Abstract
Protamylasse is a residual compound occurring during the industrial production of starch from potatoes. It contains a variety of nutrients and all necessary minerals and could be used as a carbon, nitrogen, and energy source for the growth of bacteria and also for cyanophycin (CGP) biosynthesis. Media containing protamylasse as the sole compound diluted only in water were therefore examined for their suitability in CGP production. Among various bacterial strains investigated in this study, a recombinant strain of Escherichia coli DH1 harboring plasmid pMa/c5-914::cphA6803, which carries the cyanophycin synthetase structural gene (cphA) from Synechocystis sp. strain PCC6803, was found to be most suitable. Various cultivation conditions for high CGP contents were first optimized in shake flask cultures. The optimized conditions were then successfully applied to 30- and 500-liter fermentation scales in stirred tank reactors. A maximum CGP content of 28% (wt/wt) CGP per cell dry matter was obtained in 6% (vol/vol) protamylasse medium at an initial pH of 7.0 within a cultivation period of only 24 h. The CGP contents obtained with this recombinant strain employing protamylasse medium were higher than those obtained with the same strain cultivated in mineral salts medium or in expensive commercial complex media such as Luria-Bertani or Terrific broth. It was shown that most amino acids present in the protamylasse medium were almost completely utilized by the cells during cultivation. Exceptions were alanine, tryptophan, tyrosine, and most interestingly, arginine. Furthermore, CGP was easily isolated from protamylasse-grown cells by applying the acid extraction method. The CGP exhibited a molecular mass of about 26 to 30 kDa and was composed of 50% (mol/mol) aspartate, 46% (mol/mol) arginine, and 4% (mol/mol) lysine. The use of cheap residual protamylasse could contribute in establishing an economically and also ecologically feasible process for the biotechnological production of CGP.
Cyanophycin, also known as cyanophycin granule polypeptide (CGP), is a nonribosomally synthesized protein-like copolymer composed of a polyaspartic acid backbone and arginine residues linked with the α-amino group to the β-carboxyl group of each aspartate residue (36, 37, 38). CGP occurs as insoluble inclusions in the cytoplasm and serves as a storage compound for carbon, nitrogen, and energy (20, 22). CGP is insoluble in water at physiological pH and soluble under acidic or alkaline conditions (17). CGP is synthesized naturally in cyanobacteria (6) and also in some other nonphotosynthetic bacteria (15). The key enzyme of CGP synthesis is cyanophycin synthetase (CphA), which catalyzes the ATP-dependent polymerization of aspartate and arginine. Genes encoding cyanophycin synthetases (cphA) have been identified in many cyanobacteria (1, 5, 12, 27, 44). In addition, cphA homologues have also been detected in the genomes of many noncyanobacteria, such as Acinetobacter sp. strain ADP1 (15, 45). CGP is not affected by proteases (31, 37, 38); however, it is hydrolyzed to Asp-Arg dimers by cyanophycinases (20, 25, 26, 29, 31).
Purified CGP can be chemically converted to a derivative with a reduced arginine content (14) or to completely biodegradable poly(aspartic acid) (4). Poly(aspartic acid) is used as a substitute for nonbiodegradable polyacrylates, for which many technical applications are described (35), such as water treatment (water softeners) and others (4, 32). Biomedical applications of poly(aspartic acid) have also been described (19, 30, 43). In addition, it can be also considered a source of arginine and aspartic acid, and therefore there is strong interest in producing CGP. Since for various reasons cyanobacteria are not suitable for the production of CGP (13), cyanobacterial cphA genes were heterologously expressed in various heterotrophic bacteria (3, 5, 12, 27, 44) or in plants (24). A recombinant strain of Escherichia coli harboring cphA of Synechocystis sp. strain PCC6803 accumulated CGP as up to 24% (wt/wt) of the cell dry matter (CDM); however, such high CGP contents were obtained only during cultivation in costly complex media (11). In addition, the cultivation conditions for noncyanobacteria possessing cphA homologues were optimized with regard to high CGP contents. This was successfully done with Acinetobacter sp. strain ADP1, which accumulated CGP to the highest ever reported content, 46% (wt/wt) of the cell dry mass in mineral salt medium, but only when arginine was used as the carbon source (9).
There are multiple pieces of evidence showing that provisions of CphA with their substrates aspartate, and in particular, arginine represent bottlenecks of CGP synthesis in cells (9, 39, 41). Therefore, CGP accumulation in cells cultivated in mineral salt medium is low, and complex nutrients or arginine must be added to the medium as a supplement to obtain cells with higher CGP contents. This makes the biotechnological production of CGP economically unfeasible unless a cheap source of these amino acids is found. Protamylasse, or potato juice concentrate, is an abundant residual fraction remaining during the industrial production of starch from potatoes. In The Netherlands alone, about 70,000 tons of dry weight of protamylasse is produced annually. Protamylasse contains soluble peptides, amino acids, with asparagine as the main component, organic acids, carbohydrates, salts, and minerals (Table 1) and may be a suitable substrate for biotechnological production processes. With regard to the commercial production of CGP, the high arginine, aspartate, and asparagine contents of protamylasse are of particular interest. The application of protamylasse as a sole and complete medium could make the biotechnological production of CGP economically feasible, because the costs of protamylasse are much lower than those of other complex media or mineral salt media, and also environmentally friendly, because it provides a useful application of this residual of the starch industry. Protamylasse has been applied as raw material for fermentation to obtain single cell proteins, ethanol, or beta-glucan (21). Protamylasse was also fractionated into potassium salt and basic or acidic amino acids (40). Since protamylasse is, to our knowledge, not used in biotechnological processes on an industrial scale, we investigated its suitability for CGP production.
TABLE 1.
Chemical composition of protamylassea
| Component | Concn (g/liter) |
|---|---|
| Amino acids | 183.0 |
| Arginine | 9.2 |
| Aspartic acid plus asparagine | 65.1 |
| Lysine | 7.1 |
| Phenylalanine | 4.4 |
| Tyrosine | 3.7 |
| Valine | 5.8 |
| Glutamic acid plus glutamine | 39.7 |
| Glycine | 4.2 |
| Methionine | 2.2 |
| Cysteine | 3.0 |
| Tryptophan | 1.0 |
| Histidine | 1.9 |
| Leucine | 4.7 |
| Isoleucine | 3.3 |
| Proline | 6.8 |
| Serine | 4.9 |
| γ-Aminobutyric acid | 4.5 |
| Ornithine | 2.1 |
| Alanine | 8.7 |
| Organic acids | 135.2 |
| Citric acid | 76.3 |
| Malic acid | 35.6 |
| Oxalic acid | 6.1 |
| Acetic acid | 4.1 |
| Lactic acid | 13.2 |
| Sugars | 142.3 |
| Glucose | 35.6 |
| Fructose | 35.6 |
| Saccharose | 71.2 |
| Ash | 225.7 |
| K+ | 97.6 |
| (PO4)3− | 25.4 |
| Mg2+ | 4.5 |
| Ca2+ | 0.6 |
| Na+ | 0.8 |
| (NH4)+ | 4.0 |
| Mn2+ | 0.03 |
| Zn2+ | 0.08 |
| Cu2+ | 0.04 |
| Cl− | 8.1 |
| (NO3)− | 2.2 |
| (SO4)2− | 22.4 |
Amounts of components were calculated for concentrated protamylasse.
MATERIALS AND METHODS
Bacterial strains, plasmids, and cultivation conditions.
All bacterial strains and plasmids used in this study are listed in Table 2. E. coli strains were cultivated at 30 or 37°C in Erlenmeyer flasks without baffles in either Luria-Bertani (LB) (33) or protamylasse medium. Pseudomonas putida KT2440 harboring cphA from Synechocystis sp. strain PCC6803 and Acinetobacter calcoaceticus strain ADP1 were cultivated at 30°C in 250-ml Erlenmeyer flasks containing 100 ml of protamylasse medium. Flasks (New Brunswick Scientific) were incubated at 37°C or 30°C on a rotary shaker (Finos AG, Basel, Switzerland) at 150 or 130 rpm, respectively. Antibiotics were added as indicated in the text.
TABLE 2.
Strains used for this study
| Strain | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Acinetobacter calcoaceticus strain ADP1 | Wild type | ATCC 33305 |
| Escherichia coli DH1(pMa/c5-914::cphA6803) | F−endA1 gyrA96 hsdR17 (rK− mK+) recA1 supE thi-1 λ; contains plasmid pMa/c5-914 with a 2.6-kbp PCR product from Synechocystis sp. strain PCC6803 genomic DNA harboring cphA | 11 |
| Pseudomonas putida KT2440 (pBBR1MCS-2::cphA6803) | Wild-type strain containing plasmid pBBR1MCS-2::cphA6803 harboring cphA from Synechocystis sp. strain PCC6803 | 41 |
Preparation of protamylasse.
Protamylasse material (58% [dry weight]) as received from Avebe (Veendam, The Netherlands) was first diluted 1:1 (vol/vol) using distilled H2O and then filtered by passing it through a filter cloth. The resulting solution was then centrifuged at 3,500 × g to remove water-insoluble fine particles. The pH of diluted protamylasse was about 5.5 and was adjusted to 7.5 with 10 M NaOH before sterilization.
Cultivation at the 30-liter scale.
A Biostat DL30 stainless steel reactor (B. Braun Biotech International, Melsungen, Germany) with a total volume of 42 liters (28-cm inner diameter and 71-cm height) and a d/D value ratio (ratio of stirrer diameter to vessel diameter) of 0.375, as described previously (11), was used. The dissolved oxygen, temperature, pH, foam, and optical density were measured with probes and sensors. The bioreactor was filled with 24 liters of 6% (vol/vol) protamylasse, and the pH value was set to 7.5 before in situ sterilization. After sterilization, the cultivation parameters such as temperature, stirrer speed, and airflow were adjusted, and 100 μg/ml ampicillin was added to the medium before inoculation. Carbon dioxide and oxygen concentrations in the spent gas leaving the bioreactor were measured with a URAS 10 P NDIR spectrophotometer and a Magnos 6 G oxygen analyzer (both from Mannesmann, Hartmann and Braun, Frankfurt, Germany), respectively. Fermentation was carried out at 30°C if the culture was used as a preculture to inoculate the 650-liter bioreactor (see below) or at 37°C if the culture was the main culture, with pO2 ranges in the medium of 0 to 100% and 20 to 40% saturation for batch and fed-batch cultivations, respectively. Agitation and aeration rates were varied between 150 and 350 rpm or 0.8 and 1.0 volume per volume and min (vvm), respectively, as indicated in the text. The pH in the medium was either not adjusted or held between 7.0 and 7.5 or between 6.8 and 7.0 by the addition of 4 M HCl or NaOH. Foam was controlled by a mechanical foam destroyer and by the addition of the antifoam agent Silikon Antischaum emulsion SLE (Wacker, Darwin Vertriebs GmbH, Ottobrunn, Germany), if necessary.
Cultivation at the 500-liter scale.
Cultivation at the 500-liter scale was performed in a Biostat D650 stainless steel bioreactor (B. Braun Biotech International), which had a total volume of 650 liters (64-cm inner diameter and 198-cm height) and the same d/D value as the Biostat DL30 reactor described above. All other equipment was the same as that described above for the Biostat DL30 bioreactor. Fermentations were done as described above.
Cell harvest from 30- and 500-liter cultivations.
Cells from 30- and 500-liter cultivations were harvested by centrifugation at 4°C in a CEPA type Z41 or type Z61 continuous centrifuge (Carl Padberg Zentrifugenbau GmbH, Lahr, Germany), respectively.
Analysis of ammonium.
Ammonium was determined in cell-free supernatants by using a gas-sensitive ammonium electrode (type 152303000; Mettler Toledo GmbH).
Analysis of inorganic phosphate concentration in protamylasse.
The concentrations of inorganic phosphate in cell-free supernatants were determined colorimetrically as described previously (8).
Determination of cell dry matter.
To determine the bacterial CDM, defined volumes of cultures were centrifuged in a bench centrifuge at 3,500 × g and 4°C. The supernatant was discarded, and cells were washed by centrifugation after being suspended in saline (0.9% [wt/vol] NaCl). The cells were lyophilized, and their masses were gravimetrically determined.
Purification and analysis of CGP.
CGP was isolated from cells of E. coli DH1, P. putida KT2440, and A. calcoaceticus strain ADP1 by the procedure described by Simon and Weathers (37). A recently described fast method (9) was also used for routine CGP determination. For this method, the cells were first disintegrated by sonification for 2 min per ml of cells suspended in 50 mM Tris-HCl (pH 7.5), using a Sonoplus sonifier (Bandelin Electronic, Berlin, Germany), before CGP was extracted by the fast method.
Electrophoresis and determination of protein concentration and amino acid composition.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed in 11.5% (wt/vol) gels according to a standard method (16). Proteins and cyanophycin were stained with Serva Blue R (42). Concentrations of CGP were determined as described by Bradford (7). The amino acid composition of the diluted protamylasse or CGP was determined by high-performance liquid chromatography (HPLC) analysis as described previously (11).
RESULTS
Acinetobacter sp. strain ADP1 and recombinant strains of E. coli and P. putida KT2440 expressing the cyanophycin synthetase of Synechocystis sp. strain PCC6803 were examined for the capability to use protamylasse for the growth and synthesis of CGP. Cells were cultivated aerobically in water containing three different concentrations of protamylasse as the sole component, and their CGP contents were determined. All three strains utilized protamylasse as a source of carbon and other macroelements for cell growth reasonably well, but they differed significantly regarding the CGP contents detected in cells (Table 3). Acinetobacter sp. strain ADP1, which synthesizes CGP only under phosphate starvation conditions and if the medium is supplemented with arginine (9, 15), synthesized only a little CGP, even if the concentration of phosphate in protamylasse was reduced by precipitation with CaCl2. Cells of the recombinant P. putida strain KT2440 accumulated similar small amounts of CGP, whereas the recombinant E. coli strain accumulated CGP up to about 18% ± 6.4% (wt/wt) of the cell dry matter when grown on 5% (vol/vol) protamylasse. E. coli DH1(pMa/c5-914::cphA6803) was therefore used for further optimization.
TABLE 3.
CGP biosynthesis in bacteria using protamylasse as a complex medium for growth
| Protamylasse concn (% [vol/vol])a |
A. calcoaceticus ADP1b
|
E. coli DH1(pMa/c5-914::cphA6803)c
|
P. putida KT2440(pBBR1MCS-2::cphA6803)d
|
|||
|---|---|---|---|---|---|---|
| CGP content (% of CDM) | Cell density (g CDM/liter) | CGP content (% of CDM) | Cell density (g CDM/liter) | CGP content (% of CDM) | Cell density (g CDM/liter) | |
| 1.0 | 0.60 ± 0.01 | 1.1 ± 0.38 | 5.1 ± 0.19 | 0.6 ± 0.004 | 0.7 ± 0.03 | 1.0 ± 0.01 |
| 2.5 | 0.10 ± 0.001 | 2.2 ± 0.09 | 11.2 ± 0.61 | 1.7 ± 0.02 | 0.5 ± 0.14 | 2.4 ± 0.01 |
| 5.0 | 0.07 ± 0.006 | 2.7 ± 0.01 | 18.0 ± 6.4 | 3.5 ± 0.03 | 0.1 ± 0.02 | 2.3 ± 0.03 |
All strains were cultivated in 250-ml Erlenmeyer flasks without baffles containing 100 ml of 6% (vol/vol) protamylasse at 30°C or 37°C for 44 h. Protamylasse was added to H2O, and the pH was adjusted to 7.0 before sterilization as described in Materials and Methods. Data shown represent mean values and standard deviations of three independent experiments. The corresponding initial pH values for 1%, 2.5%, and 5% protamylasse (vol/vol) were 5.7, 5.6, and 5.5, respectively, and were caused by the acidic protamylasse in unbuffered medium.
A 1.8 mM sterile solution of CaCl2 · 2H2O was added to reduce the phosphate concentration in the medium. Cultures of A. calcoaceticus ADP1 were inoculated from a preculture previously grown at 30°C.
Cultures of E. coli DH1(pMa/c5-914::cphA6803) were supplemented with 100 μg/ml penicillin and were inoculated from a preculture previously grown on protamylasse at 30°C and incubated at 37°C to induce the cells for CGP synthesis.
Cultures of P. putida KT2440 were supplemented with 50 μg/ml kanamycin and inoculated from a preculture grown at 30°C. Cells were harvested at the end of the cultivation period and analyzed for CGP contents, and the cell dry mass was determined.
Effect of protamylasse concentration on growth and CGP synthesis in E. coli DH1(pMa/c5-914::cphA6803).
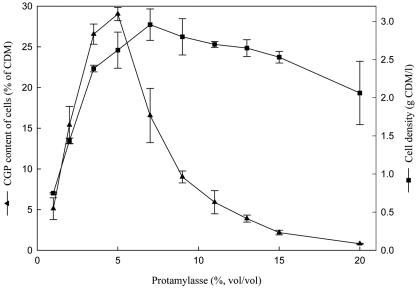
To determine the optimum concentrations of protamylasse for high CGP contents and cell densities, cells were cultivated in 1 to 20% (vol/vol) protamylasse in the presence of 100 μg/ml ampicillin to retain the plasmid. With protamylasse concentrations increasing from 1% (vol/vol), the cell density and CGP content of the cells increased (Fig. 1), reaching a maximum CGP content of 29% ± 0.8% (wt/wt) of CDM with 5% (vol/vol) protamylasse. At further increasing concentrations of protamylasse, the CGP contents of the cells decreased sharply. In contrast, the maximum cell density of 3 ± 0.2 g/liter for cells containing only 16% ± 3.3% (wt/wt) CGP was obtained with 7% (vol/vol) protamylasse. The cell density remained at a high level even if the concentration of protamylasse was further increased to 20% (vol/vol). Additional cultivation experiments applying 5 or 6% (vol/vol) protamylasse under the same cultivation conditions gave CGP contents of the cells of 25.6% ± 1.6% and 26.8% ± 1.5% (wt/wt), respectively, with almost the same high cell density. Therefore, a protamylasse concentration of 6% (vol/vol) was used in further experiments.
FIG. 1.
Effect of protamylasse concentration on growth and CGP accumulation in E. coli DH1(pMa/c5-914::cphA6803). Experiments were done in 250-ml Erlenmeyer flasks, each containing 100 ml medium with the indicated concentration of protamylasse and 100 μg ampicillin per ml. The flasks were inoculated from a preculture previously grown at 30°C. CGP biosynthesis was induced by incubating the cultures at 37°C. After an incubation period of 44 h, cells were harvested and analyzed for CGP content (▴) and cell density (▪) as described in Materials and Methods. The data shown represent mean values and standard deviations of three independent cultivation experiments.
Effect of initial pH on CGP content of E. coli DH1(pMa/c5-914::cphA6803) cells.
The initial pH value of the standard medium containing 6% (vol/vol) protamylasse was about 5.5. The initial pH of the protamylasse medium was adjusted to other values before sterilization by adding 10 M NaOH. The maximum CGP content of 27.2% ± 3.3% (wt/wt) of CDM, as well as the maximum cell density, was obtained if the initial pH was 7.5. However, the CGP contents of the cells remained at almost the same high levels at initial pH values between 5.4 and 8.0 (Table 4). The CGP contents of the cells were lower at lower initial pH values; for example, at an initial pH of 5.5, the CGP content of the cells was only 13.9% ± 0.8% (wt/wt) of CDM. The initial pH also exerted an influence on cell size and shape. Cells grown at an initial pH of 5.5 were much longer and thinner than cells grown at an initial pH of 7.5.
TABLE 4.
Effect of different initial pH values on CGP content in E. coli DH1(pMa/c5-914::cphA6803)
| Initial pHa | Final pHb | Cell density (g CDM/liter) | CGP content of cells (% of CDM) |
|---|---|---|---|
| 5.5 | 5.58 | 0.97 ± 0.07 | 13.9 ± 0.80 |
| 6.0 | 7.90 | 2.44 ± 0.08 | 15.7 ± 2.07 |
| 6.5 | 8.10 | 2.69 ± 0.08 | 25.9 ± 0.80 |
| 7.0 | 8.31 | 2.96 ± 0.01 | 25.4 ± 1.70 |
| 7.5 | 8.50 | 3.19 ± 0.02 | 27.2 ± 3.30 |
| 8.0 | 8.50 | 3.08 ± 0.02 | 25.9 ± 4.40 |
Cells of E. coli DH1(pMa/c5-914::cphA6803) were grown in 250-ml Erlenmeyer flasks without baffles which contained 100 ml 6% (vol/vol) protamylasse supplemented with 100 μg/ml ampicillin. The initial pH was adjusted with 10 M NaOH before sterilization, and the flasks were incubated at 37°C for 44 h. Cells were harvested at the end of cultivation period and analyzed for CGP content, and the cell dry mass was determined. Data shown represent mean values and standard deviations of three independent experiments.
Final pH values were measured in the supernatant at the end of the cultivation period.
Effect of incubation period on CGP accumulation in E. coli DH1(pMa/c5-914::cphA6803).
To examine the optimum incubation period, several 250-ml Erlenmeyer flasks, each containing 100 ml 6% (vol/vol) protamylasse medium at an initial pH of 7.5, were inoculated from a preculture grown at 30°C and then incubated for 3 days at 37°C. Each day, three flasks were taken, and the cell densities and CGP contents of the cells were determined as described in Materials and Methods (Table 5). The maximum CGP content of 28.9% ± 3.7% (wt/wt) of CDM was already obtained after an incubation of only 24 h. During a further 22 h of incubation, the cell density increased by 53% (wt/wt), maintaining almost the same high CGP content of the cells. Therefore, a cultivation period of about 2 days represented the optimum period to obtain reasonably high cell densities and also high CGP contents at the flask scale.
TABLE 5.
Effect of incubation period on CGP synthesis in E. coli DH1(pMa/c5-914::cphA6803)a
| Incubation period (h) | Cell density (g CDM/liter) | CGP content of cells (% of CDM) |
|---|---|---|
| 24 | 2.40 ± 0.35 | 28.9 ± 3.70 |
| 46 | 3.67 ± 0.46 | 27.7 ± 0.09 |
| 71 | 3.50 ± 0.44 | 27.6 ± 1.45 |
Cells of E. coli DH1(pMa/c5-914::cphA6803) were grown in 250-ml Erlenmeyer flasks without baffles which contained 100 ml 6% (vol/vol) protamylasse supplemented with 100 μg/ml ampicillin. The initial pH was adjusted to 7.5 before sterilization, and the flasks were incubated at 37°C. Each day, one sample was taken and analyzed for CDM and CGP content as described in Materials and Methods. Data shown represent mean values and standard deviations of three independent experiments.
Cultivation at 30-liter scale in bioreactor.
To facilitate further optimization, cultivation of E. coli DH1(pMa/c5-914::cphA6803) was then done in 24 liters of 6% (vol/vol) protamylasse with an initial pH of 7.5 in a 30-liter Biostat DL30 stirred tank reactor. The medium was inoculated with four 400-ml precultures grown for 12 h at 30°C in 6% (vol/vol) protamylasse supplemented with 100 μg/ml ampicillin in 1-liter Erlenmeyer flasks. Fermentation was done at 37°C to induce the expression of cyanophycin synthetase, and samples were taken at intervals of 2 h. The CGP contents of the cells increased rapidly within the first 4 h, to 26% ± 1% (wt/wt). While cell growth continued, the highest CGP content of 33.2% ± 0.08% and highest cell density of about 5 ± 0.3 g/liter were obtained after 19 h of incubation, and afterward the CGP content of the cells remained almost constant, at about 26% (wt/wt) of CDM. After 25 h of cultivation, the final CGP concentration was about 1.3 g liter−1, and the pH was 8.0 (data not shown). The cell densities obtained in the bioreactor were about twofold higher than those obtained in Erlenmeyer flask experiments.
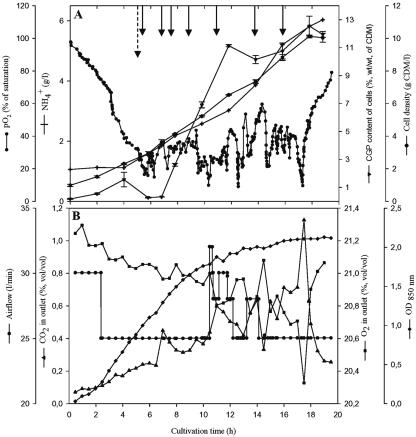
Fed-batch cultivation is one of the most often-used strategies to achieve high cell densities and may also be useful for the production of CGP. The cultivation of E. coli DH1(pMa/c5-914::cphA6803) was therefore done one more time in 24 liters of 6% (vol/vol) protamylasse, with an initial pH of 7.0. This time the pH was maintained between 6.8 and 7.0. During growth, additional protamylasse was fed as shown in Fig. 2. Airflow and stirring were changed during cultivation to keep the dissolved oxygen concentration between 20 and 40% (pO2, % of saturation) and to avoid a restricted oxygen supply. The temperature was shifted from 30 to 37°C after 6 h of cultivation to induce the expression of cyanophycin synthetase. Cells grew exponentially at a growth rate (μ) of 0.17 h−1 during the batch period at 30°C. During the fed-batch period at 37°C, the cell density continued to increase, and after 19 h of cultivation a final density of 10 ± 0.3 g/liter was obtained, which was about twofold higher than that in the previous batch cultivation experiment. However, the CGP content of the cells was only 11.9% ± 0.21% (wt/wt), and due to the lower CGP concentration in the cells, the final CGP concentration in the bioreactor could not be increased in this experiment.
FIG. 2.
Fed-batch fermentation of E. coli DH1(pMa/c5-914::cphA6803) in Biostat DL30 stirred tank reactor containing 22 liters of 6% (vol/vol) protamylasse medium. The medium was inoculated with four 400-ml precultures grown in the same medium, but at a temperature of 30°C only. The fermentation parameters and cultivation conditions in the Biostat DL30 reactor were a pH of 6.8 to 7.0, aeration at 0.8 to 1.0 vvm, and agitation at 150 to 350 rpm. Aeration and agitation were adjusted according to the oxygen demand of the culture. The temperature was raised from 30 to 37°C after a cultivation period of 6 h (dashed arrow) to induce the expression of CGP synthetase. Portions of 250 to 500 ml 25% (vol/vol) protamylasse were repeatedly added during fermentation at the indicated times (solid arrows). (A) Cell density (•), CGP content of cells (▾), pO2 (•-•), and ammonium concentration (+). (B) Optical density at 850 nm (OD850, ⧫), airflow (•), O2 in exhaust gas (▪), and CO2 in exhaust gas (▴) were determined as described in Materials and Methods.
Cultivation at 500-liter scale in bioreactor.
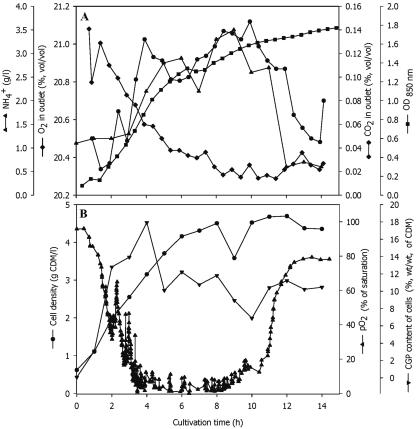
To continue towards the development of a process for large-scale CGP production from protamylasse, E. coli DH1(pMa/c5-914::cphA6803) was cultivated at the 500-liter scale. A Biostat D650 bioreactor was inoculated with cells cultivated for 12 h at 30°C in a 30-liter bioreactor in 6% (vol/vol) protamylasse medium containing 100 μg/ml ampicillin. Batch fermentation was done for 15 h in 6% (vol/vol) protamylasse medium with an initial pH of 7.5 (Fig. 3). Cells grew at a specific growth rate of 0.19 h−1 during the exponential growth phase. During this fermentation, the maximum CGP content of the cells of about 18% ± 0.9% (wt/wt) of CDM was already obtained after 4 h of cultivation. At this time, the cell density was about 3.15 ± 0.08 g/liter and continued to increase to 4.7 g CDM per liter during further cultivation, whereas the CGP content of the cells decreased to 10.5% ± 2.1% (wt/wt). Microscopic examination revealed a large percentage of cells without CGP granules, indicating that the genetic information for CGP biosynthesis was not stable under these conditions and that a significant fraction of the cells had probably lost the plasmid. During this fermentation, cells and CGP were produced at a rate of 0.24 g CDM per liter and h and about 0.03 g CGP per liter and h, respectively.
FIG. 3.
Batch fermentation of E. coli DH1(pMa/c5-914::cphA6803) in Biostat D650 stirred tank reactor containing 400 liters 6% (vol/vol) protamylasse medium. The preculture was prepared at the 30-liter fermentation scale using the same medium but an incubation temperature of 30°C only. The fermentation parameters and cultivation conditions in the Biostat D650 reactor were a pH of 7.5, aeration at 0.15 vvm, agitation at 150 to 200 rpm, and a temperature of 37°C to induce the expression of CGP synthetase. (A) Ammonium (▴-▴), optical density at 600 nm (OD600, ▪), O2 in exhaust gas (⧫), and CO2 in exhaust gas (⧫-⧫). (B) Cell density (•), CGP content of the cells (▾), and pO2 (▴) were determined as described in Materials and Methods.
Isolation and analysis of CGP.
Cells harvested from the 500-liter bioreactor were lyophilized and then used for large-scale isolation by applying a previously described acid extraction method (11). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis revealed a molecular mass of about 26 to 30 kDa for CGP produced in protamylasse medium, which was the same when E. coli DH1(pMa/c5-914::cphA6803) was cultivated in mineral salt medium or complex media (11). HPLC analysis of the individual amino acid constituents of the isolated CGP revealed that the polymer was mainly composed of aspartate and arginine and that it contained only a little lysine, not exceeding 4% (mol/mol).
Pattern of amino acid utilization.
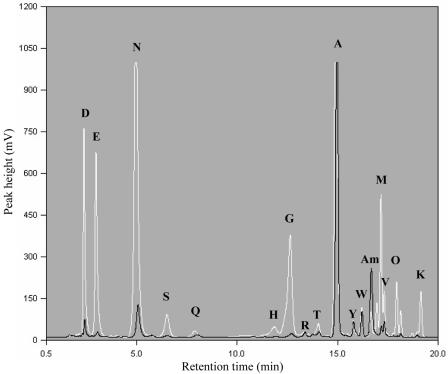
Samples from the 500-liter batch cultivation were centrifuged for 10 min at 14,000 × g to remove cells and other insoluble material and then were analyzed by HPLC without hydrolysis. The concentration of each amino acid was quantified at the beginning and at the end of the fermentation (Fig. 4). Of the aspartic acid and asparagine, which comprised about 35% of the amino acids present at the beginning of cultivation (Fig. 4), about 93% was utilized during the growth of E. coli DH1(pMa/c5-914::cphA6803). Similarly, glutamic acid and glutamine, amounting to 11.3% of the total amino acids present at the beginning, were reduced to 0.9% during cultivation. Glycine, histidine, lysine, methionine, ornithine, serine, threonine, and valine were also almost completely utilized during cultivation. In contrast, only 30% of the initial alanine was consumed. Therefore, alanine became the most abundant amino acid in the medium at the end of the cultivation period, constituting about 60% of the remaining amino acids. Beside alanine, only the concentrations of arginine, tryptophan, and tyrosine remained almost constant in the medium during fermentation, whereas that of ammonium increased during cultivation (Fig. 4).
FIG. 4.
Pattern of amino acid utilization during cultivation of E. coli DH1(pMa/c5-914::cphA6803) on 6% (vol/vol) protamylasse. Samples were obtained from the 500-liter batch cultivation experiment with the recombinant strain of E. coli and centrifuged, and the supernatant was then analyzed by HPLC as described in Materials and Methods. The white line shows the chromatogram obtained from an analysis of the sample withdrawn at the beginning of the fermentation (0 h), whereas the black line represents the chromatogram of the sample withdrawn at the end of the cultivation (14 h). Each peak represents a specific amino acid or another metabolite in relation to the retention time, as indicated in the figure. The values shown on the y axis (mV) indicate the concentrations of the respective compounds. Abbreviations: A, alanine; Am, ammonium; D, aspartic acid; E, glutamic acid; G, glycine; H, histidine; K, lysine; M, methionine; N, asparagine; O, ornithine; Q, glutamine; R, arginine; S, serine; T, threonine; V, valine; W, tryptophan; Y, tyrosine.
DISCUSSION
In this study, an economically efficient strategy for the biotechnological production of CGP using protamylasse as the sole medium component was outlined. Protamylasse represents an abundant residual of industrial starch production. So far, there have been no reports of the use of protamylasse as a nutrient for biotechnological processes; it is currently partially used in animal feed and as a source of glycoalkaloids in chemical synthesis (28). Until now, CGP could be obtained at the laboratory and technical scales and amounted to contents as high as 46% and 24% (wt/wt) of CDM in cells of A. calcoaceticus ADP1 and E. coli DH1(pMa/c5-914::cphA6803), respectively. However, such high CGP contents were only obtained if the mineral salt medium was supplemented with arginine or if expensive commercial complex media were used (9, 11).
All three bacterial strains investigated in this study grew to appropriate cell densities on protamylasse. However, Acinetobacter sp. strain ADP1 and P. putida KT2440 cells accumulated only a little CGP when grown on protamylasse under the applied conditions, which corresponded to only 2.5% of the CGP contents of E. coli DH1(pMa/c5-914::cphA6803) cells. One reason for the low CGP contents of these cells may be the high phosphate content of protamylasse (25.4 g/liter) because Acinetobacter sp. strain ADP1 only synthesizes CGP under phosphate limitation conditions. However, even if the phosphate concentration in protamylasse was reduced by precipitation in the presence of CaCl2, no significant increase in the CGP content of the cells occurred. Another explanation for the low CGP contents of the cells may be the presence of other amino acids beside arginine that could negatively interfere with CGP biosynthesis (9). Moreover, the synthesis of CGP in both bacterial strains depends on arginine feeding as the sole carbon source or on supplementation of arginine and aspartic acid, respectively (9, 41). Protamylasse contains more aspartic acid and asparagine than arginine (Table 1), which is not optimal for CGP synthesis in Acinetobacter sp. strain ADP1.
In contrast, E. coli DH1(pMa/c5-914::cphA6803) cells showed marked CGP synthesis when grown in protamylasse and when cyanophycin synthetase was induced by a shift of the cultivation temperature from 30 to 37°C (11). Cultivation experiments with different concentrations of protamylasse revealed that high CGP synthesis and a high cell density were achieved using 6% (vol/vol) protamylasse. A further increase in the protamylasse concentration reduced the cell density and CGP content by 33% and 95%, respectively. Whether some nutrients reach a toxic concentration when high concentrations of protamylasse are used has to be analyzed. Furthermore, the presence of other amino acids or organic acids could partially inhibit or reduce CGP biosynthesis. Short-chain-length peptides present in the protamylasse could act as primers for CGP biosynthesis, since the provision of amino acids or small peptides was reported in other studies to increase CGP biosynthesis (2, 3, 11).
One goal of this study was to achieve CGP production on a large scale by obtaining cells with high CGP contents. The cultivation experiments with E. coli DH1(pMa/c5-914::cphA6803) at 30- and 500-liter scales demonstrated that CGP can be produced in large amounts by using protamylasse as complete medium. Protamylasse may be pretreated to reduce the amount of organic acids, which are known to reduce the amounts of recombinant proteins formed (18) or to increase the concentration of arginine, the key amino acid in CGP synthesis in many bacterial strains (9, 41). Furthermore, application of the acid extraction method (11) for the isolation of CGP is an effective and time-saving process compared to the costly and labor-intensive method described previously by Simon and Weathers (37) and was also applicable to the cells obtained in this study. The CGP isolated by the acid extraction method resembled very much the CGP previously isolated from cells of E. coli cultivated in other media with regard to its molecular weight and polydispersity, and it exhibited a high degree of purity when analyzed by HPLC (data not shown).
Qualitative and quantitative analyses of the amino acid patterns of the protamylasse medium before and after cultivation of the recombinant E. coli strain revealed that the cells utilized most of the amino acids present in the medium to a large extent. This included the major fractions of aspartic acid, glutamic acid, asparagine, and glutamine. The consumption of 92% of the glutamic acid and glutamine during the cultivation of E. coli DH1(pMa/c5-914::cphA6803) in a medium containing a relatively high salt concentration (Table 1) may be coupled with the accumulation of potassium glutamate (10, 23), whereas it is known that the intracellular pools of alanine, arginine, and lysine remain approximately constant in cells of E. coli K-12 (34). There were only a few amino acids, such as alanine, arginine, tryptophan, and threonine, that were scarcely utilized. Interestingly, high CGP contents were obtained even though only a little arginine was present in the protamylasse medium and although the concentration of arginine in the medium did not change very much during cultivation. In all previous studies, arginine had to be fed to cultures to obtain high CGP contents in cells of recombinant strains of E. coli and other organisms. Whereas aspartic acid could be directly incorporated into CGP, E. coli obviously synthesized arginine from the other amino acids during cultivation in protamylasse medium.
The aim of this study was to investigate whether protamylasse provides a suitable medium for the biotechnological production of CGP on a large scale. The various cultivation experiments done here clearly revealed that protamylasse is suitable for CGP production. Assuming that E. coli can be cultivated to densities of 100 g/liter or higher, that CGP contents of the cells of 25% of CDM are obtained in industrial-scale fermentation within about 30 h, and that CGP can be released without mechanical cell disruption, the production costs for CGP might become very low. Beside further optimization of the process towards higher cell densities, the foreign cphA gene must be stabilized in the E. coli host.
Acknowledgments
We are very grateful to Herbert Ahlers, Ingo Voss, and Simone Diniz for their technical support and to Hans Mooibroek and Nico Oosterhuis for their assistance.
REFERENCES
- 1.Aboulmagd, E., F. B. Oppermann-Sanio, and A. Steinbüchel. 2000. Molecular characterization of the cyanophycin synthetase from Synechocystis sp. strain PCC6308. Arch. Microbiol. 174:297-306. [DOI] [PubMed] [Google Scholar]
- 2.Aboulmagd, E., F. B. Oppermann-Sanio, and A. Steinbüchel. 2001. Purification of Synechocystis sp. strain PCC 6308 cyanophycin synthetase and its characterization with respect to substrate and primer specificity. Appl. Environ. Microbiol. 67:2176-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aboulmagd, E., I. Voss, F. B. Oppermann-Sanio, and A. Steinbüchel. 2001. Heterologous expression of cyanophycin synthetase and cyanophycin synthesis in the industrial relevant bacteria Corynebacterium glutamicum and Ralstonia eutropha and in Pseudomonas putida. Biomacromolecules 2:1338-1342. [DOI] [PubMed] [Google Scholar]
- 4.Alford, D. D., A. P. Wheeler, and C. A. Pettigrew. 1994. Biodegradation of thermally synthesized polyaspartate. J. Environ. Polym. Degrad. 2:225-236. [Google Scholar]
- 5.Berg, H., K. Ziegler, K. Piotukh, K. Baier, W. Lockau, and R. Volkmer-Engert. 2000. Biosynthesis of the cyanobacterial reserve polymer multi-l-arginylpoly-l-aspartic acid (cyanophycin). Mechanism of the cyanophycin synthetase reaction studied with synthetic primers. Eur. J. Biochem. 267:5561-5570. [DOI] [PubMed] [Google Scholar]
- 6.Borzi, A. 1887. Le comunicazioni intracellulari delle Nostochinee. Malpighia 1:28-74. [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Chen, P. J., T. Y. Toribara, and H. Warner. 1956. Microdetermination of phosphorus. Anal. Chem. 28:1756-1758. [Google Scholar]
- 9.Elbahloul, Y., M. Krehenbrink, R. Reichelt, and A. Steinbüchel. 2005. Physiological conditions conductive to high cyanophycin content in biomass of Acinetobacter calcoaceticus strain ADP1. Appl. Environ. Microbiol. 71:858-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Epstein, W. 1986. Osmoregulation by potassium transport in Escherichia coli. FEMS Microbiol. Rev. 39:73-78. [Google Scholar]
- 11.Frey, K. M., F. B. Oppermann-Sanio, H. Schmidt, and A. Steinbüchel. 2002. Technical scale production of cyanophycin with recombinant strains of Escherichia coli. Appl. Environ. Microbiol. 68:3377-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hai, T., F. B. Oppermann-Sanio, and A. Steinbüchel. 1999. Purification and characterization of cyanophycin and cyanophycin synthetase from the thermophilic Synechococcus sp. MA19. FEMS Microbiol. Lett. 181:229-236. [DOI] [PubMed] [Google Scholar]
- 13.Hai, T., H. Ahlers, V. Gorenflo, and A. Steinbüchel. 2000. Axenic cultivation of anoxygenic phototrophic bacteria, cyanobacteria, and microalgae in a new closed tubular glass photobioreactor. Appl. Microbiol. Biotechnol. 53:383-389. [DOI] [PubMed] [Google Scholar]
- 14.Joentgen, W., T. Groth, A. Steinbüchel, T. Hai, and F. B. Oppermann. 1998. Polyaspartic acid homopolymers and copolymers, biotechnological production and use thereof. Patent application WO 98/39090.
- 15.Krehenbrink, M., F. B. Oppermann-Sanio, and A. Steinbüchel. 2002. Evaluation of non-cyanobacterial genome sequences for occurrence of genes encoding proteins homologous to cyanophycin synthetase and cloning of an active cyanophycin synthetase from Acinetobacter sp. strain DSM 587. Arch. Microbiol. 177:371-380. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 17.Lang, N. J., R. D. Simon, and C. P. Wolk. 1972. Correspondence of cyanophycin granules with structured granules in Anabaena cylindrica. Arch. Microbiol. 83:313-320. [Google Scholar]
- 18.Lee, S. Y. 1996. High cell-density culture of Escherichia coli. Trends Biotechnol. 14:98-105. [DOI] [PubMed] [Google Scholar]
- 19.Leopold, C. S., and D. R. Friend. 1995. In vivo pharmacokinetics study for the assessment of poly(l-aspartic acid) as a drug carrier for colon-specific drug delivery. J. Pharmacokinet. Biopharm. 4:397-406. [DOI] [PubMed] [Google Scholar]
- 20.Li, H., D. M. Sherman, S. Bao, and L. A. Sherman. 2001. Pattern of cyanophycin accumulation in nitrogen-fixing and non-nitrogen-fixing cyanobacteria. Arch. Microbiol. 176:9-18. [DOI] [PubMed] [Google Scholar]
- 21.Lotz, M. 2004. Added value from potato fruit juice. Starch 56:432-437. [Google Scholar]
- 22.Mackerras, A. H., N. M. de Chazal, and G. D. Smith. 1990. Transient accumulation of cyanophycin in Anabaena cylindrical and Synechocystis 6308. J. Gen. Microbiol. 136:2057-2065. [Google Scholar]
- 23.McLaggan, D., J. Naprstek, E. T. Buurman, and W. Epstein. 1994. Interdependence of K+ and glutamate accumulation during osmotic adaptation of Escherichia coli. J. Biol. Chem. 269:1911-1917. [PubMed] [Google Scholar]
- 24.Neumann, K., D. P. Stephan, K. Ziegler, M. Hühns, I. Broer, W. Lockau, and E. K. Pistorius. 2005. Production of cyanophycin, a suitable source for the biodegradable polymer polyaspartate, in transgenic plants. Plant Biotechnol. J. 3:249-258. [DOI] [PubMed] [Google Scholar]
- 25.Obst, M., F. B. Oppermann-Sanio, and A. Steinbüchel. 2002. Isolation of cyanophycin degrading bacteria, cloning and characterization of an extracellular cyanophycinase gene (cphE) from Pseudomonas anguilliseptica strain BI—the cphE gene from P. anguilliseptica BI encodes a cyanophycin-hydrolyzing enzyme. J. Biol. Chem. 277:25096-25105. [DOI] [PubMed] [Google Scholar]
- 26.Obst, M., A. Sallam, H. Luftmann, and A. Steinbüchel. 2003. Isolation and characterization of gram-positive cyanophycin-degrading bacteria—kinetic studies on cyanophycin depolymerase activity in aerobic bacteria. Biomacromolecules 5:153-161. [DOI] [PubMed] [Google Scholar]
- 27.Oppermann-Sanio, F. B., T. Hai, E. Aboulmagd, F. F. Hezayen, S. Jossek, and A. Steinbüchel. 1999. Biochemistry of microbial polyamide metabolism, p. 185-193. In A. Steinbüchel (ed.), Biochemical principles and mechanisms of biosynthesis and biodegradation of polymers. Wiley-VCH, Weinheim, Germany.
- 28.Patrick, J. E., K. Nadeshda, and A. de Groota. 2004. The synthesis of 16-dehydropregnenolone acetate (DPA) from potato glycoalkaloids. ARKIVOC II:24-50. [Google Scholar]
- 29.Picossi, S., A. Valladares, E. Flores, and A. Herrero. 2004. Nitrogen-regulated genes for the metabolism of cyanophycin, a bacterial nitrogen reserve polymer. J. Biol. Chem. 279:11582-11592. [DOI] [PubMed] [Google Scholar]
- 30.Pratesi, G., G. Savi, G. Pezzoni, O. Bellini, S. Penco, S. Tinelli, and F. Zunino. 1985. Poly(aspartic acid) as a carrier for doxorubicin: a comparative in vivo study of free and polymer-bounded drug. Br. J. Cancer 6:841-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richter, R., M. Hejazi, R. Kraft, K. Ziegler, and W. Lockau. 1999. Cyanophycinase, a peptidase degrading the cyanobacterial reserve material multi-l-arginyl-poly-l-aspartic acid (cyanophycin). Molecular cloning of the gene of Synechocystis sp. PCC6803, expression in Escherichia coli, and biochemical characterization of the purified enzyme. Eur. J. Biochem. 263:163-169. [DOI] [PubMed] [Google Scholar]
- 32.Roweton, S., S. J. Huang, and G. Swift. 1997. Poly(aspartic acid): synthesis, biodegradation, and current applications. J. Environ. Polym. Degrad. 5:175-181. [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Schleyer, M., R. Schmid, and E. P. Bakker. 1993. Transient, specific and extremely rapid release of osmolytes from growing cells of Escherichia coli K-12 exposed to hypoosmotic shock. Arch. Microbiol. 160:424-431. [DOI] [PubMed] [Google Scholar]
- 35.Schwamborn, M. 1998. Chemical synthesis of polyaspartates: a biodegradable alternative to currently used polycarboxylate homo- and copolymers. Polym. Degrad. Stabil. 59:39-45. [Google Scholar]
- 36.Simon, R. D. 1976. The biosynthesis of multi-l-arginyl-poly(l-aspartic acid) in the filamentous cyanobacterium Anabaena cylindrica. Biochim. Biophys. Acta 422:407-418. [DOI] [PubMed] [Google Scholar]
- 37.Simon, R. D., and P. Weathers. 1976. Determination of the structure of the novel polypeptide containing aspartic acid and arginine which is found in cyanobacteria. Biochim. Biophys. Acta 420:165-176. [DOI] [PubMed] [Google Scholar]
- 38.Simon, R. D. 1987. Inclusion bodies in the cyanobacteria: cyanophycin, polyphosphate, polyhedral bodies, p. 199-225. In P. Fay and C. van Baalen (ed.), The cyanobacteria. Elsevier, Amsterdam, The Netherlands.
- 39.Stephan, D. P., H. G. Ruppel, and E. K. Pistorius. 2000. Interrelation between cyanophycin synthesis, l-arginine catabolism and photosynthesis in the cyanobacterium Synechocystis sp. strain PCC 6803. Z. Naturforsch. 55:927-942. [DOI] [PubMed] [Google Scholar]
- 40.Vorage, M., and P. Eichner. 1997. European patent 094251, U.S. patent 6274105.
- 41.Voss, I., S. Cardoso-Diniz, E. Aboulmagd, and A. Steinbüchel. 2004. Identification of the Anabaena sp. strain PCC7120 cyanophycin synthetase as suitable enzyme for production of cyanophycin in gram-negative bacteria like Pseudomonas putida and Ralstonia eutopha. Biomacromolecules 5:1588-1595. [DOI] [PubMed] [Google Scholar]
- 42.Weber, K., and M. Osborn. 1969. The reliability of molecular weight determination by dodecylsulfate-polyacrylamide gel electrophoresis. J. Biol. Chem. 244:4406-4412. [PubMed] [Google Scholar]
- 43.Yokoyama, M., M. Miyauchi, N. Yamada, T. Okano, Y. Sakurai, K. Kataoka, and S. Inoue. 1990. Characterization and anticancer activity of the micelle-forming polymeric anticancer drug adriamycin-conjugated poly(ethylene glycol)-poly(aspartic acid) block copolymer. Cancer Res. 6:1693-1700. [PubMed] [Google Scholar]
- 44.Ziegler, K., A. Diener, C. Herpin, R. Richter, R. Deutzmann, and W. Lockau. 1998. Molecular characterization of cyanophycin synthetase, the enzyme catalyzing the biosynthesis of the cyanobacterial reserve material multi-l-arginyl-poly-l-aspartate (cyanophycin). Eur. J. Biochem. 254:154-159. [DOI] [PubMed] [Google Scholar]
- 45.Ziegler, K., R. Deutzmann, and W. Lockau. 2002. Cyanophycin synthetase-like enzymes of non-cyanobacterial eubacteria: characterization of the polymer produced by a recombinant synthetase of Desulfitobacterium hafniense. Z. Naturforsch. Sect. C 57:522-529. [DOI] [PubMed] [Google Scholar]