Abstract
The ability to utilize heme compounds as iron sources was investigated in Vibrio anguillarum strains belonging to serotypes O1 to O10. All strains, regardless of their serotype or isolation origin could utilize hemin and hemoglobin as sole iron sources. Similarly, all of the isolates could bind hemin and Congo red, and this binding was mediated by cell envelope proteins. PCR and Southern hybridization were used to assay the occurrence of heme transport genes huvABCD, which have been previously described in serotype O1. Of 23 strains studied, two serotype O3 isolates proved negative for all huvABCD genes, whereas nine strains included in serotypes O2, O3, O4, O6, O7, and O10 tested negative for the outer membrane heme receptor gene huvA. A gene coding for a novel outer membrane heme receptor was cloned and characterized in a V. anguillarum serotype O3 strain lacking huvA. The new heme receptor, named HuvS, showed significant similarity to other outer membrane heme receptors described in Vibrionaceae, but little homology (39%) to HuvA. This heme receptor was present in 9 out of 11 of the V. anguillarum strains that tested negative for HuvA. Furthermore, complementation experiments demonstrated that HuvS could substitute for the HuvA function in Escherichia coli and V. anguillarum mutants. The huvS and huvA sequences alignment, as well as the analysis of their respective upstream and downstream DNA sequences, suggest that horizontal transfer and recombination might be responsible for generating this genetic diversity.
Vibrio anguillarum is a fish pathogen that causes the disease known as vibriosis, a lethal hemorrhagic septicemia affecting a large number of mainly marine fish species, as well as bivalve mollusks and crustaceans (35, 36). Vibriosis is one of the most serious diseases affecting the mariculture industry worldwide, causing important economic losses, although vaccination programs have proved to be effective (34, 35). Sørensen and Larsen grouped V. anguillarum isolates in 10 serogroups based on antigen ‘O’ (29). The number of O-serotypes has been extended up to 23 (9, 27). However, only serotypes O1 and O2 and, to a lesser extent, serotype O3 are considered important pathogens since most vibriosis outbreaks described thus far were caused by one of these serotypes (11, 35). The other serotypes are considered environmental strains, although their pathogenic potential cannot be ruled out (26, 29, 35).
V. anguillarum expresses several virulence factors; the most important thus far recognized is the ability to produce different enzymes and toxins that notoriously contribute to cause disease (36). Another important virulence factor is the ability to scavenge the iron contained in host tissues that is bound by different iron-binding proteins. The main mechanisms of iron acquisition from these proteins are based on the synthesis of siderophores, whose production has been demonstrated in the main serotypes of V. anguillarum (3, 4, 12, 21, 32), even in those considered nonpathogenic (13). Interestingly, different mechanisms for siderophore-mediated iron acquisition seem to be present in V. anguillarum serotypes (12, 21, 32).
Siderophore-independent mechanisms for iron uptake have been described in many bacterial pathogens, one of the best known being the utilization of host heme compounds as iron sources (8, 25, 37). We have previously demonstrated that serotypes O1 and O2 of V. anguillarum can use heme groups as iron sources and that this ability can be useful for survival and to colonize fish tissues (14, 16).
Although several heme uptake systems have been described in bacteria, most of them include an outer membrane heme receptor and a TonB-dependent internalization system to transport the heme molecule into the periplasm, where a periplasmic heme-binding protein binds heme. A permease protein and an ATPase conforming an inner membrane-associated ABC transporter are necessary to further transport heme into the cytoplasm (25, 37). A genetic system consisting of nine clustered genes involved in heme uptake has been recently characterized in V. anguillarum serotype O1 strain 775 (20), which includes an iron-regulated outer membrane heme receptor HuvA, a TonB system, a periplasmic heme-binding protein HuvB, and an ABC transporter conformed by HuvC and HuvD. However, nothing is known about the presence of this mechanism of iron acquisition in other strains and serotypes of V. anguillarum. Since the siderophore-based mechanisms were found to be different among serotypes and strains of this fish pathogen (3), we sought to analyze the presence of a heme uptake mechanism in strains of V. anguillarum serotypes other than O1 in order to determine whether heme binding and heme utilization as an iron source is a species determinant of V. anguillarum and whether the genetic system for heme uptake is shared by the different serotypes and strains.
MATERIALS AND METHODS
Strains and culture conditions used.
V. anguillarum strains have been described in previous studies (3, 26), and their serotypes and origins are indicated in Table 2. The identity of strains has been confirmed by biochemical tests, and rRNA 16S sequence. Serogroup was confirmed by slide agglutination tests (26). We have included 23 strains representative of serogroups O1 to O10. Cells were routinely grown at 25°C in tryptic soy agar (Difco) supplemented with 1% NaCl (TSA-1), as well as in M9 minimal medium (19) supplemented with 0.2% Casamino Acids (Difco) (CM9). Escherichia coli HB101, used as control, was grown at 37°C in Luria-Bertani (LB) medium. All strains were stored frozen at −80°C in LB broth with 20% glycerol. Iron-deficient culture conditions were achieved by adding the nonassimilable iron chelator ethylenediamine-di-(o-hydroxyphenyl-acetic acid) (EDDA) at different concentrations to CM9.
TABLE 2.
Bacterial strains of V. anguillarum used in this study, utilization of hemin as the only iron source, and presence of huvA, huvBCD, and huvS genes
| Strain | Serotype | Source, location | Utilization of Hma | Presence ofb:
|
||
|---|---|---|---|---|---|---|
| huvA | huvBCD | huvS | ||||
| R82 | O1 | Scophthalmus maximus, Spain | + | + | + | − |
| ATCCc 43305 | O1 | Oncorhynchus mykiss, Denmark | + | + | + | − |
| 775 | O1 | Oncorhynchus kisutch, United States | + | + | + | − |
| TM-14 | O1 | Oncorhynchus mykiss, Spain | + | + | + | − |
| 96F | O1 | Morone saxatilis, United States | + | + | + | − |
| RV22 | O2 | Scophthalmus maximus, Spain | + | + | + | − |
| ATCC 43306 | O2 | Gadus morhua, Denmark | + | − | + | + |
| ATCC 14181 | O2 | Gadus morhua, Denmark | + | + | + | − |
| 43F | O2 | Morone saxatilis, United States | + | + | + | − |
| 13A5 | O3 | Seawater, Spain | + | − | − | − |
| B.1.1.2/4 | O3 | Seawater, Denmark | + | − | − | − |
| ET-208 | O3 | Anguilla japonica, Japan | + | − | + | + |
| 11008 | O3 | Dicentrarchus labrax, France | + | − | + | + |
| ATCC 43307 | O3 | Oncorhynchus mykiss, Denmark | + | − | + | + |
| PT-493 | O3 | Plecoglossus altivelis, Japan | + | − | + | + |
| RPM 41.11 | O4 | Scophthalmus maximus, Spain | + | + | + | − |
| ATCC 43308 | O4 | Gadus morhua, Denmark | + | − | + | + |
| ATCC 43309 | O5 | Gadus morhua, Denmark | + | + | + | − |
| ATCC 43310 | O6 | Gadus morhua, Denmark | + | − | + | + |
| ATCC 43311 | O7 | Anguilla anguilla, Denmark | + | − | + | + |
| ATCC 43312 | O8 | Gadus morhua, Denmark | + | + | + | − |
| ATCC 43313 | O9 | Gadus morhua, Denmark | + | + | + | − |
| ATCC 43314 | O10 | Gadus morhua, Denmark | + | − | + | + |
Utilization of hemin (Hm) at 10 μM as the only iron source tested in CM9 plates containing EDDA. All strains were also positive in the utilization of Hemoglobin as iron source.
Presence detected by PCR and confirmed by Southern blot hybridization.
ATCC, American Type Culture Collection, Manassas, Va.
Hemin and hemoglobin utilization assays.
The utilization of hemin and hemoglobin as iron sources was tested in liquid and in solid media. Stock solutions of bovine hemoglobin (Sigma-Aldrich) and bovine hemin (Sigma-Aldrich) were prepared at 5 mM in deionized water and at 10 mM in 10 mM NaOH, respectively. Both solutions were freshly prepared before use and filter sterilized. CM9 medium was supplemented with 500 μM EDDA, a concentration sufficient to cause a total growth inhibition of strains to be tested. Utilization of hemin and hemoblobin was tested in CM9 plus EDDA liquid medium by adding these compounds at concentrations between 0.1 and 10 μM. A 1/100-log-phase inoculum grown in LB broth was used to inoculate CM9 medium. Growth of the strains was monitored by measuring the optical density at 600 nm (OD600) at different time intervals. Alternatively, soft CM9 medium supplemented with EDDA was mixed with a 1/100 LB broth inoculum and poured onto plates. After solidification, sterile paper disks impregnated with 10 μM hemoglobin or hemin were placed onto the medium. After incubation at 25°C for 48 h, visible growth around disks indicates utilization of the compound as iron source.
Hemin and Congo red binding.
Hemin and Congo red binding by V. anguillarum cells was monitored in liquid medium according to the methods previously described (15, 28). Cells were incubated for 12 h in CM9 medium in duplicate with or without iron supplement (10 μM FeCl3). After incubation, cells were resuspended in M9 salts (pH 7.5), and Congo red or hemin were incorporated to a final concentration of 30 μg ml−1. The cell suspension was incubated at 25°C, and 1-ml samples were withdrawn at 30-min intervals. Samples were immediately centrifuged, and the absorbance of supernatants was measured at 488 nm for Congo red binding and at 400 nm for hemin binding.
Hemin-binding assays were also performed by using a solid-phase dot blot assay (15, 28). A total of 40 μl of a bacterial cell suspension (ca. 2 × 107 CFU), grown in CM9 or CM9 plus hemin 10 μM, was filtered onto nitrocellulose membranes (0.45-μm pore size; Millipore) and tested for hemin binding as previously described (6, 15). After immobilization, membranes were air dried and blocked with gelatin (2% in 50 mM Tris plus 0.9% NaCl) for 2 h. The membrane was washed several times in distilled water, immersed in a buffer (50 mM Tris, 0.9% NaCl, 0.5 mM EDTA, 0.05% Triton X-100) containing hemin 10 μM, and incubated for 2 h at 30°C with gentle shaking. Membrane was then washed in distilled water and stained with DMB (3,3′-dimethoxibenzidine; Sigma). A DMB solution of 10 mg ml−1 in distilled water was freshly prepared just before staining and, after being stirred for 15 min, 10 ml of a 0.5 M sodium citrate buffer (pH 4.4) and 200 μl of 30% H2O2 were added. After the mixture was stained for 1 to 3 min, the membrane was thoroughly washed with water and air dried.
The involvement of proteins in hemin binding was tested by treating cells with a protease before the hemin-binding assay. Before immobilization on nitrocellulose membranes, the cells were resuspended in phosphate-buffered saline buffer containing 0.2 mg of proteinase K (Sigma) ml−1 and incubated for 2 h at 37°C. The cells were then washed two times with CM9 medium, filtered on nitrocellulose membranes, and tested for hemin binding as described above.
Southern hybridization and PCR.
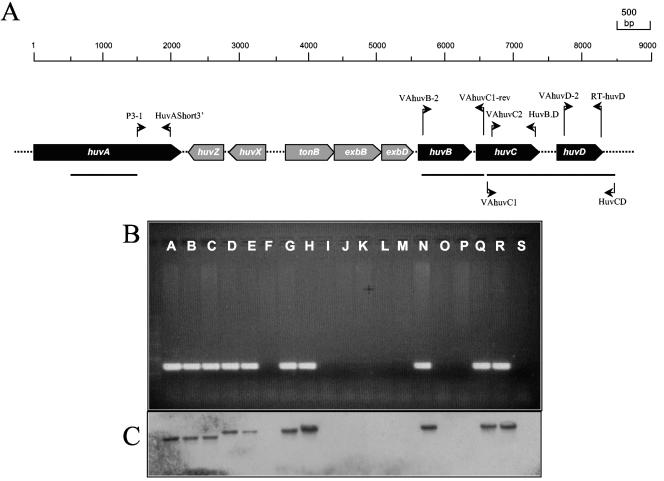
Chromosomal DNA from V. anguillarum strains was purified by using the Easy-DNA kit (Invitrogen). DNA was digested overnight with BglII, and restriction fragments were separated in 1.1% agarose gels. Southern blots were performed onto Hybond-N+ membranes (Amersham-Pharmacia) by using the ECL Direct Nucleic Acid Labeling and Detection System (Amersham-Pharmacia) according to the manufacturer's recommendations. A 1.0-kb HindIII fragment was used as a probe for the huvA gene. PCR screening for presence of huvA was accomplished with primers P3-1 and HuvAShort-3′. Probes for huvB and huvCD genes were PCR amplified and purified from agarose gels with a QIAquick gel extraction kit (QIAGEN). The oligonucleotides used to amplify the huvB probe were VAhuvB-2 and VAhuvC1-rev. The same primers were used for PCR screening of presence of huvB gene. huvCD probe was amplified with primers VAhuvC-1 and HuvC.D. To confirm the huvCD hybridization data, huvC and huvD genes were independently amplified in a PCR screening. huvC was amplified with VAhuvC-2 and HuvB.D. Primers used to amplify huvD were VAhuvD-2 and RT-huvD. Primers sequence and locations are described in Table 1 and Fig. 2A.
TABLE 1.
Oligonucleotides used as primers in PCR
| Primer | Nucleotide sequence | Amplified region |
|---|---|---|
| P3-1 | GGAATGTTGTCGCAGCACTA | huvA |
| HuvA-Short 3′ | CATGGAACAACAAAGCCAGC | |
| VAhuvB-2 | GAACGGATTATCAGTGCGGGC | huvB and huvB probe |
| VAhuvC1-rev | AAAATTGCGCCGACCAACAA | |
| VAhuvC-1 | TTGTTGGTCGGCGCAATTTT | huvCD probe |
| HuvC.D | GCGAATTCGCATTATAGCGAAGGGACGCG | |
| VAhuvC-2 | GCATTGACTACCTTGTTGGT | huvC |
| HuvB.D | GCGAATTCGTGAGTAAGAGTGCACCAAGT | |
| VAhuvD-2 | TTCTTCCTCAGCAAAGTACG | huvD |
| RT-huvD | TAGACCATTGGGAAATCCA | |
| cDNA huvZ2 | CCAAGAGTTTCGTCAAGAAC | Downstream huvZ |
| VAhuvX1 | AAGGCGCTTTCACTTCAAAT | |
| huvS3 | CGAAGACCAGCGGTTAATAT | huvS |
| huvS6-rev | GCTCTCGCAGAAGAAGTTTC |
FIG. 2.
(A) Physical map of the heme uptake cluster of V. anguillarum O1 strain 775 (see reference 20). Thick arrows denote ORFs, and the scale above indicates the length in nucleotides. Thin arrows indicate the position and direction of primers used in the PCR screening and in the amplification of huvB and huvCD DNA probes. Horizontal bars denote location of DNA probes. (B) Results of PCR screening for presence of the heme receptor huvA gene in a collection of V. anguillarum strains. (C) Southern blot confirmation of presence of huvA gene. Letters in panels B and C denote V. anguillarum strains as follows: A, 775; B, R-82; C, TM-14; D, 96-F; E, ATCC 14181; F, ATCC 43306; G, RV22; H, 43-F; I, PT-493; J, 13A5; K, 11008; L, ET-208; M, ATCC 43307; N, RPM 41.11; O, ATCC 43310; P, ATCC 43311; Q, ATCC 43312; R, ATCC 43313; S, ATCC 43314.
Cloning of a new heme receptor gene huvS.
The existence of a putative new heme receptor downstream of huvZ gene in huvA-negative V. anguillarum strains was investigated in strain ET-208 (serotype O3) by using inverse PCR. For this purpose, chromosomal DNA was cut with a single restriction enzyme (BssHII) and self-ligated. Ligation products were used as a template in a PCR with primers cDNA-huvZ2 and VAhuvX1 (Table 1), by using the Expand Long Template Kit (Roche Diagnostics). The PCR product was cloned in pGEM-T Easy (Promega) to yield pSML88 and sequenced. The complete huvS gene was excised from pSML88 as an ApaLI fragment and cloned into the ApaLI site of vector pACYC177 to yield pSML92. huvS was excised from pSML92 as a PstI-HindIII fragment and cloned into the mobilizable vector pMMB208 to yield pSML93, which was transformed into E. coli S17-1-λ-pir and further transferred to V. anguillarum ΔhuvA mutant strain (20) by conjugation.
The presence of huvS in V. anguillarum isolates was screened by PCR amplification with primers huvS3 and huvS6-rev (Table 1), which amplify a 1,231-bp internal fragment of huvS gene. The results were confirmed by Southern blot hybridization, with this 1,231-bp PCR fragment as a probe.
DNA sequence and data analysis.
The DNA sequence was determined by the dideoxy chain termination method using the CEQ DTCS-Quick Start Kit (Beckman Coulter) using a capillary DNA sequencer CEQ 8000 (Beckman Coulter). The European Bioinformatics Institute services were used to consult the EMBL and SWALL databases with the FASTA3 and BLAST algorithms. The EMBL accession number for the sequence described in this article is AM042548.
Complementation experiments.
About 100 μl of overnight cultures of E. coli 101ESD Δ(entC-entA) transformed with plasmids pSML33 (20) and pSML92 were added to 3 ml of molten soft CM9 minimal medium and plated onto appropriate prepoured CM9 or CM9 supplemented with 150 μM 2,2′-dipyridyl plates. Sterile filter paper disks were loaded with 20 μl of either 5 mM hemin or 0.1 mM hemoglobin. A disk containing 20 μl of 10 μM FeSO4 was also included as positive control. The results were annotated as positive or negative after 24 h of incubation. Overnight cultures of V. anguillarum ΔhuvA (20) transformed with pSML93 were subcultured 1:100 in tubes of CM9 medium supplemented with 5 μM EDDA, with or without hemin 10 μM or hemoglobin 1 μM as the sole iron sources. FeSO4 was also used at 10 μM as positive control. After 24 h of incubation the OD600 was measured.
RESULTS AND DISCUSSION
Utilization of hemin and hemoglobin as iron sources.
All strains, regardless of their serotype or isolation source, could utilize hemin or hemoglobin as the only iron sources when cultured in iron-restricted conditions (Table 2). Growth curves in the presence of these iron sources showed no significant differences between strains (data not shown). This suggests that utilization of heme compounds as sole iron sources is a characteristic of the species V. anguillarum, since the 23 tested strains showed the same heme utilization profile. In previous studies we reported that hemin and hemoglobin could promote the in vitro growth of V. anguillarum strains belonging to serotypes O1 and O2 (14). However, no data were available for serotypes O3 to O10. The present study confirms that a system for the uptake of heme and its utilization as an iron source is likely widespread in all V. anguillarum strains.
The presence of a heme uptake system is not a marker of virulence in V. anguillarum, since serotypes O4 to O10 are environmental and unable to cause an infection, while retaining the ability to use heme compounds as iron source. However, this does not rule out a role for heme utilization during the infective process of this fish pathogen. The ability of a bacterial pathogen to acquire iron from heme compounds present in host tissues is supposed to constitute an advantage for colonization and to establish an infection, as has been demonstrated in pathogens such as V. cholerae (10), Haemophilus ducreyi (30), Streptococcus pneumoniae (33), Neisseria meningitidis (31), and V. anguillarum serotype O1 (16). V. anguillarum strains have been reported to produce extra cellular enzymes such as hemolysins and cytolisins (7, 36). Thus, once the bacterium invades the host, these enzymes could release heme groups from blood cells and make heme iron available for its utilization by the pathogen.
Hemin binding by V. anguillarum cells surface.
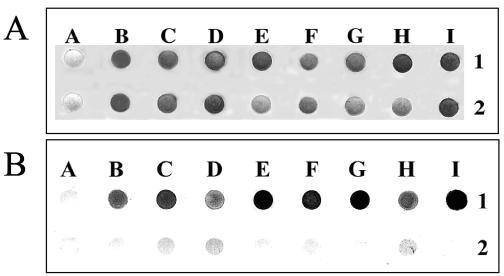
Whole cells from all strains could bind hemin in a solid-phase dot blot assay (Fig. 1A) or in liquid media. Hemin binding was confirmed by Congo red binding assays in liquid media (data not shown). No difference in binding was noted among strains tested (Fig. 1A). Binding was inhibited in all serotypes and strains tested when cells were previously treated with proteinase K (Fig. 1B), which indicates the involvement of cell surface proteins in heme binding. In addition, heme-binding ability was shown to be independent of the iron load in the culture medium, suggesting that constitutive outer membrane proteins play a role in this function.
FIG. 1.
(A) Hemin binding by whole cells from different strains of V. anguillarum cultured under iron-rich (1) and iron-deficient (2) conditions. (B) Effect of proteinase K treatment (2) on hemin binding compared to nontreated cells (1). Lanes: A, E. coli HB101 (negative control); B, 775; C, RV22; D, ET-208; E, RM 40.1; F, ATCC 43309; G, ATCC 43310; H, ATCC 43311; I, ATCC 43313.
Hemin-binding ability is widespread in many bacteria, and several cell surface molecules such as polysaccharides (1, 5) or proteins can mediate this binding. For example, at least eight membrane-associated proteins of Bartonella quintana bind hemin (2). The role of cell surface proteins in heme binding in V. anguillarum was previously reported in serotype O1 and O2 strains, where outer membrane proteins of 39 and 37 kDa were isolated, respectively (17). However, these outer membrane heme-binding proteins may not be involved in heme transport across the membrane. In previous studies, we could demonstrate that mutation of the outer membrane heme receptor (HuvA) in a serotype O1 strain does not have a significant influence on hemin binding (16), suggesting that V. anguillarum has surface molecules with hemin-binding activity but without transport function.
Differential occurrence of heme transport genes.
In a previous study, a gene cluster involved in the uptake of heme was characterized in V. anguillarum serotype O1 strain 775 (20). This cluster included genes for the outer membrane receptor HuvA, the periplasmic hemin-binding protein HuvB, the inner membrane permease HuvC, and the inner membrane ATPase HuvD proteins, the last two constituting an ABC transporter (Fig. 2A). To assess the presence of these heme transport genes among different V. anguillarum strains, we performed PCR amplification with specific primers for huvABCD genes (Table 1). The results are summarized in Table 2 and show that strains 13A5 and B1.1.2/4 (serotype O3) were negative for all genes assayed. In addition, strain ATCC 43306 (serotype O2), all serotype O3 strains, strain ATCC 43308 (serotype O4), strain ATCC 43310 (serotype O6), strain ATCC 43311 (serotype O7), and strain ATCC 43314 (serotype O10) were negative for the presence of the heme receptor gene huvA, (Table 2 and Fig. 2B), whereas all of them gave positive results for the presence of huvBCD genes. Southern blot hybridizations with probes specific for huvA, huvB, and a probe for huvCD genes confirmed the results obtained by PCR (Table 2 and Fig. 2C).
Interestingly, all serotype O3 strains included in the present study proved negative for huvA, the outer membrane receptor (Table 2 and Fig. 2). Moreover, two of these strains (13A5 and B1.1.2/4), which were isolated from seawater, tested negative for the four heme transport genes assayed (huvABCD), being the unique V. anguillarum isolates which did not share the genes for the periplasmic hemin-binding protein and the ABC transporter reported in serotype O1. All of the serotype O1 strains showed positive hybridization for huvABCD, indicating a conserved gene content in their heme uptake mechanism. In the rest of the strains, no relationship between serogroup and the presence or absence of these genes could be inferred.
In a previous study we demonstrated that huvABCD genes are essential for heme utilization in serotype O1 strain 775 since mutation of any of these genes cause the loss of ability to grow in presence of heme or hemoglobin as sole iron sources (20). This therefore indicates that V. anguillarum 775 has only one functional heme uptake system, and no additional redundant genes can substitute for the functions of either huvABCD. In contrast, in V. cholerae, redundant gene functions have been described for the homologues of huvABCD (18, 22). In the present work, we report that some non-O1 V. anguillarum strains are able to utilize heme compounds as iron source while lacking either huvA, huvBCD, or the four genes altogether. This indicates that additional, yet-uncharacterized genes must substitute for these transport functions in the strains that proved negative for the genes assayed. The differential occurrence of several heme uptake genes in different strains of V. anguillarum suggests the possible use of these genes as epidemiological markers.
Cloning and characterization of huvS, a new heme receptor gene.
In V. anguillarum, the existence of negative strains for the huvA gene suggests the likely existence of one or more additional receptors with heme transport activity. In these strains the nucleotide sequence of the putative unknown heme receptor could differ with respect to that of the previously characterized huvA (16) to prevent it from hybridizing with the huvA probe or from being amplified by PCR. We hypothesized that, if an alternative outer membrane heme receptor gene existed in the huvA-negative V. anguillarum strains and if its chromosomal location were conserved with respect to huvA, it would be feasible to clone it by chromosome walking and investigating the DNA region in the vicinity of huvZ (Fig. 3).
FIG. 3.
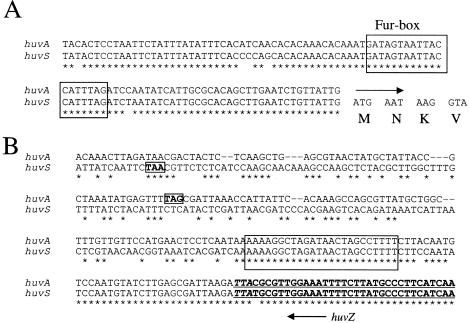
Alignment of the amino acid sequences of V. anguillarum HuvS and HuvA outer membrane heme receptors. Numbers refer to the amino acid position. Asterisk indicates amino acid positions conserved in the two proteins, and similar residues are denoted by periods. The putative TonB boxes are shown boxed.
In order to test this possibility, an Inverse-PCR aimed at amplifying DNA downstream of huvZ gene was carried out with the huvA-negative ET-208 strain (serotype O3). A PCR product was obtained and sequenced to completion, and a complete open reading frame (ORF) consisting of 2,160 bp and coding for a putative protein of 719 amino acids was inferred, which we named HuvS. The highest similarities were to V. mimicus heme receptor MhuA (63% identity), Vibrio fischeri heme receptor (59%), and Photobacterium damselae heme receptor HutA (47%). The deduced HuvS protein showed 39% identity to V. anguillarum HuvA, being remarkable the fact that the first 44 positions were identical between the two V. anguillarum heme receptors, whereas the rest of the molecule showed significant sequence divergence (Fig. 3) .
HuvS contains conserved features that are common to other TonB-dependent outer membrane heme receptor proteins. HuvS shares with HuvA a 100% identical TonB-box (Fig. 3). In addition, the amino acid sequence of HuvS shows a conserved terminal phenylalanine residue that is required for incorporation into the outer membrane and that is shared by most of the heme receptors described to date. The nucleotide sequence upstream of the huvS start codon shows 96% identity to the respective sequence of huvA (Fig. 4A) and contains a putative binding site for the Fur protein (the so-called Fur-box). This clearly points out to that huvS transcription is regulated by Fe(III) levels, as has been demonstrated for huvA by transcriptional fusion analysis (S. Mouriño, C. R. Osorio, M. L. Lemos, and J. H. Crosa, unpublished data). However, as stated above, the hemin-binding phenotype of huvA or huvS containing strains is independent of iron levels. This apparent contradiction can be explained by the multicomponent nature of the hemin-binding ability (16).
FIG. 4.
(A) Comparative sequence analysis of the nucleotide sequence of huvA and huvS predicted promoter regions. A putative conserved Fur-box is boxed. The arrow indicates the translation start site for both huvA and huvS. The N-terminal residues of HuvS and HuvA are denoted by using the single-letter code. (B) Comparative sequence analysis of the predicted termination regions of huvA and huvS. The respective stop codons are shown in bold and boxed. A region of dyad symmetry conforming a putative transcriptional terminator is boxed. Sequence of the 3′ end of the neighbor huvZ gene is shown in bold and underlined, and the huvZ stop codon is shown in italics.
Downstream of the termination codon, there is a region of nucleotide sequence that differs between the two genes, but the sequence is again highly conserved in the region close to the stop codon of the neighbor huvZ gene and nearly identical in the coding sequence of huvZ (Fig. 4B). This stretch of conserved sequence contains a region of dyad symmetry that might constitute a putative transcriptional terminator. The fact that the nucleotide sequence of huvA (from serotype O1 strain 775) and huvS (from serotype O3 ET-208 strain) is highly conserved in the regions flanking the ORF, but highly variable in the ORF itself, suggests that one of the two genes could have been gained by lateral transfer. Recombination could have occurred within the huvZ sequence and the DNA upstream of huvA/huvS, and lead to the exchange of the sequence corresponding to the heme receptor ORF.
The presence of huvS in V. anguillarum isolates was screened by PCR amplification and confirmed by Southern blot hybridization. The results demonstrated that huvS is present in nine V. anguillarum strains that have tested negative for huvA (Table 2). huvS gene was present in four of six of the serotype O3 strains tested, as well as in strains of serotypes O2, O4, O6, O7, and O10. Curiously, the two serotype O3 strains that tested negative for the four genes huvABCD also tested negative for huvS (Table 2). The nucleotide sequence of huvBCD genes in strains containing huvS was almost identical to the huvBCD genes of strains with huvA (data not shown). According to the virulence degree of each strain, as reported by Pazos et al. (26), there is not a clear correlation between presence of huvA or huvS and the virulence degree. However, it is noteworthy that the two strains in which the huvBCD genes and huvA or huvS were absent are environmental isolates with a low degree of virulence (50% lethal dose of >107).
Although heme uptake systems seem to be quite similar in different species (20, 24), diversity of specific heme transport genes within a species has also been reported previously. For example, Worst et al. (38) described the existence of several heme utilization loci in Helicobacter pylori. In V. cholerae, at least three different outer membrane heme receptor genes coexisting in the same strain have been reported (18). These three receptors have differences in their sequence, but all of them share functional characteristics in the use of heme as iron source. Similarly, strains of Pseudomonas aeruginosa containing two distinct outer membrane heme receptors have been described (23). In the present study, we have demonstrated that two different heme receptors exist within V. anguillarum but do not coexist in the same cell, which makes a difference with the situation reported in V. cholerae.
Substitution of huvA function by huvS.
To test that HuvS actually functions as an outer membrane heme receptor, a plasmid containing this gene was transformed into E. coli 101ESD previously transformed with plasmid pSML33. This strain contains all of the genes necessary for heme internalization but lacks an outer membrane heme receptor and thus cannot use heme as an iron source (20). However, when this strain was transformed with plasmid pSML92 containing huvS, it could utilize hemin and hemoglobin as sole iron sources, demonstrating that the new V. anguillarum heme receptor can substitute HuvA in the E. coli complementation model (Table 3). In addition, the huvS gene was introduced into a V. anguillarum ΔhuvA mutant strain (unable to grow with hemin or hemoglobin as sole iron sources) and tested for its ability to restore the wild-type phenotype. The ΔhuvA strain transformed with plasmid pSML93 recovered the ability to grow with heme compounds as the sole iron source and achieving growth rates similar to wild-type levels (Table 3). This confirms that HuvS functions in V. anguillarum as an outer membrane heme receptor and that it can efficiently substitute the HuvA function.
TABLE 3.
Utilization of hemin or hemoglobin as an iron source by E. coli and V. anguillarum heme uptake-deficient mutants complemented with outer membrane heme receptors huvA and huvS genes
| Strain | Gene(s) present in plasmids | Utilization ofa:
|
||
|---|---|---|---|---|
| Hm at 10 μM | Hb at 1 μM | FeSO4 at 10 μM | ||
| E. coli | ||||
| 101ESD | None | − | − | + |
| 101ESD/pCAR121 | huvA | − | − | + |
| 101ESD/pSML33 | tonB-exbBD-huvBCD | − | − | + |
| 101ESD/pSML33/pCAR121 | tonB-exbBD-huvBCD huvA | + | + | + |
| 101ESD/pSML92 | huvS | − | − | + |
| 101ESD/pSML33/pSML92 | tonB-exbBD-huvBCD huvS | + | + | + |
| V. anguillarumb | ||||
| H775-3 | None | + (1.27) | + (1.31) | + (1.20) |
| H775-3 ΔhuvA | None | − (0.18) | − (0.18) | + (1.20) |
| H775-3 ΔhuvA/pCAR121 | huvA | + (1.17) | + (1.10) | + (1.18) |
| H775-3 ΔhuvA/pSML93 | huvS | + (1.15) | + (1.06) | + (1.19) |
+ or −, growth or no growth in the plate assays. Hm, hemin; Hb, hemoglobin.
Numbers in parentheses indicate the OD600 mean values in the liquid medium assays (see Materials and Methods) reached after 24 h of incubation.
Conclusion.
The results obtained in the present study reveal that the heme uptake system of V. anguillarum shows genetic diversity. The sequence analysis of huvS and huvA and their respective upstream and downstream DNA sequences suggest that horizontal transfer and recombination might be responsible for generating this variability. Still, two V. anguillarum strains showed to lack huvABCD and huvS genes, suggesting that the genetic diversity of the heme uptake system of this species can be higher that expected. Studies to analyze what genes substitute for huvABCD roles in these strains are currently under way. Furthermore, the fact that all V. anguillarum strains examined have a heme uptake system, although encoded by different genetic determinants, suggest that this mechanism must have a basic role in the cell physiology, although their role in virulence is still unclear.
Acknowledgments
This study was supported by grant AGL2003-00086 from the Ministry of Science and Technology of Spain (cofunded by the FEDER Programme from the European Union) and grants PGIDIT04PXIC23501PN and PGIDIT04RMA261014PR-3 from Xunta de Galicia to M.L.L.
REFERENCES
- 1.Bélanger, M., C. Bégin, and M. Jacques. 1995. Lipopolysaccharides of Actinobacillus pleuropneumoniae bind pig hemoglobin. Infect. Immun. 63:656-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carroll, J. A., S. A. Coleman, L. S. Smitherman, and M. F. Minnick. 2000. Hemin-binding surface proteins from Bartonella quintana. Infect. Immun. 68:6750-6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conchas, R. F., M. L. Lemos, J. L. Barja, and A. E. Toranzo. 1991. Distribution of plasmid- and chromosome-mediated iron uptake systems in Vibrio anguillarum strains of different origins. Appl. Environ. Microbiol. 57:2956-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Lorenzo, M., M. Stork, A. F. Alice, C. S. López, and J. H. Crosa. 2004. Vibrio, p. 241-255. In J. H. Crosa, A. R. Mey, and S. M. Payne (ed.), Iron transport in bacteria. ASM Press, Washington, D.C.
- 5.Do Vale, A., B. Magariños, J. L. Romalde, M. L. Lemos, A. E. Ellis, and A. E. Toranzo. 2002. Binding of haemin by the fish pathogen Photobacterium damselae subsp. piscicida. Dis. Aquat. Org. 48:109-115. [DOI] [PubMed] [Google Scholar]
- 6.Fouz, B., R. Mazoy, M. L. Lemos, M. J. del Olmo, and C. Amaro. 1996. Utilization of hemin and hemoglobin by Vibrio vulnificus biotype 2. Appl. Environ. Microbiol. 62:2806-2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.García, T., K. Otto, S. Kjelleberg, and D. R. Nelson. 1997. Growth of Vibrio anguillarum in salmon intestinal mucus. Appl. Environ. Microbiol. 63:1034-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Genco, C. A., and D. W. Dixon. 2001. Emerging strategies in microbial haem capture. Mol. Microbiol. 39:1-11. [DOI] [PubMed] [Google Scholar]
- 9.Grisez, L., and F. Ollevier. 1995. Comparative serology of the marine fish pathogen Vibrio anguillarum. Appl. Environ. Microbiol. 61:4367-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henderson, D. P., and S. M. Payne. 1994. Vibrio cholerae iron transport systems: roles of haeme and siderophore iron transport in virulence and identification of a gene associated with multiple iron transport systems. Infect. Immun. 62:5120-5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larsen, J. L., K. Pedersen, and I. Dalsgaard. 1994. Vibrio anguillarum serovars associated with vibriosis in fish. J. Fish Dis. 17:259-267. [Google Scholar]
- 12.Lemos, M. L., P. Salinas, A. E. Toranzo, J. L. Barja, and J. H. Crosa. 1988. Chromosome-mediated iron-uptake system in pathogenic strains of Vibrio anguillarum. J. Bacteriol. 170:1920-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lemos, M. L., R. Mazoy, R. F. Conchas, and A. E. Toranzo. 1991. Presence of iron uptake mechanisms in environmental non-pathogenic strains of Vibrio anguillarum. Bull. Eur. Assoc. Fish Pathol. 11:150-152. [Google Scholar]
- 14.Mazoy, R., and M. L. Lemos. 1991. Iron-binding proteins and heme compounds as iron sources for Vibrio anguillarum. Curr. Microbiol. 23:221-226. [Google Scholar]
- 15.Mazoy, R., and M. L. Lemos. 1996. Identification of heme-binding proteins in the cell membranes of Vibrio anguillarum. FEMS Microbiol. Lett. 135:265-270. [DOI] [PubMed] [Google Scholar]
- 16.Mazoy, R., C. R. Osorio, A. E. Toranzo, and M. L. Lemos. 2003. Isolation of mutants of Vibrio anguillarum defective in haeme utilization and cloning of huvA, a gene coding for an outer membrane protein involved in the use of haeme as iron source. Arch. Microbiol. 179:329-338. [DOI] [PubMed] [Google Scholar]
- 17.Mazoy, R., F. Vázquez, and M. L. Lemos. 1996. Isolation of heme-binding proteins from Vibrio anguillarum using affinity chromatography. FEMS Microbiol. Lett. 141:19-23. [DOI] [PubMed] [Google Scholar]
- 18.Mey, A. R., and S. M. Payne. 2001. Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol. Microbiol. 42:835-849. [DOI] [PubMed] [Google Scholar]
- 19.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Mouriño, S., C. R. Osorio, and M. L. Lemos. 2004. Characterization of the heme uptake cluster genes in the fish pathogen Vibrio anguillarum. J. Bacteriol. 186:6159-6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muiño, L., M. L. Lemos, and Y. Santos. 2001. Presence of high-affinity iron uptake systems in fish-isolated and environmental strains of Vibrio anguillarum serotype O3. FEMS Microbiol. Lett. 202:79-83. [DOI] [PubMed] [Google Scholar]
- 22.Occhino, D. A., E. E. Wyckoff, D. P. Henderson, T. J. Wrona, and S. M. Payne. 1998. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol. Microbiol. 29:1493-1507. [DOI] [PubMed] [Google Scholar]
- 23.Ochsner, U. A., Z. Johnson, and M. L. Vasil. 2000. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology 146:185-198. [DOI] [PubMed] [Google Scholar]
- 24.O'Malley, S. M., S. L. Mouton, D. A. Occhino, M. T. Deanda, J. R. Rashidi, K. L. Fuson, C. E. Rashidi, M. Y. Mora, S. M. Payne, and D. P. Henderson. 1999. Comparison of the heme iron utilization systems of pathogenic vibrios. J. Bacteriol. 181:3594-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osorio, C. R., and M. L. Lemos. 2002. Haeme iron acquisition mechanisms in Vibrionaceae, p. 419-436. In S. G. Pandalai (ed.), Recent research developments in microbiology, vol. 6. Research Signpost, Kerala, India. [Google Scholar]
- 26.Pazos, F., Y. Santos, B. Magariños, I. Bandín, S. Núñez, and A. E. Toranzo. 1993. Phenotypic characteristics and virulence of Vibrio anguillarum-related organisms. Appl. Environ. Microbiol. 59:2969-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pedersen, K., L. Grisez, R. van Houdt, T. Tiainen, F. Ollevier, and J. L. Larsen. 1999. Extended serotyping scheme for Vibrio anguillarum with the definition and characterization of seven provisional O-serogroups. Curr. Microbiol. 38:183-189. [DOI] [PubMed] [Google Scholar]
- 28.Smalley, J. W., A. J. Birss, A. S. McKee, and P. D. Marsh. 1995. Congo red binding by Porphyromonas gingivalis is mediated by a 66-kDa outer-membrane protein. Microbiology 141:205-211. [DOI] [PubMed] [Google Scholar]
- 29.Sørensen, U. B. S., and J. L. Larsen. 1986. Serotyping of Vibrio anguillarum. Appl. Environ. Microbiol. 51:593-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevens, M. K., S. Porcella, J. Klesney-Tait, S. Lumbley, S. E. Thomas, M. V. Norgard, J. D. Radolf, and E. J. Hansen. 1996. A hemoglobin-binding outer membrane protein is involved in virulence expression by Haemophilus ducreyi in an animal model. Infect. Immun. 64:1724-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stojiljkovic, I., V. Hwa, L. de SaintMartin, P. O'Gaora, X. Nassif, F. Heffron, and M. So. 1995. The Neisseria meningitidis hemoglobin receptor: its role in iron utilization and virulence. Mol. Microbiol. 15:531-541. [DOI] [PubMed] [Google Scholar]
- 32.Stork M., M. Di Lorenzo, T. J. Welch, L. M. Crosa, and J. H. Crosa. 2002. Plasmid-mediated iron uptake and virulence in Vibrio anguillarum. Plasmid 48:222-228. [DOI] [PubMed] [Google Scholar]
- 33.Tai, S. S., C.-J. Lee, and R. E. Winter. 1993. Hemin utilization is related to virulence of Streptococcus pneumoniae. Infect. Immun. 61:5401-5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toranzo, A. E., and J. L. Barja. 1990. A review of the taxonomy and seroepizootiology of Vibrio anguillarum, with special reference to aquaculture in the northwest of Spain. Dis. Aquat. Org. 9:73-82. [Google Scholar]
- 35.Toranzo, A. E., and J. L. Barja. 1993. Virulence factors of bacteria pathogenic for cold water fish. Annu. Rev. Fish Dis. 3:5-36. [Google Scholar]
- 36.Toranzo, A. E., Y. Santos, and J. L. Barja. 1997. Immunization with bacterial antigens: Vibrio infections. Dev. Biol. Stand. 90:93-105. [PubMed] [Google Scholar]
- 37.Wandersman, C., and I. Stojiljkovic. 2000. Bacterial heme sources: the role of heme, hemoprotein receptors, and hemophores. Curr. Opin. Microbiol. 3:215-220. [DOI] [PubMed] [Google Scholar]
- 38.Worst, D. J., J. Maaskant, C. M. J. E. Vandenbroucke-Grauls, and J. G. Kusters. 1999. Multiple haem-utilization loci in Helicobacter pylori. Microbiology 145:681-688. [DOI] [PubMed] [Google Scholar]