Abstract
One hundred twenty-one strains of the Bacillus cereus complex, of which 80 were isolated from a variety of sources in Brazil, were screened by PCR for the presence of sequences (bceT, hblA, nheBC, plc, sph, and vip3A) encoding putative virulence factors and for polymorphisms in variable-number tandem repeats (VNTR), using a variable region of the vrrA open reading frame as the target. Amplicons were generated from isolates of B. cereus and Bacillus thuringiensis for each of the sequences encoding factors suggested to play a role in infections of mammals. Intriguingly, the majority of these sequences were detected more frequently in Bacillus thuringiensis than in B. cereus. The vip3A sequence, which encodes an insecticidal toxin, was detected exclusively in B. thuringiensis. VNTR analysis demonstrated the presence of five different fragment length categories in both species, with two of these being widely distributed throughout both taxa. In common with data generated from previous studies examining European, Asian, or North American populations, our investigation of Brazilian isolates supports the notion that B. cereus and B. thuringiensis should be considered to represent a single species.
The Bacillus cereus group comprises six valid species, including Bacillus cereus, Bacillus thuringiensis, Bacillus mycoides, and Bacillus anthracis, which show a high degree of phenotypic similarity (5). Classification into these species is based on pathogenicity (to mammals or insects), plasmid content, and gross morphological characteristics (11, 13, 34). The genetic similarity between the members of the B. cereus group has been extensively studied by means of various molecular methods, including multilocus enzyme electrophoresis (41), fluorescent amplified fragment length polymorphism analysis (24), and sequence analysis of 16S rRNA, 23S rRNA, gyrB, and a variety of housekeeping genes (6, 9, 28, 32). The data generated by the bulk of these studies indicated that B. cereus and B. thuringiensis are virtually indistinguishable genotypically, which has led a number of researchers to suggest that these taxa may constitute a single species (6, 8, 9, 23).
Some strains of B. cereus cause food poisoning and other infections (29). Two principal types of food poisoning, emetic and diarrheal, have been described (38). The emetic type is induced by a small, cyclic, heat-stable peptide, cereulide, which causes vomiting a few hours after ingestion (2, 31). Evidence for a clonal population structure of cereulide-producing emetic B. cereus has recently been presented (14). Diarrheal food poisoning has been attributed to the production of enterotoxins during vegetative growth of B. cereus within the small intestine (18, 31). Five enterotoxins have been reported to play a role in diarrheal food poisoning: three single gene products encoded by the sequences entFM, cytK, and bceT and two multicomponent protein complexes, namely, hemolysin BL (HBL) and nonhemolytic enterotoxin (NHE) (7, 18, 19).
It has been reported that both B. cereus and B. thuringiensis are capable of growth within the hemocoel of various insect species and may provoke septicemia, although the former species is not generally considered to be a true pathogen of insects (37). The entomopathogenic characteristics of B. thuringiensis result from the production of toxins (Cry, Cyt, and Vip3A) encoded by genes carried on plasmids (37, 40). If these plasmids are lost, B. thuringiensis can no longer be clearly distinguished from B. cereus. Expression of the Cry and Cyt crystal proteins, which are synthesized when the bacterium enters the stationary phase or the sporulation phase, has never been reported for any B. cereus isolate (37). On the other hand, little is known concerning the interspecies distribution of the plasmid carrying the vip3A sequence, which is expressed during vegetative growth (15). The limited data produced to date from PCR- and colony hybridization-based investigations have shown that the vip3A sequence is present in a low number (15 to 30%) of B. thuringiensis isolates (15, 20, 35).
Analysis of repetitive-element sequence polymorphism has been suggested as a means of differentiating between different members of the B. cereus complex. Indeed, PCR based analysis of a variable-number tandem repeat (VNTR) sequence in the vrrA gene was demonstrated to allow the differentiation of a limited number of B. cereus, B. thuringiensis, B. mycoides, and B. anthracis isolates simply by comparing the sizes of amplicons on agarose gels (4, 27).
The first objective of this study was to assess the distribution of genes encoding putative virulence factors, specifically, the Vip3a, BceT, PclA, Sph, NheB, and NheC components of the NHE complex and HblA of the HBL complex, in a panel of B. cereus and B. thuringiensis strains comprised mainly of Brazilian isolates. A second objective was to examine the variable region of the vrrA sequence for fragment length polymorphisms, which might prove useful for the specific identification of B. cereus and B. thuringiensis. The results of this study are discussed in the context of the ongoing debate over the relationship between these two taxa.
MATERIALS AND METHODS
Strains and growth conditions.
The isolates described in Table 1 were obtained from the Collection of Cultures of the Genus Bacillus and Correlated Species of the Department of Bacteriology, Institute Oswaldo Cruz; from the Oswaldo Cruz Culture Collection of the National Institute of Health Quality Control; or were from our own laboratory collection. Strains were grown routinely on brain heart infusion medium. Cultures for DNA extraction were produced in nutrient broth containing 0.1% glucose. Stock cultures were maintained as suspensions of spores and cells at −20°C in 20% (vol/vol) glycerol.
TABLE 1.
Distributions of virulence genes, VNTR types, and origins of the Bacillus species examined
| Strain | Species | Origin | Country | hblA | nheBC | bceT | vip3A | plcA | sph | VNTR type |
|---|---|---|---|---|---|---|---|---|---|---|
| LFB406 | B. cereus | NCTC 2599 | NDa | + | − | + | − | − | + | 5 |
| LSB208 | B. cereus | Food (ATCC 33018) | Colombia | + | + | − | − | − | + | 1 |
| LFB1122 | B. cereus | Food (ATCC 33019) | Colombia | − | + | + | − | − | + | 3 |
| LSB269 | B. cereus | Food (unspecified) | Brazil | + | + | + | − | − | + | 4 |
| LFB287 | B. cereus | Food (infant milk formula) | Brazil | + | − | − | − | + | + | 0 |
| LFB439 | B. cereus | Food (food preservative) | Brazil | + | − | + | − | − | + | 3 |
| LFB505 | B. cereus | Food (infant milk formula) | Brazil | + | − | + | − | − | + | 3 |
| LFB511 | B. cereus | Food (infant milk formula) | Brazil | + | − | + | − | − | + | 2 |
| LFB513 | B. cereus | Food (rice mousse) | Brazil | + | − | + | − | − | + | 3 |
| LFB693 | B. cereus | Food (ice cream) | Brazil | − | − | − | − | − | + | 4 |
| LFB695 | B. cereus | Food (soup) | Brazil | + | − | + | − | − | + | 3 |
| LFB696 | B. cereus | Food (milk) | Brazil | + | − | − | − | − | + | 2 |
| LFB697 | B. cereus | Food (chocolate mousse) | Brazil | + | − | − | − | − | + | 3 |
| LFB698 | B. cereus | Food (tapioca flour) | Brazil | + | − | − | − | − | + | 2 |
| LFB699 | B. cereus | Food (carrot sauce) | Brazil | + | − | − | − | − | + | 2 |
| LFB700 | B. cereus | Food (powdered milk) | Brazil | − | − | + | − | − | + | 3 |
| LFB701 | B. cereus | Food (frozen fruit pulp) | Brazil | + | − | + | − | − | + | 2 |
| LFB703 | B. cereus | Food (black beans) | Brazil | + | − | + | − | − | + | 3 |
| LFB704 | B. cereus | Food (soup) | Brazil | + | − | − | − | − | + | 5 |
| LFB705 | B. cereus | Food (raw chicken) | Brazil | + | − | − | − | − | + | 3 |
| LFB737 | B. cereus | Food (unspecified) | Brazil | + | + | + | − | − | + | 3 |
| LFB585 | B. cereus | Clinical (urinary infection) | Brazil | + | + | + | − | − | + | 3 |
| LFB586 | B. cereus | Clinical (urinary infection) | Brazil | + | + | + | − | − | + | 4 |
| LFB589 | B. cereus | Clinical (urinary infection) | Brazil | + | + | + | − | − | + | 3 |
| LFB590 | B. cereus | Clinical (urinary infection) | Brazil | + | + | + | − | − | + | 2 |
| LFB591 | B. cereus | Clinical (urinary infection) | Brazil | + | − | − | − | − | + | 2 |
| LFB593 | B. cereus | Clinical (urinary infection) | Brazil | + | + | + | − | − | + | 4 |
| LFB595 | B. cereus | Clinical (diarrheal stools) | Brazil | + | + | + | − | − | + | 3 |
| LFB596 | B. cereus | Clinical (diarrheal stools) | Brazil | + | − | − | − | − | + | 5 |
| LFB598 | B. cereus | Clinical (diarrheal stools) | Brazil | + | − | − | − | − | + | 4 |
| LFB599 | B. cereus | Clinical (diarrheal stools) | Brazil | + | + | + | − | − | + | 3 |
| LFB600 | B. cereus | Clinical (diarrheal stools) | Brazil | + | − | + | − | − | + | 3 |
| LFB601 | B. cereus | Clinical (diarrheal stools) | Brazil | + | + | + | − | − | + | 2 |
| LFB602 | B. cereus | Clinical (diarrheal stools) | Brazil | + | + | + | − | − | + | 2 |
| LFB603 | B. cereus | Clinical (diarrheal stools) | Brazil | + | + | + | − | − | + | 3 |
| LFB605 | B. cereus | Clinical (diarrheal stools) | Brazil | + | + | + | − | − | + | 2 |
| LFB607 | B. cereus | Clinical (diarrheal stools) | Brazil | + | + | − | − | − | + | 4 |
| LFB608 | B. cereus | Clinical (diarrheal stools) | Brazil | + | + | + | − | − | + | 3 |
| LFB609 | B. cereus | Clinical (diarrheal stools) | Brazil | + | + | + | − | − | + | 3 |
| LFB1181 | B. cereus | Clinical (avian septicemia) | Brazil | + | + | + | − | + | + | 0 |
| LFB1123 | B. cereus | Environment (Lepidoptera) | Madagascar | − | − | + | − | − | + | 5 |
| LFB1124 | B. cereus | Environment (Lepidoptera) | Madagascar | + | − | + | − | − | + | 2 |
| LFB1125 | B. cereus | Environment (Homoptera) | Madagascar | + | − | + | − | − | + | 3 |
| LFB1126 | B. cereus | Environment (Coleoptera) | Madagascar | + | + | + | − | − | + | 4 |
| LFB1127 | B. cereus | Environment (Coleoptera) | Madagascar | + | − | + | − | − | + | 3 |
| LFB1128 | B. cereus | Environment (Lepidoptera) | Yugoslavia | + | + | + | − | − | + | 5 |
| LFB1129 | B. cereus | Environment (Lepidoptera) | Bulgaria | + | + | + | − | − | + | 2 |
| LFB1131 | B. cereus | Environment (Lepidoptera) | Czechoslovakia | − | + | + | − | − | + | 5 |
| LFB1130 | B. cereus | Environment (Lepidoptera) | Egypt | + | + | + | − | − | + | 5 |
| LFB1141 | B. cereus | Environment (Coleoptera) | Egypt | + | + | + | − | − | + | 3 |
| LFB1134 | B. cereus | Environment (Lepidoptera) | Russia | + | + | + | − | − | + | 4 |
| LFB1135 | B. cereus | Environment (Coleoptera) | Indonesia | + | + | + | − | − | + | 3 |
| LFB1136 | B. cereus | Environment (Lepidoptera) | Pakistan | + | + | + | − | − | + | 3 |
| LFB1133 | B. cereus | Environment (Lepidoptera) | United States | + | + | + | − | − | + | 4 |
| LFB1137 | B. cereus | Environment (Diptera) | United States | + | + | + | − | − | + | 3 |
| LFB1138 | B. cereus | Environment (Diptera) | United States | + | + | + | − | − | + | 3 |
| LFB1139 | B. cereus | Environment (Diptera) | France | + | + | + | − | − | + | 3 |
| LFB1140 | B. cereus | Environment (Coleoptera) | France | + | + | + | − | − | + | 2 |
| LFB579 | B. cereus | Environment (soil) | Brazil | + | − | + | − | − | + | 3 |
| LFB694 | B. cereus | Environment (soil) | Brazil | + | + | + | − | − | + | 2 |
| LFB770 | B. cereus | Environment (soil) | Brazil | + | + | − | − | − | + | 1 |
| LFB771 | B. cereus | Environment (soil) | Brazil | − | + | + | − | − | + | 0 |
| LFB773 | B. cereus | Environment (soil) | Brazil | − | + | + | − | − | + | 3 |
| LFB1120 | B. mycoides | ATCC 11778 (ND) | ND | − | + | + | − | − | + | 2 |
| LFB855 | B. thuringiensis serotype oswaldocruzi | Food (black pepper) | Brazil | + | + | − | − | + | + | 3 |
| LFB856 | B. thuringiensis serotype oswaldocruzi | Food (black pepper) | Brazil | − | + | − | − | − | + | 2 |
| LFB857 | B. thuringiensis serotype israelensis | Food (black pepper) | Brazil | + | + | + | − | − | + | 3 |
| LFB858 | B. thuringiensis serotype aizawai | Food (black pepper) | Brazil | + | + | + | − | + | + | 3 |
| LFB860 | B. thuringiensis serotype israelensis | Food (black pepper) | Brazil | + | + | + | − | + | + | 5 |
| LFB861 | B. thuringiensis serotype israelensis | Food (black pepper) | Brazil | + | − | + | − | − | + | 5 |
| LFB862 | B. thuringiensis serotype israelensis | Food (black pepper) | Brazil | + | + | + | − | − | + | 2 |
| LFB864 | B. thuringiensis serotype israelensis | Food (black pepper) | Brazil | + | + | + | − | + | + | 3 |
| LFB867 | B. thuringiensis serotype israelensis | Food (black pepper) | Brazil | + | − | + | − | − | + | 3 |
| LFB868 | B. thuringiensis serotype israelensis | Food (black pepper) | Brazil | + | + | + | − | + | + | 3 |
| LFB869 | B. thuringiensis serotype brasiliensis | Food (black pepper) | Brazil | + | − | + | − | − | + | 3 |
| LFB891 | B. thuringiensis serotype aizawai | Food (black pepper) | Brazil | + | + | + | − | + | + | 4 |
| LFB895 | B. thuringiensis serotype israelensis | Food (black pepper) | Brazil | + | + | + | − | − | + | 2 |
| LFB109 | B. thuringiensis serotype sotto | Clinical (vaginal secretion) | Brazil | − | − | + | − | + | + | 1 |
| LFB584 | B. thuringiensis serotype israelensis | Environment (Diptera) | Israel | + | + | + | − | + | + | 2 |
| LFB471 | B. thuringiensis serotype sotto | Environment (Lepidoptera) | Pakistan | + | − | + | − | + | + | 3 |
| LFB472 | B. thuringiensis serotype sotto | Environment (Lepidoptera) | Pakistan | + | + | + | − | + | + | 1 |
| LFB721 | B. thuringiensis serotype tohokuensis | Environment (Lepidoptera) | ND | + | + | + | − | − | + | 4 |
| LFB725 | B. thuringiensis serotype nigeriensis | Environment (cocoon) | ND | + | + | + | − | − | + | 3 |
| LFB727 | B. thuringiensis serotype kyushuensis | Environment (Lepidoptera) | ND | + | + | + | − | + | + | 3 |
| LFB715 | B. thuringiensis serotype toumanoffi | Environment (Lepidoptera) | ND | + | + | + | − | + | + | 3 |
| LFB476 | B. thuringiensis serotype morrisoni | Environment (soil) | ND | − | + | − | − | + | + | 2 |
| LFB562 | B. thuringiensis serotype tolworthi | Environment (soil) | ND | + | + | + | − | + | + | 2 |
| LFB563 | B. thuringiensis serotype pakistani | Environment (soil) | ND | + | − | + | − | + | + | 4 |
| LFB677 | B. thuringiensis serotype canadenses | Environment (soil) | ND | + | + | + | − | + | + | 3 |
| LFB680 | B. thuringiensis serotype subtoxicus | Environment (soil) | ND | + | + | + | − | − | + | 2 |
| LFB681 | B. thuringiensis serotype aizawai | Environment (soil) | ND | + | + | − | − | + | + | 3 |
| LFB682 | B. thuringiensis serotype subtoxicus | Environment (soil) | ND | + | + | + | − | − | + | 2 |
| LFB683 | B. thuringiensis serotype finitimus | Environment (soil) | ND | + | + | − | − | + | + | 3 |
| LFB717 | B. thuringiensis serotype ostriniae | Environment (soil) | ND | − | − | + | − | + | + | 3 |
| LFB730 | B. thuringiensis serotype japonensis | Environment (soil) | ND | + | − | + | − | + | + | 2 |
| LFB731 | B. thuringiensis serotype darmstadiensis | Environment (soil) | ND | + | + | + | − | + | + | 2 |
| LFB724 | B. thuringiensis serotype thompsoni | Environment (soil) | ND | + | + | + | − | + | + | 2 |
| LFB99 | B. thuringiensis serotype israelensis | Environment (soil) | Brazil | − | + | − | − | − | + | 3 |
| LFB257 | B. thuringiensis serotype kurstaki | Environment (soil) | Brazil | + | + | + | − | + | + | 3 |
| LFB263 | B. thuringiensis serotype kurstaki | Environment (soil) | Brazil | + | + | + | + | + | + | 3 |
| LFB475 | B. thuringiensis serotype kurstaki | Environment (soil) | Brazil | + | + | + | − | + | + | 3 |
| LFB679 | B. thuringiensis serotype morrisoni | Environment (soil) | Brazil | + | + | + | + | + | + | 3 |
| LFB710 | B. thuringiensis serotype israelensis | Environment (soil) | Brazil | + | + | + | − | − | + | 3 |
| LFB756 | B. thuringiensis serotype morrisoni | Environment (soil) | Brazil | + | + | + | − | − | + | 2 |
| LFB761 | B. thuringiensis serotype israelensis | Environment (soil) | Brazil | + | + | − | − | − | + | 2 |
| LFB769 | B. thuringiensis serotype israelensis | Environment (soil) | Brazil | + | + | + | − | − | + | 5 |
| LFB772 | B. thuringiensis serotype israelensis | Environment (soil) | Brazil | + | + | + | − | + | + | 3 |
| LFB775 | B. thuringiensis serotype israelensis | Environment (soil) | Brazil | + | − | + | − | + | + | 2 |
| LFB776 | B. thuringiensis serotype israelensis | Environment (soil) | Brazil | + | + | + | − | + | + | 2 |
| LFB780 | B. thuringiensis serotype morrisoni | Environment (soil) | Brazil | + | − | + | + | + | + | 2 |
| LFB781 | B. thuringiensis serotype morrisoni | Environment (soil) | Brazil | + | + | + | + | + | + | 3 |
| LFB784 | B. thuringiensis serotype israelensis | Environment (soil) | Brazil | + | + | + | − | + | + | 2 |
| LFB854 | B. thuringiensis serotype israelensis | Environment (soil) | Brazil | + | + | + | − | − | + | 2 |
| LFB890 | B. thuringiensis serotype yunnanensis | Environment (soil) | Brazil | + | − | + | − | − | + | 2 |
| LFB912 | B. thuringiensis serotype israelensis | Environment (soil) | Brazil | + | + | + | − | − | + | 2 |
| LFB913 | B. thuringiensis serotype israelensis | Environment (soil) | Brazil | + | + | + | − | + | + | 3 |
| LFB914 | B. thuringiensis serotype israelensis | Environment (soil) | Brazil | + | + | + | − | − | + | 5 |
| LFB915 | B. thuringiensis serotype israelensis | Environment (soil) | Brazil | + | + | + | − | + | + | 2 |
| LFB1016 | B. thuringiensis serotype oswaldocruzi | Environment (soil) | Brazil | + | + | + | − | + | + | 3 |
| LFB1021 | B. thuringiensis serotype oswaldocruzi | Environment (soil) | Brazil | − | + | − | − | + | + | 2 |
| LFB1065 | B. thuringiensis serotype israelensis | Environment (soil) | Brazil | + | + | + | − | − | + | 3 |
ND, not determined.
Oligonucleotides and conditions for PCR.
Template DNA for PCR screening was prepared by processing 1 ml of culture grown for 18 h at 37°C, using the GenomicPrep cell and tissue DNA kit (Amersham Pharmacia Biotech Inc.). Twenty-five nanograms of DNA was used for each reaction. Ultrapure water (Gibco) was used in all negative control reactions (without DNA) and for the preparation of the PCR mixture. All reaction mixtures for amplification of sequences encoding putative virulence factors contained 5 μl of template DNA (25 ng), 10 mM Tris-HCl (pH 8.3), 10 mM KCl, 0.2 mM of each deoxynucleoside triphosphate, 2.5 mM MgCl2, 1 μM of each primer, and 0.5 U of Taq DNA polymerase (Pharmacia). Table 2 provides information concerning the primers used for the amplification of each gene and the cycling conditions used in the amplification reactions, which were conducted using a model 9600 thermocycler (Perkin-Elmer/Applied Biosystems). To validate the results, all PCR amplifications were performed a minimum of three times. Amplification products were analyzed by separating 12 μl of reaction mixture on 2.0% (wt/vol) agarose (Sigma type V) gels at 3.2 V cm−1 in TBE buffer (40 mM Tris-acetate, 1 mM EDTA [pH 8.0]), followed by staining with ethidium bromide and examination under UV light. Negative controls (without DNA) were included in all amplifications. As a control against poor quality of genomic DNA causing amplification failure, the quality of any DNA extract that failed to amplify in a specific reaction was examined by attempting amplification with a pair of universal primers designed to amplify a region of the 16S rRNA gene.
TABLE 2.
Primers and amplification conditions used in this study
| Gene | Primer name | Oligonucleotide sequence (5′→3′) | Amplification conditionsa
|
Position | Product (bp) | Reference | |||
|---|---|---|---|---|---|---|---|---|---|
| D | A | E | No. of cycles | ||||||
| plcA | Bt/cPCPLC1 | AAC GGT ATT TAT GCT GCT GAC TAT | 94°C, 45 s | 51°C, 45 s | 72°C, 60 s | 25 | 259-282 | 405 | 20 |
| Bt/cPCPLC2 | CGC TAC TAC TGC CGC TCC AT | 663-664 | |||||||
| sph | Ph1 | CGT GCC GAT TTA ATT GGG GC | 94°C, 20 s | 58°C, 20 s | 72°C, 60 s | 35 | 1007-1026 | 558 | 25 |
| Ph2 | CAA TGT TTT AAA CAT GGA TGC G | 1564-1543 | |||||||
| hblA | HBLA1 | GCT AAT GTA GTT TCA CCT GTA GCA AC | 94°C, 30 s | 65°C, 60 s | 72°C, 60.5 s | 30 | 121-147 | 883 | 30 |
| HBLA2 | AAT CAT GCC ACT GCG TGG ACA TAT AA | 968-994 | |||||||
| bceT | BCET1 | GAG TTA GTT TCA ACA GCG CG | 95°C, 30 s | 56°C, 30 s | 72°C, 120 s | 30 | 1173-1192 | 593 | 17 |
| BCET2 | TCA GGC TGA TCT AGT AGA CC | 1746-1765 | |||||||
| vip3A | VIP3A1 | CCG CCA AGT GGT TTT ATT AGC | 94°C, 30 s | 55°C, 60 s | 72°C, 60 s | 30 | 1706-1727 | 536 | This study |
| VIP3A2 | GTG AAA TTC CTC CGT CCT TAT G | 1842-1864 | |||||||
| nheBC | NHE1 | ATG TAT CGT CTG TTG ATG CG | 95°C, 30 s | 51°C, 30 s | 72°C, 120 s | 30 | 1741-1760 | 1,200 | 17 |
| NHE2 | TAC TTG ATG TCG TTT TGT CC | 2953-2972 | |||||||
| VNTRb | EW1 | TAT GGT TGG TAT TGC T | 94°C, 120 s | 55°C, 60 s | 72°C, 60 s | 35 | Vc | V | 26 |
| EW2 | ATG GTT CCG CCT TATCG | ||||||||
All PCR protocols used an initial denaturation step at 94°C for 2 min, followed by 25 to 35 cycles at the temperatures and times indicated (D, denaturation; A, annealing; E, extension. A final extension step (10 min at 72°C) was also included.
The final extension step was for 1 min at 72°C.
V, binding sites and product size were variable.
Evaluation of VNTR polymorphism in the vrrA operon.
Amplification of the region of the vrrA operon reported to contain VNTR employed the primers EWA1 and EWA2 described by Andersen et al. (4). The PCR mixture and cycling conditions were as reported by Kim et al. (27). Amplicons were analyzed following electrophoresis on 3.0% Ultrapure agarose (Invitrogen) gels in 0.5× TBE buffer. Electrophoresis was carried out for 2.5 h at 80 V. VNTR types were defined according to the presence or absence of polymorphic bands, indicated by amplicons of variable size. A 50-bp DNA ladder (Sigma Chemical Co.) was used as molecular size marker.
RESULTS AND DISCUSSION
The discussion concerning the classification of B. cereus and B. thuringiensis as a single species or as two species is primarily a philosophical one. Yet, the outcome of such a discussion may have economic implications, particularly with regard to the use of B. thuringiensis as a biological control agent. Data from previous studies (7, 12, 17, 20, 21, 25, 30, 33) have clearly demonstrated that members of both taxa possess sequences encoding the putative virulence factors examined in the current study. However, isolates from South America did not form a significant portion of any of the strain panels examined to date. To address this discrepancy, the present study examined a total of 121 bacteria from the B. cereus group, with approximately two-thirds of the strains having been isolated in Brazil.
The rationale for selecting the particular sequences examined was based first on the fact that the presence of these genes in a variety of strains of both taxa from other geographical locations had been established previously and second on our hope to extend upon previous data concerning the distribution of the vip3A sequence within these taxa. This issue required clarification, given that data from colony screening experiments with unidentified Bacillus isolates (15) suggested that the vip3A sequence could be present in some B. cereus strains.
As reported in previous PCR-based screening studies, the enterotoxin genes examined herein were widespread throughout both species and were present in strains isolated from food, clinical material, insects, and soil environments (Table 1). The presence of the HBL operon, examined via amplification of the hblA gene, encoding the B component of the tripartite enterotoxin, was detected at a level of 90% in both the total B. thuringiensis population and the Brazilian subset of strains (Table 1). The common occurrence of the HBL operon in B. thuringiensis had been demonstrated previously via PCR-based detection of the hblCD genes in 87% (65 of 74) of the isolates examined by Gaviria Rivera et al. (17) and in 92% (38 of 41) of the isolates investigated by Hansen and Hendriksen (21). Unfortunately, no information was provided as to the geographical origin of the strains used in either of those studies.
Amplicons of the expected size (883 bp) were detected for 86% of the total B. cereus population examined and for 90% of the Brazilian isolates (Table 1). These values were markedly higher than the levels recorded, using the same primers, by Prüb et al. (33) or Mäntynen and Lindström (30), who reported detection frequencies of 43% (10 of 23 isolates) and 52% (26 of 50 isolates), respectively. It is pertinent to note that in the aforementioned studies, the bulk of the strains examined were of European origin, indicating that geographic origin may influence the degree to which the hblA sequence is maintained among populations of B. cereus. It is of interest that in spite of previous studies showing a relatively limited distribution of sequences encoding the HBL complex protein among B. cereus isolates (21, 30, 31), this molecule has been classified as a hemolysin based on its ability to lyse mammalian erythrocytes. Insects do not have red blood cells, and the near universal carriage of these genes by the insect pathogen B. thuringiensis and by most of the B. cereus strains isolated from insects (18/19) in this study suggests that the actual biological function of this molecule is linked to the establishment of infections in insects, rather than to the lysis of red blood cells in potential mammalian hosts.
The sequence denoted bceT was reported to encode a single-component toxin (BceT) which exhibited Vero cell cytotoxicity and to which has also been attributed a role in the diarrheal syndrome (3). More recently, it has been suggested that the bceT sequence actually represents a cloning artifact, formed by the ligation of four individual restriction fragments derived from B. cereus genomic DNA (22). An additional study indicated that the BceT toxin was most likely an experimental artifact that probably could not contribute to food poisoning (10). Nevertheless, other workers have demonstrated that it is possible to detect at least part of the original published sequence in both B. cereus and B. thuringiensis, and for that reason, we decided to include that region of the sequence as a target in our study using previously described primers (17). In common with our data concerning the hblA sequence, the frequency of distribution of the bceT gene was found to be broadly similar throughout both taxa (Table 1). Specifically, we detected the expected 593-bp amplicon in 86% of the entire B. thuringiensis population, in 84% of the Brazilian isolates, in 78% of the total B. cereus population, and in 69% of the Brazilian B. cereus strains. Interestingly, a higher level of detection (95%) was observed among the non-Brazilian B. cereus strains, which had been isolated predominantly from insects (Table 1). Thus, in common with previous studies (17, 21), we would conclude that this sequence is broadly distributed throughout both taxa.
In contrast, the nheBC sequence was detected at a markedly lower frequency among the B. cereus strains analyzed (60% in the total test population and 50% in Brazilian strains) than among the B. thuringiensis samples (81% in both the total test and Brazilian populations) (Table 1). The level of carriage observed in our panel of B. thuringiensis isolates is consistent with the studies of Gaviria et al. (17) and Hansen and Hendriksen (21), who reported frequencies of 100% and 80%, respectively, within their smaller strain panels. Conversely, the observation that only 60% of our B. cereus isolates possessed these sequences seemed at odds with a previous investigation which demonstrated the production of the NheA protein in a substantial proportion (92%) of the 194 strains examined (36). Yet, the level seen in our study is similar to that reported by Hansen and Hendriksen (21), who detected the genes nheA, nheB, and nheC in only 59% of the 41 strains used in their study. A more profound analysis of the distribution of these sequences among our strain panel revealed that the apparently limited presence of these genes was strongly influenced by the extremely low level of detection (11%) among the Brazilian strains isolated from foodstuffs (Table 1). Moreover, the levels of detection noted among the environmental and clinical strains, both Brazilian and non-Brazilian, were in the range of 79% (Table 1), a value that falls between the levels reported in the previous studies. Taken together, these data suggested that despite having been defined as a toxin involved in diarrheal food poisoning in B. cereus, the nonhemolytic enterotoxin is probably not essential for virulence. On the other hand, the widespread distribution of these sequences among B. thuringiensis strains of diverse geographical and environmental origins indicated that this molecule fulfils an important biological function for this bacterium, as speculated for the BL hemolysin.
In comparison to the other sequences reported to occur in isolates of both taxa, the distribution among B. thuringiensis isolates of the plc gene, which encodes phosphatildylcholine-specific phospholipase C, has received relatively little attention. Data from the only other study to have investigated this topic showed the presence of this sequence in 100% of the 16 strains examined (20). Employing the same primer set and amplification conditions as used previously, we observed the presence of the plc gene in 64% of our B. thuringiensis strains. This sequence was detected more frequently in the non-Brazilian isolates (15/19; 78.9%) than in the Brazilian subset (21/38; 55.2%). Interestingly, the level in the Brazilian isolates is substantially lower than that reported previously, which questions the notion that this sequence has a universal distribution within this taxon. In the case of B. cereus, we detected the plc sequence in only two isolates, both of which were Brazilian. The first of these (LFB287), was isolated from contaminated infant milk formula, while the second strain (LFB1181) was recovered from an outbreak of avian septicemia at a zoo in São Paulo (Table 1). This result was somewhat unexpected, since this gene was detected in 100% (3/3) of the B. cereus isolates studied previously (20). It is possible that the PCR-negative strains lacked the plc sequence. Alternatively, the lack of amplification may have arisen from sequence variation in the region selected for primer annealing. Further study, using primers designed to amplify alternative regions of this sequence, will be necessary to clarify this issue.
The sph sequence, encoding sphingomyelinase, was present in all strains analyzed by PCR. This observation supported the detection of this sequence as a good indicator of the presence of B. cereus group strains, as previously proposed (25), and suggested that this molecule fulfils an essential biological function for members of both taxa. It is of considerable interest that the expression of a number of extracellular virulence factors identified in both B. thuringiensis and B. cereus, including sphingomyelinase, HBL, NHE, and phosphatildylcholine-specific phospholipase C, has been shown to be pleiotropically regulated by the PlcR protein (1). The sequences encoding these putative virulence factors are dispersed throughout the chromosome, rather than being concentrated within a pathogenicity island. This has led to the suggestion that these species are derived from a common ancestral bacterium containing the plcR regulon and associated regulated sequences (1), which further supports the notion that these bacteria represent variants of the same species.
Using PCR-based detection methods, it was demonstrated that the vip3A gene showed a limited distribution (23 to 37%) among B. thuringiensis isolates of undefined geographical origin (20, 35). However, a more recent study reported a significantly greater distribution level of 75% among a population of 40 Chinese strains (40). Using our predominantly Brazilian strain panel, we detected the vip3A sequence in only 7% of the total test population (Table 1). All four isolates were Brazilian and belonged to the Morrisoni (LFB679, -780, and -781) and Kurstaki (LFB263) serotypes, both of which have been reported to present toxicity towards lepidopteran insects (37). However, a correlation between serotype and the presence of this sequence was not established, given that two additional isolates from each serotype tested negative for the presence of the gene. In the case of B. cereus, the vip3A sequence was not detected in any of the 63 strains examined in our study. Given the absence of any other published studies examining the presence of this sequence in B. cereus, we are unable to state that our data represent conclusive evidence that this sequence is found only in B. thuringiensis. However, we do not consider it unreasonable to make such a suggestion, in the hope that it may stimulate additional research on this topic.
It has been reported recently that the vip3A sequence is carried on a 31.8-kb plasmid (40). Conversely, evidence indicating that the gene is chromosomally located has been presented (16), although that conclusion was based on the examination of a single isolate. The widely divergent values recorded for carriage of this gene among strains of B. thuringiensis may indicate that the acquisition of this gene, possibly via horizontal gene transfer from other soil microbes, was a relatively recent event in the evolution of B. thuringiensis. Furthermore, the absence of this sequence, as well as those encoding for insecticidal crystal proteins, in B. cereus indicates that the inability to effectively separate these taxa genotypically, using increasingly complex molecular techniques, may be a reflection of our inability to identify the genetic basis for both the acquisition and maintenance of the plasmids carrying such sequences.
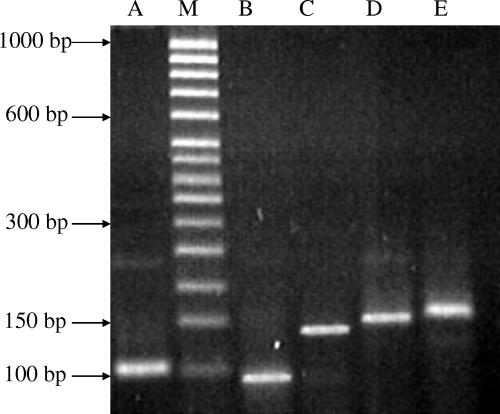
The presence of sequence polymorphism in the vrrA gene, resulting from variability in the number of tandem repeats, had previously been reported to allow the separation of different members of the B. cereus complex and even allowed differentiation between individual isolates of B. anthracis and B. cereus (4, 27). In the present study, we amplified this region of the genome and observed a total of five differently sized amplicons (Fig. 1), designated VNTR types. The most commonly encountered VNTR type in both B. cereus and B. thuringiensis was type 3 (150 bp), which was detected in approximately 45% of strains from both taxa (Table 1). Type 2 (135 bp) was observed in 38% of the B. thuringiensis isolates and at a level of 22% among the B. cereus strains (Table 1). The three other VNTR types were detected at far lower levels among the strain panel, with type 1 being the least frequently detected (Table 1). Interestingly, an amplicon of the same size (90 bp) was also detected in the single B. mycoides isolate included in the study (Table 1). No correlation between VTNR type and the origin of any strain could be demonstrated, and the suitability of variation in this region for separating the two taxa was clearly not established. Indeed, our finding that the same two VNTR types were more or less equally distributed between the two taxa could be interpreted as further evidence in support of the hypothesis that these two taxa represent the same species.
FIG. 1.
Agarose gel electrophoresis of representative VNTR amplicons in DNA extracted from Bacillus strains. Lane M, 50-bp DNA ladder (Sigma) (molecular sizes are indicated on the left); lane A, VNTR profile 5 (LFB596); lane B, VNTR profile 1 (LFB770); lane C, VNTR profile 2 (LFB856); lane D, profile 3 (LFB677); lane E, VNTR profile 4 (LFB1126).
The data from this study have raised a number of issues forconsideration. First, we recorded the same genetic profile (possession of genes encoding putative virulence factors and VNTR types) among Brazilian strains of Bacillus thuringiensis from food (black pepper) and from soil environments, indicating that these microorganisms have a clonal origin and that the same strain can be transferred from the soil to the crop. Furthermore, over 30% of the food isolates (5/13) possessed all five of the genes encoding factors considered to be involved in virulence towards mammals. In light of these observations, we would recommend that more attention should be paid to the microbial quality of the plants from which these products are derived. Although the safety of B. thuringiensis as a commercial bioinsecticide has been demonstrated, our data support the previous suggestion (17, 33) that the biocontrol industry should exercise caution with regard to the introduction of live B. thuringiensis cells into the human food chain as a result of the application of bioinsecticides to crops near the time of harvesting.
Second, our data extend the observations of various other research groups who have assessed the distribution of the putative virulence factors among these two closely related taxa. Numerous studies have attributed roles to these products from the viewpoint of their action upon animal cells or whole animals. Intriguingly, almost all studies performed to date have observed that the genes encoding the so-called mammalian virulence factors occur more frequently in B. thuringiensis, which is considered to be nonpathogenic towards mammals. Taken as a whole, these data suggest that the majority of B. cereus isolates could be considered to be nonpathogenic and that those strains which can cause infections do so because theyhave retained the majority of the sequences found in B. thuringiensis. However, the dearth of documented cases of mammalian infections caused by B. thuringiensis (26) clearly demonstrates that possession of these sequences does not necessarily correlate with virulence.
Finally, our examination of polymorphisms due to VNTR in the variable region of the vrrA sequence clearly demonstrated the close taxonomic proximity of B. cereus and B. thuringiensis, based on the theory that analysis of VNTR provides functional and evolutionary information concerning genetic relationships in microbial species (39). This observation will undoubtedly contribute to the continuing debate over whether these bacteria should actually be regarded as variants of the same species rather than as two distinct taxa.
Acknowledgments
This work was supported by the Fundação de Amparo à Pesquisa, FAPERJ. Diana Aparecida Cabral was a recipient of a grant from Conselho Nacional de Pesquisa e Desenvolvimento Tecnológico, CNPq/PIBIC.
We are grateful to Alberto Dávila (DBBM/IOC/FIOCRUZ) for hishelp in designing the primers for the amplification of the vip3A sequence.
REFERENCES
- 1.Agaisse, H., M. Gominet, O. A. Okstad, A. B. Kolsto, and D. Lereclus. 1999. PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol. Microbiol. 32:1043-1053. [DOI] [PubMed] [Google Scholar]
- 2.Agata, N., M. Mori, M. Ohta, M., S. Suwan, I. Ohtani, I., and M. Isobe. 1994. A novel duodeca—depsipeptide, cereulide, isolated from Bacillus cereus causes vacuole formation in Hep-2 cells. FEMS Microbiol. Lett. 121:31-34. [DOI] [PubMed] [Google Scholar]
- 3.Agata, N., M. Ohta, Y. Arakawa, and M. Mori. 1995. The bceT gene of Bacillus cereus encodes an enterotoxic protein. Microbiology 141:983-988. [DOI] [PubMed] [Google Scholar]
- 4.Andersen, G. L., J. M. Simchock, and K. H. Wilson. 1996. Identification of a region of genetic variability among Bacillus anthracis strains and related species. J. Bacteriol. 178:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ash, C., J. A. E. Farrow, S. Wallbanks, and M. D. Collins. 1991. Phylogenetic heterogeneity of genus Bacillus revealed by comparative analysis of small subunit-ribosomal RNA sequences. Lett. Appl. Microbiol. 13:202-206. [Google Scholar]
- 6.Bavykin, S. G., Y. P. Lysov, V. Zakhariev, J. J. Kelly, J. Jackman, D. A. Stahl, and A. Cherni. 2004. Use of 16S rRNA, 23S rRNA, and gyrB gene sequence analysis to determine phylogenetic relationships of Bacillus cereus group microorganisms. J. Clin. Microbiol. 42:3711-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beecher, D. J., J. L. Schoeni, and A. C. L. Wong. 1995. Enterotoxin activity of hemolysin BL from Bacillus cereus. Infect. Immun. 63:4423-4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlson, C. R., D. Caugant, and A. B. Kolsto. 1994. Genotypic diversity among Bacillus cereus and Bacillus thuringiensis strains. Appl. Environ. Microbiol. 60:1719-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, M. L., and H. Y. Tsen. 2002. Discrimination of Bacillus cereus and Bacillus thuringiensis with 16S rRNA and gyrB gene based PCR primers and sequencing of their annealing sites. J. Appl. Microbiol. 92:912-919. [DOI] [PubMed] [Google Scholar]
- 10.Choma, C., and P. E. Granum. 2002. The enterotoxin T (BcET) from Bacillus cereus can probably not contribute to food poisoning. FEMS Microbiol. Lett. 217:115-119. [DOI] [PubMed] [Google Scholar]
- 11.Claus, D., and R. C. W. Berkeley. 1986. Endospore-forming Gram positive rods and cocci: Bacillus, p. 1105-1138. In P. H. A. Sneath et al. (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams and Wilkins Co., Baltimore, Md. [Google Scholar]
- 12.Damgaard, P. H. 1995. Diarrhoeal enterotoxin production by strains of Bacillus thuringiensis isolated from commercial Bacillus thuringiensis based insecticides. FEMS Immunol. Med. Microbiol. 12:245-250. [DOI] [PubMed] [Google Scholar]
- 13.Drobniewski, F. 1993. Bacillus cereus and related species. Clin. Microbiol. 32:1280-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehling-Schulz, M., B. Svensson, M. H. Guinebretiere, T. Lindback, M. Andersson, A. Schulz, M. Fricker, A. Christiansson, P. E. Granum, E. Martlbauer, C. Nguyen-The, M. Salkinoja-Salonen, and S. Scherer. 2005. Emetic toxin formation of Bacillus cereus is restricted to a single evolutionary lineage of closely related strains. Microbiology 151:183-197. [DOI] [PubMed] [Google Scholar]
- 15.Estruch, J. J., G. W. Warren, M. A. Mullins, G. J. Nye, J. A. Craig, and M. G. Koziel. 1996. Vip3A, a novel Bacillus thuringiensis vegetative insecticidal protein with a wide spectrum of activities against lepidopteran insects. Proc. Natl. Acad. Sci. USA 93:5389-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franco-Rivera, A., G. Benintende, J. Cozzi, V. M. Baizabal-Aguirre, J. J. Valdez-Alarcon, and J. E. Lopez-Meza. 2004. Molecular characterization ofBacillus thuringiensis strains from Argentina. Antonie Leeuwenhoek 86: 87-92. [DOI] [PubMed] [Google Scholar]
- 17.Gaviria Rivera, A., P. E. Granum, and F. G. Priest. 2000. Common occurrence of enterotoxin genes and enterotoxicity in Bacillus thuringiensis. FEMS Microbiol. Lett. 190:151-155. [DOI] [PubMed] [Google Scholar]
- 18.Granum, P. E., and T. Lund. 1997. Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Lett. 157:223-228. [DOI] [PubMed] [Google Scholar]
- 19.Granum, P. E., K. O′Sullivan, and T. Lund. 1999. The sequence of the non-haemolytic enterotoxin operon from Bacillus cereus. FEMS Microbiol. Lett. 177:225-229. [DOI] [PubMed] [Google Scholar]
- 20.Guttmann, D. M., and D. J. Ellar. 2000. Phenotypic and genotypic comparisons of 23 strains from the Bacillus cereus complex for a selection of known and putative B. thuringiensis virulence factors. FEMS Microbiol. Lett. 188:7-13. [DOI] [PubMed] [Google Scholar]
- 21.Hansen, B. M., and N. B. Hendriksen. 2001. Detection of enterotoxic Bacillus cereus and Bacillus thuringiensis strains by PCR analysis. Appl. Environ. Microbiol. 67:185-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen, B. M., P. E. Hoiby, G. B. Jensen, and N. B. Hendriksen. 2003. The Bacillus cereus bceT enterotoxin sequence reappraised. FEMS Microbiol. Lett. 223:21-24. [DOI] [PubMed] [Google Scholar]
- 23.Helgason, E., O. A. Okstad, D. A. Caugant, H. A. Johansen, A. Fouet, M. Mock, I. Hegna, and A. B. Kolsto. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis: one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill, K. K., L. O. Ticknor, R. T. Okinaka, M. Asay, H. Blair, K. A. Bliss, M. Laker, P. E. Pardington, A. P. Richardson, M. Tonks, D. J. Beecher, J. D. Kemp, A. B. Kolsto, A. C. Wong, P. Keim, and P. J. Jackson. 2004. Fluorescent amplified fragment length polymorphism analysis of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis isolates. Appl. Environ. Microbiol. 70:1068-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsieh, Y. M., S. J. Sheu, Y. L. Chen, and H. Y. Tsen. 1999. Enterotoxigenic profiles and polymerase chain reaction detection of Bacillus cereus group cells and Bacillus cereus strains from food and food-borne outbreaks. J. Appl. Microbiol. 87:481-490. [DOI] [PubMed] [Google Scholar]
- 26.Jackson, S. G., R. B. Goodbrand, R. Ahmed, and S. Kasatiya. 1995. Bacillus cereus and Bacillus thuringiensis isolated in a gastroenteritis outbreak investigation. Lett. Appl. Microbiol. 21:103-105. [DOI] [PubMed] [Google Scholar]
- 27.Kim, W., Y. Hong, J. Yoo, W. Lee, C. Choi, and S. Chung. 2001. Genetic relationships of Bacillus anthracis and closely related species based on variable-number tandem repeat analysis and BOX-PCR genomic fingerprinting. FEMS Microbiol. Lett. 207:21-27. [DOI] [PubMed] [Google Scholar]
- 28.Ko, K. S., J. W. Kim, J. M. Kim, W. Kim, S. Chung, I. J. Kim, and Y. H. Kook. 2004. Population structure of the Bacillus cereus group as determined by sequence analysis of six housekeeping genes and the plcR gene. Infect. Immun. 72:5253-5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kramer, J. M., and R. J. Gilbert. 1989. Bacillus cereus and other Bacillus species, p. 21-70 In M. P. Doyle (ed.), Foodborne bacterial pathogens. Marcel Dekker, New York, N.Y.
- 30.Mäntynen, V., and K. Lindström. 1998. A rapid PCR-based DNA test for enterotoxic Bacillus cereus. Appl. Environ. Microbiol. 64:1634-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKillip, J. L. 2000. Prevalence and expression of enterotoxins in Bacillus cereus and other Bacillus spp., a literature review. Antonie Leeuwenhoek 77:393-399. [DOI] [PubMed] [Google Scholar]
- 32.Priest, F. G., M. Barker, L. W. Baillie, E. C. Holmes, and M. C. Maiden. 2004. Population structure and evolution of the Bacillus cereus group. J. Bacteriol. 18:7959-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prüb, B. M., R. Dietrich, B. Nibler, E. Märtlbauer, and S. Scherer. 1999. The haemolytic enterotoxin HBL is broadly distributed among species of the Bacillus cereus group. Appl. Environ. Microbiol. 65:5436-5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rasko, D. A., M. R. Altherr, C. S. Han, and J. Ravel. 2005. Genomics of the Bacillus cereus group of organisms. FEMS Microbiol. Rev. 29:303-329. [DOI] [PubMed] [Google Scholar]
- 35.Rice, W. C. 1999. Specific primers for the detection of vip3A insecticidal gene within a Bacillus thuringiensis collection. Lett. Appl. Microbiol. 28:378-382. [Google Scholar]
- 36.Rusul, G., and N. H. Yaacob. 1995. Prevalence of Bacillus cereus in selected foods and detection of enterotoxin using TECRA-VIA and BCET-RPLA. Int. J. Food Microbiol. 25:131-139. [DOI] [PubMed] [Google Scholar]
- 37.Schnepf, E., N, Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Zeigler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schoeni, J. L., and A. C. Wong. 2005. Bacillus cereus food poisoning and its toxins. J. Food Prot. 68:636-648. [DOI] [PubMed] [Google Scholar]
- 39.Van Belkum, A. 1999. Short sequence repeats in microbial pathogenesis and evolution. Cell. Mol. Life Sci. 56:729-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu, Z. L., W. Y. Guo, J. Z. Qiu, T. P. Huang, X. B. Li, and X. Guan. 2004. Cloning and localization of vip3A gene of Bacillus thuringiensis. Biotechnol. Lett. 26:1425-1428. [DOI] [PubMed] [Google Scholar]
- 41.Zahner, V., H. Momen, C. A. Salles, and L. Rabinovitch. 1989. A comparative study of enzyme variation in Bacillus cereus and Bacillus thuringiensis. J. Appl. Bacteriol. 67:275-282. [DOI] [PubMed] [Google Scholar]