Abstract
Dehalococcoides species have a highly restricted lifestyle and are only known to derive energy from reductive dehalogenation reactions. The lipid fraction of two Dehalococcoides isolates, strains BAV1 and FL2, and a tetrachloroethene-to-ethene-dechlorinating Dehalococcoides-containing consortium were analyzed for neutral lipids and phospholipid fatty acids. Unusual phospholipid modifications, including the replacement of unsaturated fatty acids with furan fatty acids, were detected in both Dehalococcoides isolates and the mixed culture. The following three furan fatty acids are reported as present in bacterial phospholipids for the first time: 9-(5-pentyl-2-furyl)-nonanoate (Fu18:2ω6), 9-(5-butyl-2-furyl)-nonanoate (Fu17:2ω5), and 8-(5-pentyl-2-furyl)-octanoate (Fu17:2ω6). The neutral lipids of the Dehalococcoides cultures contained unusually large amounts of benzoquinones (i.e., ubiquinones [UQ]), which is unusual for anaerobes. In particular, the UQ-8 content of Dehalococcoides was 5- to 20-fold greater than that generated in aerobically grown Escherichia coli cultures relative to the phospholipid fatty acid content. Naphthoquinone isoprenologues (MK), which are often found in anaerobically grown bacteria and archaea, were also detected. Dehalococcoides shows a difference in isoprenologue pattern between UQ-8 and MK-5 that is atypical of other bacteria capable of producing both quinone types. The difference in UQ-8 and MK-5 isoprenologue patterns strongly suggests a special function for UQ in Dehalococcoides, and Dehalococcoides may utilize structural modifications in its lipid armamentarium to protect against free radicals that are generated in the process of reductive dechlorination.
Tetrachloroethene (PCE) and trichloroethene (TCE) are ideal solvents for degreasing and dry cleaning, and their widespread use has led to massive groundwater pollution. Many impacted subsurface environments are cocontaminated with more readily degradable petroleum hydrocarbons at many military sites or with starch at dry-cleaning sites. Microbes readily utilize aerobically degradable substrates, thus depleting oxygen in the groundwater and inducing anaerobiosis. Biotic and abiotic processes acting on these chlorinated solvents facilitate the formation and accumulation of toxic intermediates such as the dichloroethenes (DCEs) and the human carcinogen vinyl chloride (VC). Of the 1,430 contaminated sites listed on the U.S. EPA National Priorities List, 64% contain TCE, 54% contain PCE, and 35% contain VC (51).
A variety of subsurface bacteria are capable of anaerobic reductive dehalogenation, which represents the initial attack on these chlorinated solvents (43). Sulfate-reducing and acetogenic bacteria as well as methanogenic archaea can fortuitously catalyze reductive dehalogenation by free or enzyme-bound, metal ion-containing, heat-stable tetrapyrroles (9). Calculations of redox potentials for corresponding Cl-alkyl/alkyl couples range from +250 to +487 mV, suggesting that chloroorganic compounds are favorable electron acceptors (7). The first organism described to couple the reductive dehalogenation of 3-chlorobenzoate to energy conservation, and thus to growth, was Desulfomonile tiedjei, and the concept of metabolic reductive dechlorination, also known as (de)chlororespiration or chloridogenesis, was born. This stimulated a search for other organisms linking energy conservation and growth to reductive dechlorination.
Isolation efforts with chlorinated ethenes provided as terminal electron acceptors yielded numerous bacterial cultures that partially dechlorinated PCE and TCE to mainly cis-1,2-dichloroethene (cis-DCE) (summarized by Smidt and de Vos [44]). These organisms were largely gram-negative delta or epsilon proteobacteria or firmicutes (low-GC gram-positive bacteria) with no direct connection between substrate specificity and phylogeny (27). Apparently, only members of the strictly anaerobic Dehalococcoides group, which is not closely affiliated with any of the other aforementioned bacterial groups, are able to continue dechlorination beyond DCE. The first isolate described, Dehalococcoides ethenogenes strain 195, dechlorinated PCE completely to ethene, but this organism failed to derive energy from VC reductive dechlorination, and the critical last dechlorination step was slow and cometabolic (30, 31). Subsequently, a TCE-to-ethene-dechlorinating Dehalococcoides isolate, strain FL2, was obtained from pristine river sediment, but similar to the case in strain 195, the VC dechlorination step was slow and did not support growth (17, 27).
A milestone in chloroethene detoxification was the isolation of Dehalococcoides sp. strain BAV1, an organism coupling growth to the dechlorination of all DCE isomers and VC to produce biomass, nontoxic ethene, and inorganic chloride (15). The Dehalococcoides cluster is a deeply branching clade of anaerobic organisms within the Chloroflexi (green nonsulfur bacteria) (27). Dehalococcoides species are small (diameter, <0.5 μm), apparently nonmotile bacteria with an unusual flattened-disk morphology containing a depressed center (15). Dehalococcoides strains have a highly restricted metabolism and require hydrogen as an electron donor and particular aliphatic and/or aromatic chloroorganic compounds as electron acceptors. The known Dehalococcoides isolates contain multiple nonidentical reductive dehalogenase genes, although some reductive dehalogenase genes are shared among strains (23). Thus, distinct differences in substrate range and dechlorination patterns exist among the known Dehalococcoides strains. These physiological differences, however, are not reflected at the 16S rRNA gene level, and strains with different dechlorination activities share highly similar or identical 16S rRNA genes (16).
The importance of these remarkable microorganisms for bioremediation at the numerous sites impacted with chloroorganic pollutants prompted an examination for lipid biomarkers that could be utilized to specifically detect Dehalococcoides species and perhaps determine their metabolic activity. This study characterizes the phospholipid fatty acid (PLFA) patterns and the neutral lipid quinones of pure and mixed cultures of Dehalococcoides.
MATERIALS AND METHODS
Bacterial cultures.
Dehalococcoides sp. strain FL2 (GenBank accession number AF357918.2) was isolated from uncontaminated river sediment and utilizes TCE, cis-DCE, and trans-DCE as terminal electron acceptors (17). Dehalococcoides sp. strain BAV1 (GenBank accession number AY165308) was isolated from chloroethene-contaminated Bachman Road site aquifer material (Oscoda, Mich.) (16). Isolate BAV1 utilizes all DCE isomers, VC, vinyl bromide, and 1,2-dichloroethane as growth-supporting electron acceptors (15). Bio-Dechlor INOCULUM (BDI) is a commercially available PCE-to-ethene-dechlorinating consortium that has been successfully employed for bioaugmentation at field sites to promote complete detoxification (i.e., ethene formation). The culture contains multiple Dehalococcoides species, including strain BAV1 and strain FL2 (15, 26).
Media and growth conditions.
The pure Dehalococcoides cultures were grown in 160-ml serum bottles containing 100 ml of reduced, bicarbonate-buffered basal salt medium as described previously (15, 50). Before the bottles were autoclaved, l-cysteine, Na2S · 9 H2O (0.2 mM each), and 5 mM acetate were added, and the pH was adjusted to 7.1 to 7.2 by varying the flow of CO2 in the N2 purge gas. The culture vessels were closed with black butyl rubber stoppers secured with aluminum crimps. Following sterilization, TCE (10 μl) was added with a gastight Hamilton syringe, or VC (3 ml) was added with a plastic syringe. Hydrogen was added by syringe at a threefold excess of the amount required for complete dechlorination. Culture vessels received 2% (vol/vol) inocula from dechlorinating cultures, which were grown with TCE or VC. The PCE-to-ethene-dechlorinating BDI consortium was grown with TCE as the electron acceptor under the same conditions, except that dithiothreitol (0.5 mM) was added as an additional reductant. The consumption of hydrogen and chlorinated ethenes and the formation of ethene were monitored by gas chromatography as described previously (15, 50). The cultures were harvested when the majority of the choroethenes (<90%) were converted to ethene (strain BAV1 and BDI consortium) or a mixture of VC and ethene (strain FL2).
Lipid extraction and fractionation.
Bacterial biomass was collected by centrifugation in exploratory experiments and then by filtration onto 0.2-μm Anodisc filters (Whatman filters; Fisher Scientific, Pittsburgh, PA). In the preliminary experiments, pelleted FL2 cells were removed from the tubes by three washes with 1 ml each of 50 mM phosphate buffer (pH 7.5) and extracted as outlined below. To collect cells from the Anodisc filters, the filter material was separated from the polypropylene ring, placed in a 50-ml glass tube, and extracted using a modified Bligh and Dyer extraction method (2, 54) with 4 ml of phosphate buffer, 5 ml of chloroform, and 10 ml of methanol. The samples were sonicated for 2 min and then shaken for 4 h. After the shaking step, 5 ml of chloroform and 5 ml of preextracted Optima water (Fisher Scientific Co., Fair Lawn, NJ) were added, and the mixture was allowed to separate overnight. The next day, the organic layer was recovered and placed in a new glass tube, and the solvent was removed with a gentle stream of dry nitrogen. The total lipid was dissolved in chloroform and then fractionated with elution solvent, using 300-mg silicic acid columns, into neutral lipid, glycolipid, and phospholipid fractions (11), observing the precautions described by White and Ringelberg (55). The neutral lipids, recovered in chloroform from the column, were analyzed for respiratory quinones. The phospholipid fraction, eluted from the silicic acid column with methanol, was analyzed for PLFA.
The phospholipids were transesterified to fatty acid methyl esters using a mild alkaline methanolysis (11). The phospholipids were dissolved in 1 ml methanol-chloroform (1:1 [vol/vol]), to which 1 ml freshly prepared methanolic KOH (0.02 M) was added, and the mixture was heated at 37°C for 30 min. After cooling to room temperature, the methanolysate was diluted with 2 ml hexane-chloroform and neutralized with 200 μl 1 N acetic acid, and the methyl esters of the phospholipids (PLFA) were recovered after the addition of an equal volume of water. The mixture was vortexed and centrifuged, and the upper organic layer was transferred to a clean glass tube. The solvent was removed under a stream of dry nitrogen at 37°C, and the fatty acid methyl esters were analyzed by gas chromatography-mass spectrometry (GC/MS).
GC/MS.
GC/MS confirmation of PLFA profiles was accomplished with a Hewlett Packard 5890 GC (in constant flow mode) coupled to an HP5973 quadrupole mass selective detector (MSD). Samples were diluted in hexane with a 50-pmol/μl methyl-nonadecanoic acid (19:0) or methyl-heneicosanoic acid (21:0) standard. One microliter was injected from a 10-μl syringe by an HP 7683 autosampler. The GC inlet was operated in the splitless mode at 230°C. A 60-m Restek RTX-1 column (Restek, Bellafonte, PA) with an internal diameter of 0.25 mm and a film thickness of 25 μm was used, with helium as a carrier gas at 8.3 ml/min at a pressure of 13 lb/in2. The GC oven heated the column with a program that started at 60°C, held this temperature for 2 min, ramped at 10°C/min to 150°C, held this temperature for 0 min, ramped at 3°C/min to 312°C, and held this temperature for 0 min. The solvent delay was 8.5 min, and the total run time was 65 min. Components exiting the column were transferred by a heated transfer line (280°C) to the quadrupole MSD. After electron ionization at 70 eV, the positive ions were scanned as the total ion current, with a range of 41 to 450 m/z at 1.84 scans/s. Signals were collected, identified, and quantified with an HP Chemstation G1701BA, version B.01.00 (Hewlett-Packard).
Fatty acid nomenclature and identification.
Fatty acids were named according to the convention X:YωZ, where “X” stands for the number of carbon atoms in the chain, “Y” stands for the number of unsaturated positions, and “Z” is the number of carbon atoms from the methyl end of the molecule to the first unsaturated carbon atom. The following prefixes were used: i, iso branched; a, anteiso branched; nMe, methyl branch on the n carbon from the carboxylate end; br, branched at an unknown location; and cy, cyclopropyl. The suffixes “c” and “t” stand for the cis and trans geometric isomers of unsaturation, respectively.
In this paper, the furan fatty acids are designated Fu18:2ω7, indicating an 18-carbon chain length with two unsaturation points and showing that unsaturation occurs seven carbons from the alkyl end of the molecule. Fu18:2ω7 is methyl 9,12-epoxy-9,11-diene octadecanoate, or in IUPAC designation, methyl 8-(5-hexyl-2-furyl)-octanoate. Methyl branching is shown with the prefixes “MeFu” and “diMeFu,” for methyl and dimethyl furans, respectively.
PLFA were identified by GC/MS, equivalent chain length analysis, comparison with authentic internal standards (19:0 or 21:0), and the utilization of a calibration mixture of weighed authentic methyl esters (Matreya Inc., Pleasant Gap, PA). The calibration included the following methyl esters: 11:0 to 24:0; 2-OH 10:0, 12:0, 14:0, and 16:0; 3-OH 12:0 and 14:0; i14:0, 15:0, 16:0, and 17:0; 14:1w5c; a15:0; 16:1ω7c; 16:1ω7t; 18:3ω6; 18:2ω6; 18:3ω3; 18:1ω9c; 18:1ω7c; 18:1ω7t; 18:ω5c; 10Me18:0; 19:1ω12c; 19:1ω12t; Cy19:0; 20:2ω3; 20:2ω6; 20:3ω3; 20:1ω9c; 20:1ω9t; 22:6ω3; 22:1ω9c; and 22:1ω9t.
To group PLFA for comparisons, the following terms were utilized: normal sat., the sum of the mole percentages of saturated straight-chain PLFA; terminally br., the sum of the mole percentages of saturated terminally branched (iso and anteiso) PLFA; monounsat., the sum of the mole percentages of monounsaturated PLFA; mid-chain br., the sum of the mole percentages of mid-chain-branched (br15:0 and 10Me16:0) PLFA; cyclopropyl, the mole percentage of cyclopropyl (Cy17:0) PLFA; furan FA, the sum of the mole percentages of the furan fatty acids Fu18:2ω7 and Fu18:2ω6; percent modified, the sum of the mole percentages of all monounsaturated fatty acids, plus those further modified by methyl (mid-chain-branched and cyclopropyl) or oxygen (furan) addition; and avg. length, the average length of the lipid molecule in methylene units. For average length calculations, 16:0, i17:0, 10Me16:0, and Cy17:0 all have a length of 16 carbons. Shannon's H index was calculated according to the equation −Σ [pi × log10 (pi)], where pi is the proportion of lipid. Shannon's H index provides another way to compare PLFA profiles (19). In Microsoft Excel, this value is calculated from the mole percent data, using the array formula −SUM(IF(B5. .B35=0,0,(B5. .B35/100)*LOG(B5. .B35/100))), where B5…B35 is the range on the spreadsheet containing the data.
Nomenclature of respiratory quinones.
Respiratory quinones are isoprenoids found in bacterial cytoplasmic membranes. The major structural groups of bacterial isoprenoid quinones are ubiquinones (UQ; 1-methyl-2-isoprenyl-3,4-dimethoxyparabenzoquinone), menaquinones (MK; 1-isoprenyl-2-methyl-naphthoquinone), and 2-desmethylmenaquinones (1-isoprenylnaphthoquinone). Homologous series differing in the side chain length of each component are designated by numbers representing the total number of isoprene units in the side chain (e.g., UQ-10 has 10 isoprene units and 50 carbon atoms in its side chain).
Analysis of respiratory quinones by FIA/APCI/tandem quadrupole mass spectrometry.
Flow injection analysis (FIA) of respiratory quinones has been previously described in detail (10). The neutral lipid fraction was dissolved in 50 to 100 μl methanol with care to prevent exposure to UV light or air. Quinone standards, i.e., ubiquinones UQ6, UQ7, UQ9, and UQ10 and the menaquinone MK4 (vitamin K2) (Sigma, St. Louis, MO), were dissolved in methanol and kept in amber vials at 4°C. Aliquots were used to obtain a calibration curve prior to sample analysis. An Agilent 1100 quaternary high-performance liquid chromatography pump (Agilent Technologies, Palo Alto, CA) delivered a constant flow of methanol-chloroform (80:20 [vol/vol]) at 100 μl/min from the autosampler to the atmospheric pressure chemical inlet (APCI) for tandem quadrupole mass spectrometry. The tandem mass spectrometer was a Sciex 365 triple quadrupole mass spectrometer (MDS SCIEX, Concord, Ontario, Canada) utilized in the positive mode, with the nebulizer probe maintained at 425°C. Progenitor ions [M + H]+ generating product ions of m/z 197, the benzylium ion, represented ubiquinone isoprenologues. Positive progenitor ions generating the product ion at m/z 187.0 represented the 2-methyl-naphthoquinone core and were utilized for the identification of menaquinones. The product ion at m/z 174 represented the core of 2-desmethylmenaquinone. Mass transitions for multiple reaction monitoring (MRM), tabulated by Geyer et al. (10), were used for the detection of 11 major UQs and MKs. The multiple reaction monitoring transitions were optimized individually for each of the available quinone standards to yield the maximum signal intensity in the positive mode with the APCI source by adjusting the voltages between the orifice and skimmer (ground) and between the skimmer and the ring potential, the collision cell entrance potential, and the energy for collision-induced dissociation. For quinones without authentic standards, optimum mass-dependent parameters such as voltages on skimmer, orifice, and collision energy and the cell entrance potential were calculated, assuming a linear change with increasing numbers of isoprene units in the side chain to maintain approximately the same center-of-mass collision energy of 1.6 eV for UQn and 1.8 eV for MKn. Standards were also injected at five concentrations, ranging from 10 ppm to 480 ppm, before and after sample measurement to obtain and verify the calibration curve. Quinone homologs quantified by FIA were additionally verified by chromatographic separation of a sample on a DASH-8 high-performance liquid chromatography column (Hypersil Keyston, Palo Alto, CA), and precursor ions that generated product ions at m/z 197 and m/z 187 were scanned.
RESULTS
The Dehalococcoides cultures examined displayed unusual properties in their lipid and quinone profiles.
PLFA.
Table 1 presents the mol% values for PLFA from the Dehalococcoides cultures. Parameters that further characterize the PLFA were derived from Table 1 and are presented in Table 2. The PLFA profiles of Dehalococcoides sp. strain FL2, strain BAV1, and the BDI consortium were very similar to each other and contained saturated normal and mid-chain-branched fatty acids. The PLFA from pure cultures contained no monounsaturated fatty acids. Furan fatty acids were detected in samples from all cultures. The BDI consortium had a significant proportion (22%) of monounsaturated PLFA and larger proportions of terminally branched and cyclopropyl PLFA than the pure cultures. The PLFA profiles of Dehalococcoides strain FL2 and strain BAV1 were dominated by even-carbon-number straight-chain saturates comprising 59.5 to 68.8% of the total PLFA, with lesser proportions of furan and mid-chain-branched fatty acids. Terminally branched and cyclopropyl fatty acids were only detected in the consortium. Traces (<1%) of the monounsaturated fatty acid 18:1ω9c were detected in strains BAV1 and FL2. The total monounsaturated PLFA represented 22.25% of the total PLFA in the consortium. The percentage of modified fatty acids ranged from 31.1 to 40.5%, the average chain length varied between 15.8 and 16.4, and the fatty acid diversity, expressed as Shannon's H index (19), varied from 0.61 to 0.91 for both isolates and the consortium.
TABLE 1.
Means and standard deviations (SD) of phospholipid fatty acids of Dehalococcoides sp. strain FL2 (n = 4), strain BAV1 (n = 4), and the BDI consortium (n = 5)
| PLFA | Amt of PLFA (mean mol% [SD]) in indicated culture
|
|||
|---|---|---|---|---|
| FL2a | BAV1 | FL2 | BDI | |
| 12:0 | 2.9 | 0.91 (0.46) | 1.53 (0.55) | 0.63 (0.39) |
| i13:0 | 0.08 (0.05) | |||
| a13:0 | 0.10 (0.06) | |||
| 13:0 | 0.20 (0.06) | |||
| i14:0 | 0.55 (0.18) | |||
| 14:1ω5c | 1.12 (1.28) | |||
| 14:0 | 13.4 | 7.44 (2.33) | 8.55 (2.50) | 6.47 (1.74) |
| br15:0 | 1.3 | 1.54 (0.44) | 0.32 (0.46) | 0.14 (0.08) |
| i15:0 | 1.27 (0.77) | |||
| a15:0 | 3.42 (1.39) | |||
| 15:1 | 0.46 (0.20) | |||
| 15:0 | 1.95 (0.52) | |||
| br16:0 | 0.05 (0.04) | |||
| i16:0 | 0.15 (0.13) | |||
| 16:1ω9c | 0.23 (0.07) | |||
| 16:1ω7c | 11.54 (0.96) | |||
| 16:1ω7t | 0.12 (0.08) | |||
| 16:1ω5c | 6.04 (1.76) | |||
| 16:0 | 39.5 | 41.85 (4.01) | 49.79 (5.10) | 42.22 (2.82) |
| 10Me16:0 | 11.0 | 21.26 (1.25) | 24.42 (1.99) | 4.92 (2.88) |
| i17:0 | 0.05 (0.06) | |||
| a17:0 | 0.75 (0.21) | |||
| Cy17:0 | 2.45 (1.49) | |||
| 17:0 | 0.50 (0.14) | |||
| 18:1ω9c | 0.35 (0.22) | 0.53 (0.38) | 0.44 (0.24) | |
| 18:1ω7c | 2.30 (0.74) | |||
| 18:0 | 7.2 | 18.19 (2.77) | 8.77 (6.61) | 10.33 (1.61) |
| Fu18:2ω7 | 13.7 | 4.33 (1.50) | 3.71 (1.51) | 1.25 (1.12) |
| Fu18:2ω6 | 10.9 | 3.64 (1.75) | 2.37 (1.03) | 0.29 (0.21) |
| 20:0 | 0.49 (0.37) | |||
These FL2 data are from a preliminary experiment using a culture characterized by a long incubation time and cell harvesting by centrifugation.
TABLE 2.
Parameters derived from the phospholipid fatty acid mol % values in for Dehalococcoides strains FL2 and BAV1 and the BDI consortium
| Parametera | Value (mean [SD]) for indicated culture
|
|||
|---|---|---|---|---|
| FL2 | BAV1 | FL2 | BDI | |
| Biomass (nmol/liter) | 41.7 | 4.4 (2.3) | 3.1 (2.1) | 2.0 (0.8) |
| Fatty acid structural groups (mol %) | ||||
| Straight sat. | 63.0 | 68.87 (3.71) | 68.64 (1.93) | 62.30 (1.54) |
| Terminally br. | 6.36 (2.41) | |||
| Monounsat. | 0.35 (0.22) | 0.53 (0.38) | 22.25 (3.10) | |
| Mid-chain br. | 12.4 | 22.80 (1.36) | 24.75 (1.88) | 5.11 (2.95) |
| Cyclopropyl | 2.45 (1.49) | |||
| Furan FA | 24.6 | 7.98 (3.02) | 6.08 (2.38) | 1.53 (1.32) |
| Other derived parameters | ||||
| % Modified | 37.0 | 31.1 (3.7) | 31.4 (1.9) | 31.3 (1.6) |
| Avg length | 16.2 | 16.1 (0.1) | 15.9 (0.1) | 15.8 (0.1) |
| Shannon's H index | 0.76 | 0.69 (0.04) | 0.61 (0.04) | 0.91 (0.04) |
Straight sat., straight-chain saturated fatty acids; terminally br., terminally branched (iso- and anteiso-branched) fatty acids; monounsat., monounsaturated fatty acids; mid-chain br., mid-chain-branched saturated fatty acids (10Me16:0); cyclopropyl, cyclopropyl fatty acids (Cy17:0). For furan FA, see Fig. 1 and 2. % Modified, sum of monounsaturated, mid-chain-branched, cyclopropyl, and furan fatty acids; Shannon's H, fatty acid diversity (see Materials and Methods).
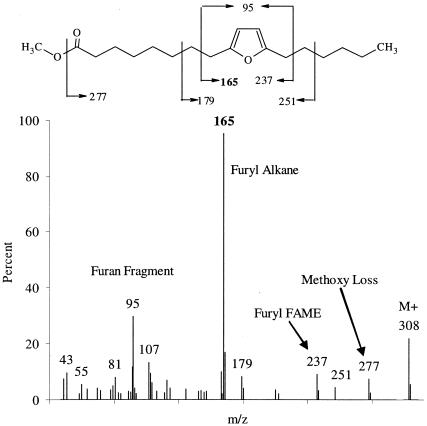
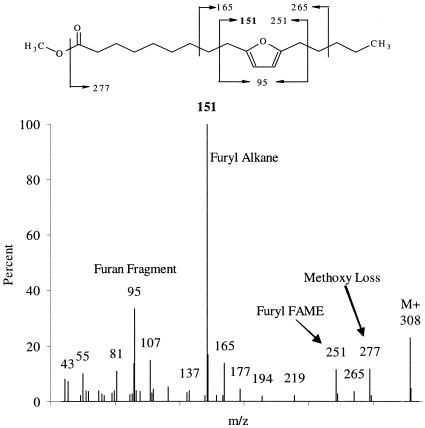
In all cultures analyzed, two 18-carbon furan PLFA were identified by GC/MS as Fu18:2ω7 and Fu18:2ω6 (Fig. 1 and 2), and comparison with published spectra confirmed their identity (4, 41, 53). The mass spectra of furan fatty acids are characterized by five peaks in the fragmentogram (Fig. 1 and 2 and Table 3). The molecular ion is usually apparent at m/z 56 + 14n, where n is the carbon chain length plus the number of methyl branches on the furan fatty acid. Evidence of the loss of a methoxy group is seen at M-31 at m/z 277 (Fig. 1 and 2). Beta-cleavage at either side of the furanyl moiety forms a furanyl alkane and a furanyl fatty acid methyl ester, and beta-cleavage at both sides of the furan yields a furan fragment. The furan fragment is at m/z 95, 109, or 123, depending on whether there are 0, 1, or 2 methyl groups on the furan, respectively.
FIG. 1.
Mass spectrum of Fu18:2w7, also called methyl 8-(5-hexyl-2-furyl)-octanoate or methyl 9,12-epoxy-9,11-octadecadienoate, recovered from FL2 and BAV1 biomass. FAME, fatty acid methyl ester.
FIG. 2.
Mass spectrum of Fu18:2ω6, also called methyl 9-(5-pentyl-2-furyl)-nonanoate or methyl 10,13-epoxy-10,12-octadecadienoate, recovered from FL2 and BAV1 biomass. FAME, fatty acid methyl ester.
TABLE 3.
Furan fatty acids identified in Dehalococcoides sp. strain FL2
| Furan fatty acid | IUPAC name | ECL | Molecular ion (m/z) | Methanol loss (m/z) | Furyl alkane (m/z) | Furyl-FAME (m/z) | Furan fragment (m/z) |
|---|---|---|---|---|---|---|---|
| Fu17:2ω6 | Methyl 8-(5-pentyl-2-furyl)-octanoate, methyl 9,12-epoxy-9,11-heptadecadienoate | 17.21 | 294 | 263 | 151 | 237 | 95 |
| Fu17:2ω5 | Methyl 9-(5-butyl-2-furyl)-nonanoate, methyl 10,13-epoxy-10,12-heptadecadienoate | 17.29 | 294 | 263 | 137 | 251 | 95 |
| Fu18:2ω7 | Methyl 8-(5-hexyl-2-furyl)-octanoate, methyl 9,12-epoxy-9,11-octadecadienoate | 18.20 | 308 | 277 | 165 | 237 | 95 |
| Fu18:2ω6 | Methyl 9-(5-pentyl-2-furyl)-nonanoate, methyl 10,13-epoxy-10,12-octadecadienoate | 18.25 | 308 | 277 | 151 | 251 | 95 |
Two additional furan fatty acids, Fu17:2ω6 and Fu17:2ω5, were detected in strain FL2 (Table 3). These additional furan fatty acids were <0.02% of the total PLFA and could not be accurately quantified.
Respiratory quinones.
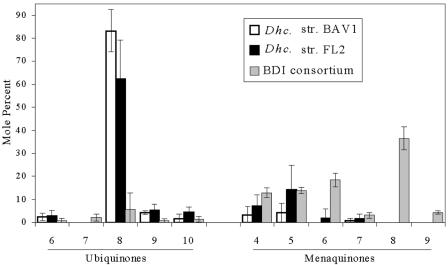
The respiratory quinone profiles of Dehalococcoides sp. strains FL2 and BAV1 and the BDI consortium are presented in Fig. 3. Both Dehalococcoides isolates had most of their respiratory quinones in the form of ubiquinones, specifically UQ-8, but they also contained menaquinones. The situation was reversed in the consortium, with most of the respiratory quinones being menaquinones, with MK-8 being the most abundant. No progenitor ions yielding product ions at m/z 174 were detected, indicating that 2-desmethylmenaquinones were not present. The UQ/MK molar ratios are 25.5, 5.3, and 0.1 for BAV1, FL2, and BDI, respectively. The amounts of respiratory quinone in Dehalococcoides strains BAV1 and FL2 were at least 5 times larger than that in aerobic, oxygen-respiring Escherichia coli (35), and 20 times larger amounts were detected in the BDI consortium (Table 4). The UQ/PLFA ratios are 26, 46, and 25 mmol/mol for BAV1, FL2, and the consortium, respectively. The MK/PLFA ratios of BAV1, FL2, and the BDI consortium are 3, 18, and 225 mmol/mol, respectively, and these values are higher than the MK/PLFA ratio of anaerobically growing E. coli of 2.5 mmol/mol (36). Dehalococcoides contained UQ-8 and MK-5 as major isoprenologues, thus showing a different major isoprenologue for each quinone class. UQ-8 predominated among the ubiquinones, and MK-5 predominated among the menaquinones. Not only do the predominant isoprenologues differ in chain length, but they also differ in their relative abundance. On a molar basis, about six- to eightfold more UQ-8 than MK-5 was detected in Dehalococcoides.
FIG. 3.
Quinone profiles for Dehalococcoides sp. strains BAV1 and FL2, and for the BDI consortium.
TABLE 4.
Biological parameters calculated from the respiratory quinones of Dehalococcoides strains BAV1 and FL2 and the BDI consortiuma
| Culture | Q (pmol/liter) | Q/PLFA (mmol/mol) | UQ/Q (mol/mol) | Avg N for UQ | Avg N for MK | % Even UQ | % Even MK |
|---|---|---|---|---|---|---|---|
| BAV1 | 151 (44) | 29 (12) | 0.9 (0.1) | 8.0 (0.0) | 4.9 (0.5) | 95 (1) | 28 (35) |
| FL2 | 177 (128) | 59 (17) | 0.7 (0.2) | 8.1 (0.1) | 4.8 (0.3) | 92 (6) | 41 (9) |
| BDI consortium | 435 (172) | 253 (174) | 0.1 (0.1) | 7.8 (0.5) | 6.6 (0.1) | 58 (34) | 76 (2) |
Q, total respiratory quinones; PLFA, phospholipid fatty acids; UQ, total ubiquinones; N, number of isoprene units in the quinone side chain; % even, percentage of quinones with an even number of isoprene units in the side chain. Q is expressed as pmol/liter of culture medium. Data are means (SD).
DISCUSSION
Lack of monounsaturated PLFA.
The PLFA profiles of Dehalococcoides sp. strains BAV1 and FL2 were unusual for their very large proportions of even-carbon-number, straight-chain saturated PLFA. The pure cultures had no detectable odd-carbon-number, iso-branched, or anteiso-branched PLFA. Monounsaturated PLFA typically represent 20 to 60% of the total PLFA of bacteria and eukaryotes living at mesothermic temperatures. Monounsaturated PLFA are typically part of the adaptation of the membrane to temperature and can serve as precursors of polyunsaturated, mid-chain-branched, and cyclopropyl fatty acids in many bacteria. Both isolates BAV1 and FL2, however, have only trace levels of monounsaturated PLFA. Dehalococcoides species are deeply rooted in the bacterial phylogenetic tree and are members of the Chloroflexi (green nonsulfur bacteria). Roseiflexus castenholzii, a phototrophic member of the Chloroflexi, also has no detectable monounsaturates (14), and only saturated straight-chain, even-carbon-number fatty acids were found in two other members of the Chloroflexi, Anaerolinea thermophila and Caldilinea aerophila (39). This is not a general characteristic of the Chloroflexi, as strains of Chloroflexus aurantiacus were reported to have 47 to 72% monounsaturates (25), and 38% monounsaturates were detected in Oscillochloris trichoides (14). Dehalococcoides sp. strain BAV1 and strain FL2 had detectable monounsaturated PLFA as <1% of the total PLFA (Table 2) and hence utilize another mechanism to modify membrane fluidity.
Furan fatty acids.
More remarkable than the lack of monounsaturated PLFA is the presence of furan PLFA derivatives in Dehalococcoides. Mid-chain-branched and furan fatty acids are synthesized from monounsaturated PLFA in other bacteria (1, 41, 42). Furan fatty acids have been detected at low levels in plants (33), animals, fungi (53), and bacteria (41, 42). Fu18:2ω6 from native cherry plants (Exocarpus cupressiformis, Santalaceae) was the first furan fatty acid described (33). The furan fatty acids found in these Dehalococcoides strains differ from those previously reported in not having one or two methyl groups attached to the furan ring carbons. Other Dehalococcoides isolates subsequently analyzed showed furan fatty acids together with a methylated furan fatty acid (molecular ion m/z 322, fragment ions at m/z 291, 265, 165, 109, and 95) and significant amounts of MK-7, MK-8, and MK-9 in addition to UQ-8, UQ-9, and MK-5 (R. Geyer, unpublished results). To our knowledge, Fu17:2ω6, Fu17:2ω5, and Fu18:2ω7 have not been reported in the literature before, and this is the first evidence of odd-carbon-number furan fatty acids in living cells.
Furan fatty acids are potent scavengers of hydroxyl radicals (34), inhibit erythrocyte hemolysis induced by singlet oxygen (35), and are found exclusively at the sn1 position of phosphatidylcholine in salmon roe (24), which is a favored structure for methylation, cyclopropylation, and furan ring formation. The proportions of furan PLFA in the pure Dehalococcoides cultures differed between the two batches of cultures analyzed. Two batches of FL2 produced 6% and 25% furan PLFA, compared to 8% for strain BAV1. The two batches of strain FL2 biomass generated 12.4% and 24.8% mid-chain-branched fatty acids, compared to 23% for strain BAV1. The culture conditions for both batches of FL2 biomass were identical; however, the time of incubation was longer for the first batch. The biomass of the first batch was harvested by centrifugation, and the pellet was immediately frozen in a plastic tube before the extraction procedures. The second batch of cells was harvested by filtration onto a membrane, followed immediately by the extraction procedure. Hence, differences in sample handling or the time of incubation may explain the observed shift in mid-chain-branched PLFA and furan fatty acid abundance.
Respiratory quinones.
The most abundant respiratory quinone in Dehalococcoides strains BAV1 and FL2 was UQ-8 (Fig. 4). Three other Chloroflexi isolates for which quinones have been reported have only menaquinones—MK-10 in Chloroflexus auriantiacus (14), MK-10 and a small amount of MK-4 in Chloroflexus aggregans (12), and MK-11 in Roseiflexus castenholzii (13). The redox potentials, E0, for the cis-DCE/VC, NO3/NO2−, and Fe(III)/Fe(II) couples are +0.35, +0.42, +0.2 V, respectively (52). The E0 of the ubiquinone-ubiquinol couple of +0.11 V suggests that UQ-8 could mediate electron transport to the chloroorganic electron acceptor; however, the unusually high concentration of UQ-8 in this organism is perplexing (18, 20, 47, 48). The anaerobe Dehalobacter restrictus utilizes MK in electron transport from hydrogen to PCE (38). Desulfomonile tiedjei requires 1,4-naphthoquinone for respiratory growth with 3-chlorobenzoate and forms a quinone without an isoprenoid side chain or MK and UQ structures (28). The PCE-to-cis-DCE-respiring, hydrogen-oxidizing, gram-negative organism Dehalobacter restrictus contains MK-7 and MK-8, with lesser amounts of MK-6 and MK-9 (22), and Sulfurospirillum (formerly named Dehalospirillum) multivorans, a PCE-to-cis-DCE dechlorinator utilizing hydrogen, formate, lactate, pyruvate, ethanol, and glycerol as electron donors, contains MK isoprenologues (37).
Prior to this report, UQs have only been reported for gram-negative Proteobacteria, some purple photosynthetic bacteria growing anaerobically, and a few Actinobacteria, most of which have MKs. In the Proteobacteria, UQs are formed during growth with high-potential terminal electron acceptors like oxygen and nitrate. UQs are also commonly found in eukaryotes with mitochondria. Anaerobically grown gram-negative bacteria, gram-positive bacteria, and archaea form MK isoprenologues (18). Some gram-negative bacteria grown anaerobically with nitrate form UQs, such as Novosphingobium pentaromativorans, which forms UQ-10 (49). Many others form MKs under these conditions. In E. coli, UQ biosynthesis is induced when oxygen is supplied (the UQ/PLFA ratio of aerobically respiring E. coli is about 5.2 mmol/mol) (36), but the UQ concentration did not increase following exposure to hydrogen peroxide or the redox cycling agent paraquat. Both compounds initiate lipid peroxidation by generating superoxide radicals and singlet oxygen (3, 45, 47). A knockout mutant of E. coli unable to synthesize UQs grew normally with nitrate or fumarate as a terminal electron acceptor but showed extremely diminished aerobic growth (46). Experiments with membrane preparations from this mutant suggested strongly that UQs donated electrons to cytochrome b. A function of respiratory quinones in oxidative stress protection, in addition to aerobic electron transport, is supported by multiple lines of evidence (47). Reduced UQ serves as a lipid-soluble antioxidant preventing lipid peroxidation. Although rates of superoxide and hydrogen peroxide production were higher in wild-type E. coli, the accumulated levels of both molecules were higher in the membranes of the UQ knockout mutant (48). Exposure of the mutant to hydrogen peroxide caused increased catalase activity and sensitivity to copper ion generation by superoxide. Water-soluble UQ-0 or UQ-2 added to the knockout mutant membranes significantly decreased hydrogen peroxide and superoxide accumulation. These experiments demonstrate a role for UQs in protecting E. coli cells from radicals generated by oxidative stress and indicate that these compounds fulfill similar roles for protecting Dehalococcoides species from radicals generated during its metabolism.
Another unusual feature of the pure Dehalococcoides cultures' quinone profiles is that the most abundant UQ homolog is UQ-8, while the most abundant MK is MK-5. Other organisms that synthesize both UQs and MKs typically have very similar proportions of different homologues of both quinones due to a shared biosynthetic pathway for producing the isoprenyl side chain for both quinones (5). For example, E. coli grown under microaerophilic conditions formed the isoprenologues UQ-8 and MK-8 as the largest proportions of isoprenologues of both quinones.
The formation of highly protective furan PLFA and UQ in response to oxidative stress (i.e., the generation of free radicals due to oxygen exposure) during cultivation is unlikely because reductants poised the redox potential at low levels (resazurin clear), and no air was introduced into the cultures during cultivation. The only exposure of the cells to oxygen occurred during harvesting; however, it is unlikely that this brief exposure can explain the observed furan PLFA and the UQ patterns. The exposure that Dehalococcoides most likely experiences is due to anaerobically generated free radicals. There is evidence that anaerobic reductive dechlorination generates radicals. The anaerobic dehalogenation of halothane anesthetics involves radical formation stabilized by spin trapping and detected by electron spin resonance (8). Recent studies found that the dehalogenation of chlorinated phenols is the result of an oxidative coupling reaction resulting in the generation of unstable free radicals (6). The known PCE-reductive dehalogenases contain corrinoid cofactors (29), and PCE-reductive dechlorination may involve a radical mechanism (21, 32). Reduced corrinoids are also implicated to have a function in Dehalococcoides reductive dehalogenases, and it is likely that chloroethene-reductive dehalogenation proceeds via one-electron transfer generation of free radicals (21, 32).
UQs have heretofore not been found in obligate anaerobes (5, 18, 20). The six- to eightfold greater amount of UQ-8 suggests a special function of UQs in Dehalococcoides. Typically, bacteria that make both quinones show parallel isoprenologue patterns, and the difference in proportions of the isoprenologues UQ-8 and MK-5 in Dehalococcoides again suggests a special function of UQ. The genome of Dehalococcoides ethenogenes reveals the presence of a superoxide dismutase (DET0956), but no other free radical oxidative stress-protective genes were detected, which is consistent with the observation that pure Dehalococcoides cultures are sensitive to oxygen (15, 30, 40). The genome sequence suggests that D. ethenogenes contains proteins for quinone modification, but none have yet been identified for quinone ring biosynthesis (40; S. H. Zinder, personal communication).
In conclusion, this research identified the following two potential Dehalococcoides lipid biomarkers: the presence of furan fatty acids in the phospholipids and high levels of high-potential benzoquinones in anaerobic venues. Both furan fatty acids and high-potential benzoquinones appear in other bacteria as protective compensation for oxidative stress. The growth yield of chlororespiring species is much lower than one would predict from the energetics of the reductive dechlorination process. Our findings hint that the energy investments in protective measures against free radicals are a possible cause for the lower-than-expected growth yields of chlororespiring bacteria, although a role of benzoquinones in electron transfer to the chlorinated electron acceptor cannot be excluded.
The detection of high levels of UQ-8 and furan PLFA as biomarkers in Dehalococcoides and the utilization of acetate as a carbon source suggest that in situ exposure of these organisms to 13C-labeled acetate and the subsequent detection of 13C in these biomarker lipids provide means for monitoring the growth and putative metabolic activities of these organisms in the field.
Acknowledgments
This research was partially supported by grant DE-FG02-04ER63939 from the Department of Energy, Office of Science, NABIR Field Program, and a grant from Regenesis Bioremediation Products, San Clemente, CA. Additional support was provided by the National Science Foundation under grant 0090496 (CAREER award to F.E.L.).
We thank S. H. Zinder for helpful discussions.
REFERENCES
- 1.Batna, A., J. Scheinkonig, and G. Spiteller. 1993. The occurrence of furan fatty acids in Isochrysis sp. and Phaeodactylum tricornutum. Biochim. Biophys. Acta 1166:171-176. [DOI] [PubMed] [Google Scholar]
- 2.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Phys. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 3.Bus J. S., S. D. Aust, and J. E. Gibson. 1976. Paraquat toxicity: proposed mechanism of action involving lipid peroxidation. Environ. Health Perspect. 16:139-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christie, W. W. 2005. Methyl esters of epoxy, oxy, and hydroxyl fatty acids. [Online.] http://www.lipidlibrary.co.uk/masspec.html.
- 5.Collins, M. D., and D. Jones. 1981. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implications. Microbiol. Rev. 45:316-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dec, J., K. Haider, and J. M. Bollag. 2003. Release of substituents from phenolic compounds during oxidative coupling reactions. Chemosphere 52:549-556. [DOI] [PubMed] [Google Scholar]
- 7.Dolfing, J. 2003. Thermodynamic considerations for dehalogenation, p. 88-114. In M. M. Häggblom and I. D. Bossert (ed.), Dehalogenation microbial processes and environmental applications. Kluwer Academic, New York, N.Y.
- 8.Fuji, K., M. Morio, H. Kikuchi, S. Ishihara M. Okida, and F. Ficor. 1984. In vivo spin-trap study on anaerobic dehalogenation of halothane. Life Sci. 35:463-468. [DOI] [PubMed] [Google Scholar]
- 9.Gantzer, J. C., and L. P. Wackett. 1991. Reductive dechlorination catalyzed by bacterial transition-metal coenzymes. Environ. Sci. Technol. 25:715-722. [Google Scholar]
- 10.Geyer, R., A. D. Peacock, D. C. White, C. Lytle, and G. J. Van Berkel. 2004. Atmospheric pressure chemical ionization and atmospheric pressure photoionization for simultaneous mass spectrometric analysis of microbial respiratory ubiquinones and menaquinones. J. Mass Spectrom. 39:922-929. [DOI] [PubMed] [Google Scholar]
- 11.Guckert, J. B., C. P. Antworth, P. D. Nichols, and D. C. White. 1985. Phospholipid, ester-linked fatty acid profiles as reproducible assays for changes in prokaryotic community structure of estuarine sediments. FEMS Microbiol. Ecol. 31:147-158. [Google Scholar]
- 12.Hanada, S., A. Hiraishi, K. Shimada, and K. Matsuura. 1995. Chloroflexus aggregans sp. nov., a filamentous phototrophic bacterium which forms dense cell aggregates by active gliding movement. Int. J. Syst. Bacteriol. 45:676-681. [DOI] [PubMed] [Google Scholar]
- 13.Hanada, S., S. Takaichi, K. Matsuura, and K. Nakamura. 2002. Roseiflexus castenholzii gen. nov., sp. nov., a thermophilic, filamentous, photosynthetic bacterium that lacks chlorosomes. Int. J. Syst. Evol. Microbiol. 52:187-193. [DOI] [PubMed] [Google Scholar]
- 14.Hanada, S., and B. K. Pierson. 2002. The family Chloroflexaceae. In M. Dworkin (ed.), The prokaryotes: an evolving electronic resource for the microbiology community, 3rd ed., release 3.11. Springer-Verlag, New York, N.Y. [Online.] http://141.150.157.117:8080/prikPUB/index.htm.
- 15.He, J., K. M. Ritalahti, Y. L. Yang, S. S. Koenigsberg, and F. E. Löffler. 2003. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424:62-65. [DOI] [PubMed] [Google Scholar]
- 16.He, J., K. M. Ritalahti, M. R. Aiello, and F. E. Löffler. 2003. Complete detoxification of vinyl chloride by an anaerobic enrichment culture and identification of the reductively dechlorinating population as a Dehalococcoides species. Appl. Environ. Microbiol. 69:996-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He, J., Y. Sung, R. Krajmalnik Brown, K. M. Ritalahti, and F. E. Löffler. 2005. Isolation and characterization of Dehalococcoides sp. strain FL2, a trichloroethene (TCE)- and 1,2-dichloroethene-respiring anaerobe. Environ. Microbiol. 7:1442-1450. [DOI] [PubMed] [Google Scholar]
- 18.Hedrick, D. B., and D. C. White. 1986. Microbial respiratory quinones in the environment. I. A sensitive liquid chromatographic method. J. Microbiol. Methods 5:243-254. [Google Scholar]
- 19.Hedrick, D. B., A. Peacock, J. R. Stephen, S. J. Macnaughton, J. Brüggemann, and D. C. White. 2000. Measuring soil microbial community diversity using polar lipid fatty acid and denaturing gradient gel electrophoresis data. J. Microbiol. Methods 41:235-248. [DOI] [PubMed] [Google Scholar]
- 20.Hiraishi, A. 1999. Isoprenoid quinones as biomarkers of microbial populations in the environment. J. Biosci. Bioeng. 88:449-460. [DOI] [PubMed] [Google Scholar]
- 21.Holliger, C., G. Wohlfarth, and G. Diekert. 1999. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol. Rev. 22:383-398. [Google Scholar]
- 22.Holliger, C., D. Hahn, H. Harmsen, W. Ludwig, W. Schumacher, B. Tindall, F. Varquez, N. Weiss, and A. J. B. Zehnder. 1998. Dehalobacter restrictus gen. nov. and sp. nov., a strictly anaerobic bacterium that reductively dechlorinates tetra- and trichloroethene in anaerobic respiration. Arch. Microbiol. 169:313-321. [DOI] [PubMed] [Google Scholar]
- 23.Hölscher, T., R. Krajmalnik-Brown, K. M. Ritalahti, F. von Wintzingerode, H. Grisch, F. E. Löffler, and L. Adrian. 2004. Multiple nonidentical reductive-dehalogenase homologous genes are common in Dehalococcoides. Appl. Environ. Microbiol. 70:5290-5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishii, K., H. Okajima, Y. Okada, and H. Watanabe. 1988. Studies on furan fatty acids of salmon roe phospholipids. J. Biochem. 103:836-839. [DOI] [PubMed] [Google Scholar]
- 25.Kenyon, C. N., and A. M. Grey. 1974. Preliminary analysis of lipids and fatty acids of green bacteria and Chloroflexus aurantiacus. J. Bacteriol. 120:131-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lendvay, J. M., F. E. Löffler, M. Dollhopf, M. R. Aiello, G. Daniels, B. Z. Fathepure, M. Gebhard, R. Heine, R. Helton, J. Shi, R. Krajmalnik-Brown, C. L. Major, Jr., M. J. Barcelona, E. Petrovskis, R. Hickey, J. M. Tiedje, and P. Adriaens. 2003. Bioreactive barriers: a comparison of bioaugmentation and biostimulation for chlorinated solvent remediation. Environ. Sci. Technol. 37:1422-1431. [Google Scholar]
- 27.Löffler, F. E., R. M. Cole, K. M. Ritalahti, and J. M. Tiedje. 2003. Diversity of dechlorinating bacteria, p. 58-87. In M. Häggblom and I. D. Bossert (ed.), Dehalogenation microbial processes and environmental applications. Kluwer Academic, New York, N.Y.
- 28.Louie, T. M., and W. W. Mohn. 1999. Evidence for a chemiosmotic model of dehalorespiration in Desulfomonile tiedjei DCB-1. J. Bacteriol. 181:40-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magnuson, J. K., R. V. Stern, J. M. Gossett, S. H. Zinder, and D. R. Burris. 1998. Reductive dechlorination of tetrachloroethene to ethene by a two-component enzyme pathway. Appl. Environ. Microbiol. 64:1270-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maymó-Gatell, X., Y. Chien, J. M. Gossett, and S. H. Zinder. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568-1571. [DOI] [PubMed] [Google Scholar]
- 31.Maymó-Gatell, X., I. Nijenhuis, and S. H. Zinder. 2001. Reductive dechlorination of cis-1,2-dichloroethene and vinyl chloride by “Dehalococcoides ethenogenes.” Environ. Sci. Technol. 35:516-521. [DOI] [PubMed] [Google Scholar]
- 32.McCauley, K. M., D. A. Pratt, S. R. Wilson, J. Shey, T. J. Burkey, and W. A. van der Donk. 2005. Properties and reactivity of chlorovinylcobalamin and vinylcobalamin and their implications for vitamin B12-catalized reductive dechlorination of chlorinated alkenes. J. Am. Chem. Soc. 127:1126-1136. [DOI] [PubMed] [Google Scholar]
- 33.Morris, L. J., M. O. Marshall, and V. Kelly. 1966. A unique furanoid fatty acid from Exocarpus seed oil. Tetrahedron Lett. 7:4249-4253. [Google Scholar]
- 34.Okada, Y., M. Kaneko, and H. Okajima. 1996. Hydroxyl radical scavenging activity of naturally occurring furan fatty acids. Biol. Pharm. Bull. 19:1607-1610. [DOI] [PubMed] [Google Scholar]
- 35.Okada, Y., H. Okamjima, M. Terauchi, H. Konishi, I. M. Liu, and H. Watanabe. 1990. Inhibitory effects of naturally occurring furan fatty acids on hemolysis of erythrocytes induced by singlet oxygen. Yakugaku Zasshi 110:665-672. (In Japanese.) [DOI] [PubMed] [Google Scholar]
- 36.Polglase, W. J., W. T. Pun, and J. Withaar. 1966. Lipoquinones of Escherichia coli. Biochim. Biophys. Acta 118:425-426. [DOI] [PubMed] [Google Scholar]
- 37.Scholz-Muramatsu, H., A. Neumann, M. Messmer, E. Moore, and G. Diekert. 1995. Isolation and characterization of Dehalospirillum multivorans gen. nov., sp. nov., a tetrachloroethene-utilizing, strictly anaerobic bacterium. Arch. Microbiol. 163:48-56. [Google Scholar]
- 38.Schumacher, W., and C. Holliger. 1996. The proton/electron ratio of the menaquinone-dependent electron transport from dihydrogen to tetrachloroethylene in “Dehalobacter restrictus.” J. Bacteriol. 178:2328-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sekiguchi, Y., T. Yamada, S. Hanada, A. Ohashi, H. Harada, and Y. Kamagata. 2003. Anaerolinea thermophila gen. nov., sp. nov., and Caldilinea aerophila gen. nov., sp. nov., novel filamentous thermophiles that represent a previously uncultured lineage of the domain Bacteria at the subphylum level. Int. J. Syst. Evol. Microbiol. 53:1843-1851. [DOI] [PubMed] [Google Scholar]
- 40.Seshadri, R., L. Adrian, D. E. Fouts, J. A. Eisen, A. M. Phillippy, B. A. Methe, N. L. Ward, W. C. Nelson, R. T. Deboy, H. M. Khouri, J. F. Kolonay, R. J. Dodson, S. C. Daugherty, L. M. Brinkac, S. A. Sullivan, R. Madupu, K. E. Nelson, K. H. Kang, M. Impraim, K. Tran, J. M. Robinson, H. A. Forberger, C. M. Fraser, S. H. Zinder, and J. F. Heidelberg. 2005. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307:105-108. [DOI] [PubMed] [Google Scholar]
- 41.Shirasaka, S., K. Nishi, and S. Shimizu. 1995. Occurrence of a furan fatty acid in marine bacteria. Biochim. Biophys. Acta 1258:225-227. [DOI] [PubMed] [Google Scholar]
- 42.Shirasaka, S., K. Nishi, and S. Shimizu. 1997. Biosynthesis of furan fatty acids (F-acids) by a marine bacterium, Shewanella putrefaciens. Biochim. Biophys. Acta 1346:253-260. [DOI] [PubMed] [Google Scholar]
- 43.Smidt, H., A. D. Akkermans, J. van der Oost, and W. M. de Vos. 2000. Halorespiring bacteria—molecular characterization and detection. Enzyme Microb. Technol. 27:812-820. [DOI] [PubMed] [Google Scholar]
- 44.Smidt, H., and W. M. de Vos. 2004. Anaerobic microbial dehalogenation. Annu. Rev. Microbiol. 58:43-73. [DOI] [PubMed] [Google Scholar]
- 45.Søballe, B., and R. K. Poole. 1997. Aerobic and anaerobic regulation of the ubiCA operon, encoding enzymes for the first two committed steps of ubiquinone biosynthesis in Escherichia coli. FEBS Lett. 414:373-376. [DOI] [PubMed] [Google Scholar]
- 46.Søballe, B., and R. K. Poole. 1998. Requirement for ubiquinone downstream of cytochrome(s) b in the oxygen-terminated respiratory chains of Escherichia coli K-12 revealed using a null mutant allele of ubiCA. Microbiology 144:361-373. [DOI] [PubMed] [Google Scholar]
- 47.Søballe, B., and R. K. Poole. 1999. Microbial ubiquinones: multiple roles in respiration, gene regulation and oxidative stress management. Microbiology 145:1817-1830. [DOI] [PubMed] [Google Scholar]
- 48.Søballe, B., and R. K. Poole. 2000. Ubiquinone limits oxidative stress in Escherichia coli. Microbiology 146:787-796. [DOI] [PubMed] [Google Scholar]
- 49.Sohn, J. H., K. K. Kwon, J. H. Kang, H. B. Jung, and S. J. Kim. 2004. Novosphingobium pentaromativorans sp. nov., a high-molecular-mass polycyclic aromatic hydrocarbon-degrading bacterium isolated from estuarine sediment. Int. J. Syst. Evol. Microbiol. 54:1483-1487. [DOI] [PubMed] [Google Scholar]
- 50.Sung, Y., K. M. Ritalahti, R. A. Sanford, J. W. Urbance, S. J. Flynn, J. M. Tiedje, and F. E. Löffler. 2003. Characterization of two tetrachloroethene-reducing, acetate-oxidizing anaerobic bacteria and their description as Desulfuromonas michiganensis sp. nov. Appl. Environ. Microbiol. 69:2964-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.U.S. Environmental Protection Agency. 1996. Agency for toxic substances and disease registry. ToxFAQs for chlorinated ethenes. [Online.] http://www.atsdr.cdc.gov/tfacts70.html.
- 52.Vogel, T. M., C. S. Criddle, and P. L. McCarty. 1987. Transformations of halogenated aliphatic compounds. Environ. Sci. Technol. 21:722-736. [DOI] [PubMed] [Google Scholar]
- 53.Wahl, H. G., A. Chrzanowski, H. M. Liebich, and A. Hoffmann. 1994. Identification of furan fatty acids in nutritional oils and fats by multidimensional GC-MSD. AppNote 6/1994. GERSTEL GmbH & Co. KG, Mülheim an der Ruhr, Germany.
- 54.White, D. C., W. M. Davis, J. S. Nickels, J. D. King, and R. J. Bobbie. 1979. Determination of the sedimentary microbial biomass by extractable lipid phosphate. Oecologia 40:51-62. [DOI] [PubMed] [Google Scholar]
- 55.White, D. C., and D. B. Ringelberg. 1998. Signature lipid biomarker analysis, p. 255-272. In R. S. Burlage, R. Atlas, D. Stahl, G. Geesey, and G. Sayler (ed.), Techniques in microbial ecology. Oxford University Press, New York, N.Y.