in this work the functionality and usefulness of the pART2 and pART3 vectors for protein overproduction in Arthrobacter spp. These vectors are particularly valuable for the production of proteins, which cannot be produced in their active form in heterologous systems. These plasmids will be used to overproduce active nicotine dehydrogenase and ketone dehydrogenase for biochemical studies with their natural host, A. nicotinovorans. These enzymes are inactive when overproduced in E. coli (P. Sachelaru and R. Brandsch, unpublished) because the molybdopterin cytosine dinucleotide cofactor required for their activity is not made in E. coli, which instead synthesizes the molybdopterin guanine dinucleotide cofactor. In addition, these plasmids will be used as essential tools in genetic complementation studies with Arthrobacter spp.
Nucleotide sequence accession numbers.
The DNA sequences of the two plasmids described in this study have been deposited in GenBank under accession numbers DQ191047 (pART2) and DQ191048 (pART3).
Acknowledgments
This work was supported by a grant from the Deutsche Forschungsgemeinschaft to R.B.
We thank K. H. Gartemann, Bielefeld, Germany, for providing plasmid p25435.
REFERENCES
- 1.Baitsch, D., C. Sandu, R. Brandsch, and G. L. Igloi. 2001. Gene cluster on pAO1 of Arthrobacter nicotinovorans involved in degradation of the plant alkaloid nicotine: cloning, purification, and characterization of 2,6-dihydroxypyridine 3-hydroxylase. J. Bacteriol. 183:5262-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernauer, H., L. Mauch, and R. Brandsch. 1992. Interaction of the regulatory protein NicR1 with the promoter region of the pAO1-encoded 6-hydroxy-d-nicotine oxidase gene of Arthrobacter oxidans. Mol. Microbiol. 6:1809-1820. [DOI] [PubMed] [Google Scholar]
- 3.Brandsch, R., W. Faller, and K. Schneider. 1986. Plasmid pAO1 of Arthrobacter oxidans encodes 6-hydroxy-d-nicotine oxidase: cloning and expression of the gene in Escherichia coli. Mol. Gen. Genet. 202:96-101. [DOI] [PubMed] [Google Scholar]
- 4.Chiribau, C. B., C. Sandu, M. Fraaije, E. Schiltz, and R. Brandsch. 2004. A novel gamma-N-methylaminobutyrate demethylating oxidase involved in catabolism of the tobacco alkaloid nicotine by Arthrobacter nicotinovorans pAO1. Eur. J. Biochem. 271:4677-4684. [DOI] [PubMed] [Google Scholar]
- 5.Eberwein, H., F. A. Gries, and K. Decker. 1961. On the decomposition of nicotine by bacterial enzymes. II. Isolation and characterization of a nicotine-splitting soil bacterium. Hoppe-Seyler's Z. Physiol. Chem. 323:236-248. [DOI] [PubMed] [Google Scholar]
- 6.Gartemann, K. H., and R. Eichenlaub. 2001. Isolation and characterization of IS1409, an insertion element of 4-chlorobenzoate-degrading Arthrobacter sp. strain TM1, and development of a system for transposon mutagenesis. J. Bacteriol. 183:3729-3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall, A. T. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 8.Igloi, G. L., and R. Brandsch. 2003. Sequence of the 165-kilobase catabolic plasmid pAO1 from Arthrobacter nicotinovorans and identification of a pAO1-dependent nicotine uptake system. J. Bacteriol. 185:1976-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leret, V., A. Trautwetter, A. Rince, and C. Blanco. 1998. pBLA8, from Brevibacterium linens, belongs to a gram-positive subfamily of ColE2-related plasmids. Microbiology 144:2827-2836. [DOI] [PubMed] [Google Scholar]
- 10.Loviny-Anderton, T., P. C. Shaw, M. K. Shin, and B. S. Hartley. 1991. d-Xylose (d-glucose) isomerase from Arthrobacter strain N.R.R.L. B3728. Gene cloning, sequence and expression. Biochem. J. 277:263-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maeda, S., and K. Takuro. 1981. Microbial degradation of nicotine-1′-N-oxide. II. Identification of nicotine-1′-N-oxide degrading bacteria. Agric. Biol. Chem. 45:565-569. [Google Scholar]
- 12.Mauch, L., V. Bichler, and R. Brandsch. 1990. Functional analysis of the 5′ regulatory region and the UUG translation initiation codon of the Arthrobacter oxidans 6-hydroxy-d-nicotine oxidase gene. Mol. Gen. Genet. 221:427-434. [DOI] [PubMed] [Google Scholar]
- 13.Mauch, L., V. Bichler, and R. Brandsch. 1989. Site-directed mutagenesis of the FAD-binding histidine of 6-hydroxy-d-nicotine oxidase. Consequences on flavinylation and enzyme activity. FEBS Lett. 257:86-88. [DOI] [PubMed] [Google Scholar]
- 14.Morikawa, M., H. Daido, S. Pongpobpibool, and T. Imanaka. 1994. Construction of a new host-vector system in Arthrobacter sp. and cloning of the lipase gene. Appl. Microbiol. Biotechnol. 42:300-303. [DOI] [PubMed] [Google Scholar]
- 15.Oguma, T., T. Kurokawa, K. Tobe, S. Kitao, and M. Kobayashi. 1999. Cloning and sequence analysis of the gene for glucodextranase from Arthrobacter globiformis T-3044 and expression in Escherichia coli cells. Biosci. Biotechnol. Biochem. 63:2174-2182. [DOI] [PubMed] [Google Scholar]
- 16.Ohashi, H., Y. Katsuta, M. Nagashima, T. Kamei, and M. Yano. 1989. Expression of the Arthrobacter viscosus penicillin G acylase gene in Escherichia coli and Bacillus subtilis. Appl. Environ. Microbiol. 55:1351-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okushima, M., D. Sugino, Y. Kouno, S. Nakano, J. Miyahara, H. Toda, S. Kubo, and A. Matsushiro. 1991. Molecular cloning and nucleotide sequencing of the Arthrobacter dextranase gene and its expression in Escherichia coli and Streptococcus sanguis. Jpn. J. Genet. 66:173-187. [DOI] [PubMed] [Google Scholar]
- 18.Parschat, K., B. Hauer, R. Kappl, R. Kraft, J. Huttermann, and S. Fetzner. 2003. Gene cluster of Arthrobacter ilicis Ru61a involved in the degradation of quinaldine to anthranilate: characterization and functional expression of the quinaldine 4-oxidase qoxLMS genes. J. Biol. Chem. 278:27483-27494. [DOI] [PubMed] [Google Scholar]
- 19.Roberts, A. N., L. Barnett, and S. Brenner. 1987. Transformation of Arthrobacter and studies on the transcription of the Arthrobacter ermA gene in Streptomyces lividans and Escherichia coli. Biochem. J. 243:431-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sandu, C., C. B. Chiribau, and R. Brandsch. 2003. Characterization of HdnoR, the transcriptional repressor of the 6-hydroxy-d-nicotine oxidase gene of Arthrobacter nicotinovorans pAO1, and its DNA-binding activity in response to l- and d-nicotine derivatives. J. Biol. Chem. 278:51307-51315. [DOI] [PubMed] [Google Scholar]
- 21.Shaw, P. C., and B. S. Hartley. 1988. A host-vector system for an Arthrobacter species. J. Gen. Microbiol. 134:903-911. [DOI] [PubMed] [Google Scholar]
- 22.Trautwetter, A., and C. Blanco. 1991. Structural organization of the Corynebacterium glutamicum plasmid pCG100. J. Gen. Microbiol. 137:2093-2101. [DOI] [PubMed] [Google Scholar]