Abstract
Clostridium acetobutylicum and Clostridium aminovalericum, both obligatory anaerobes, grow normally after growth conditions are changed from anoxic to microoxic, where the cells consume oxygen proficiently. In C. aminovalericum, a gene encoding a previously characterized H2O-forming NADH oxidase, designated noxA, was cloned and sequenced. The expression of noxA was strongly upregulated within 10 min after the growth conditions were altered to a microoxic state, indicating that C. aminovalericum NoxA is involved in oxygen metabolism. In C. acetobutylicum, genes suggested to be involved in oxygen metabolism and genes for reactive oxygen species (ROS) scavenging were chosen from the genome database. Although no clear orthologue of C. aminovalericum NoxA was found, Northern blot analysis identified many O2-responsive genes (e.g., a gene cluster [CAC2448 to CAC2452] encoding an NADH rubredoxin oxidoreductase-A-type flavoprotein-desulfoferrodoxin homologue-MerR family-like protein-flavodoxin, an operon [CAC1547 to CAC1549] encoding a thioredoxin-thioredoxin reductase-glutathione peroxidase-like protein, an operon [CAC1570 and CAC1571] encoding two glutathione peroxidase-like proteins, and genes encoding thiol peroxidase, bacterioferritin comigratory proteins, and superoxide dismutase) whose expression was quickly and synchronously upregulated within 10 min after flushing with 5% O2. The corresponding enzyme activities, such as NAD(P)H-dependent peroxide (H2O2 and alkyl hydroperoxides) reductase, were highly induced, indicating that microoxic growth of C. acetobutylicum is associated with the expression of a number of genes for oxygen metabolism and ROS scavenging.
Bacteria belonging to the genus Clostridium are classified as obligatory anaerobes (26, 62) and are widely used in the field of solvent fermentation, biodegradation, and microbial energy production. Oxygen has a crucial effect on the growth of clostridia, but the mechanisms of growth inhibition, as well as the existence of O2 metabolic systems, remain unknown. Some hypotheses to explain aerobic growth inhibition in anaerobes were proposed, such as the possibility that oxygen attacks oxygen-sensitive enzymes causing metabolic cessation or that anaerobes lack the ability to decompose active oxygen species, such as catalase, which cause irreversible oxidative damage to DNA and lipid molecules (2, 27, 49, 63, 67). O'Brien and Morris proposed that NAD(P)H oxidation systems react with oxygen to cause oxidation of the electron donor, i.e., NAD(P)H, which is required for the central pathway for anaerobic metabolism; this then leads to the eventual inability of clostridia to maintain their internal redox balance (51, 55). However, many questions remain about the mechanisms of aerobic growth inhibition in clostridia (50).
Most Clostridium species do not form colonies in the presence of 1% oxygen (2, 62); however, they can accept microoxic conditions when grown in liquid medium (32-35, 39, 51, 55). Based on physiological examination, clostridia possess systems to metabolize O2, and oxygen itself does not have an irreversible effect on growth. Clostridium butyricum, a type species of the genus Clostridium, has the ability to resume growth after oxygen consumption without any lasting damage (33). Küsel etal. reported that Clostridium glycolicum, an acetogenic bacterium, can grow in the presence of oxygen (under up to 6% headspace oxygen in static culture) with oxygen-consuming activities. Under these conditions, this acetogen switches to a fermentative metabolism that is not as sensitive to oxygen as acetogenesis (39). Karnholz et al. reported that Clostridium magnum, an acetogenic bacteria, can grow normally in nonreduced liquid culture medium in the presence of oxygen (1% headspace oxygen) with oxygen-consuming activity and with normal acetogenesis (32). Clostridium acetobutylicum and Clostridium aminovalericum, both typical Clostridium species, grow normally under continuous microoxic conditions by consuming oxygen (34, 35). These observations suggest that clostridia should possess some systems to metabolize O2 as well as to scavenge active oxygen species derived from O2 reduction; however, the existence of these systems has not been well investigated.
Based on the recent molecular-based approach to finding clostridial oxygen metabolic systems, some oxygen response genes have been identified. In the pathogen Clostridium perfringens, several genes (CPE0781, encoding a probable flavoprotein; CPE0782, encoding an alkyl hydrogen peroxide reductase; and CPE0782, encoding a superoxide dismutase [SOD]) respond to air at the transcriptional level (28). In C.acetobutylicum, novel oxygen response proteins (A-type flavoprotein and rubrerythrins) were identified by two-dimensional electrophoresis, and the transcripts encoding these proteins were found to be upregulated quickly and strongly within minutes after the cells were exposed to microoxic aeration (35). Although the above-mentioned enzyme activities and their roles in vivo have not been determined, it is obvious that clostridia possess systems to respond to O2 at the transcriptional level.
In this study, we investigated the expression of genes required for oxygen metabolic systems using two species, C. aminovalericum and C. acetobutylicum. In C. aminovalericum, an H2O-forming NADH oxidase was purified for the first time from a member of the Clostridium genus and was characterized as a kinetically functional oxidase in vivo (34). Expression studies of the C. aminovalericum noxA gene revealed that the enzyme is involved in oxygen metabolism. In C. acetobutylicum, many genes were identified as oxygen stress responsive for the first time in the genus Clostridium. The possible functions of these O2-responsive gene products are discussed.
MATERIALS AND METHODS
Bacterial strains and media.
Clostridium aminovalericum NRIC0223 (= DSM1283) and Clostridium acetobutylicum DSM792 (= ATCC 824) were used in this study. Growth conditions and aeration studies were described previously (34, 35). Briefly, anoxic cultures were achieved by flushing the medium with O2-free nitrogen gas in a jar fermenter (10-liter jar fermenter, model MDL1000; Marubishi-Bioenge, Tokyo, Japan). Aeration was achieved by flushing with 3% O2-97% N2 mixed gas or with 5% O2-95% N2 mixed gas (a gas flow meter was used to fix the ratio of each gas) into the anaerobically growing strain at the mid-exponential phase.
Cloning of a C. aminovalericum H2O-forming NADH oxidase.
We previously determined the N-terminal 13 amino acids of C. aminovalericum NADH oxidase (34). To obtain genomic DNA encoding C. aminovalericum NADH oxidase, we performed long N-terminal sequencing again. The methods for sequencing were described previously. The N-terminal amino acid sequence (40 amino acids from the N terminus) was determined by the Edman degradation method with a peptide sequencer (model 92HT; Perkin-Elmer Applied Biosystems, Foster City, CA). We first isolated the partial genome region by using degenerated sense and antisense primers based on the N-terminal amino acid sequence. The degenerated sense primer (5′-ATGAARATHGTNGTNATHGG-3′ [where H is A, T, or C; N is A, C, G, or T; and R is A or G]) and antisense primers (5′-TCRAADATNGTDATYTCNGC-3′ and 5′-ATNGTDATYTCNGCYTCNGG [where D is A, G, or T and Y is C or T]) were used for the first PCR using C. aminovalericum genomic DNA as a template. Sequence analysis indicated that 96 bp of the DNA fragment obtained from the first PCR encoded the N-terminal part of the target protein. Using an in vitro cloning kit (Takara-bio, Tokyo, Japan), a DNA fragment of 489 bp including the 96-bp first PCR DNA fragment was obtained. The 489-bp fragment was used as a probe to screen a C. aminovalericum genomic library in the Lambda Zap bacteriophage. C. aminovalericum genomic DNA partially digested with the enzyme Sau3AI was used to construct a genomic DNA library. All steps for library construction and in vitro packing were performed according to the manufacturer's instructions (Lambda ZAP Express library construction kit and Gigapack III Gold Packing extracts; Stratagene). The probe was labeled by random priming using [32P]dCTP as described previously by Sambrook et al. (59). The pBluescript plasmids containing chromosomal inserts were excised from positive plaques according to the manufacturer's instructions. One plasmid, pNoxA-10, was chosen for sequencing of the noxA genome region. Multiple sequence alignment was achieved with the program Clustal W.
Northern hybridization.
All steps were performed according to methods described previously (34, 35). Briefly, total RNAs from cell lysates harvested at several time points of aeration were isolated using TRIzol (Invitrogen, CA) according to the instruction manual. Northern blotting was carried out by standard procedures (35). The RNA (15 μg) was electrophoresed in 1.0% agarose gels and blotted onto nylon membranes (Hybond N+; Amersham, Japan). The membranes were probed with the PCR-amplified DNA fragment encoding the target region. The identity of the amplified DNA fragment was confirmed by size and nucleotide sequence. After the RNA was subjected to prehybridization at 60°C for 30 min, the 32P-labeled DNA probe was hybridized to RNA on the membrane at 60°C for 12 h.
Enzyme assays.
C. aminovalericum and C. acetobutylicum cells harvested at several time points of aeration were suspended in 50 mM potassium phosphate buffer (pH 7.0) and disrupted by treatment with a French pressure cell at 140 MPa. Cell extracts were obtained by removal of cell debris by centrifugation at 15,000 × g for 15 min.
The activities of NADH oxidase and NADPH oxidase were assayed spectrophotometrically in 1 ml of air-saturated 50 mM sodium phosphate buffer (pH 7.0) containing 0.15 mM NAD(P)H at 37°C. The reaction was started by the addition of enzyme solution, and the decrease in absorbance at 340 nm was monitored with a spectrophotometer (model U-160; Shimadzu, Kyoto, Japan). One unit of activity was defined as the amount of enzyme that catalyzes the oxidation of 1 μmol NAD(P)H per min.
The activities of NADH- and NADPH-dependent H2O2 or alkyl hydroperoxide reductase were assayed anaerobically in anaerobic glass cuvettes in 2 ml of Ar gas-saturated 50 mM sodium phosphate buffer (pH 7.0) containing 0.15 mM NAD(P)H at 37°C. The reaction was started by the addition of Ar gas-treated enzyme solution, and the decrease in absorbance at 340 nm after the addition of 1 mM H2O2, t-butyl hydroperoxide, or cumen hydroperoxide was monitored with a spectrophotometer (model U-160; Shimadzu, Kyoto, Japan). One unit of activity was defined as the amount of enzyme that catalyzes the oxidation of 1 μmol NAD(P)H per min.
Glutathione (GSH) peroxidase activities were measured indirectly by examining a coupled reaction with glutathione reductase (18). The principle is that the oxidized glutathione produced by the reduction of H2O2 by GSH peroxidase is recycled to reduced GSH in the presence of excess glutathione reductase and NADPH. The reaction mixture contained 50 mM sodium phosphate buffer (pH 7.0) containing 1 mM GSH plus 0.15 mM NADPH, 1 U glutathione reductase, and the enzyme, and the reaction was started by the addition of Ar gas-treated enzyme solution. The decrease in absorbance at 340 nm after the addition of 1 mM H2O2 was measured spectroscopically. One unit of activity was defined as the amount of enzyme that catalyzes the oxidation of 1 μmol NAD(P)H per min.
Superoxide dismutase activity was determined using the xanthine oxidase-cytochrome c system as described previously by McCord and Fridovich (49).
Nucleotide sequence accession number.
The nucleotide sequence data reported in this study have been deposited in the DDBJ/EMBL/GenBank database under accession number AB219226.
RESULTS AND DISCUSSION
Cloning and characterization of the gene encoding H2O-forming NADH oxidase from C. aminovalericum.
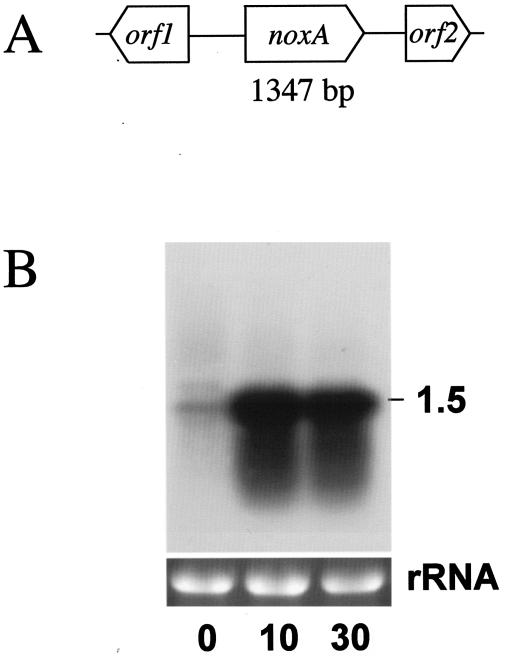
C. aminovalericum NADH oxidase, which reduces O2 to H2O with a comparatively high affinity for O2 (the Km for oxygen is 61.9 μM), was purified and characterized (34). The sequence of the 40 amino acids from the N terminus was determined to be MKIVVIGCTHAGTAAVKTILKENPEAEITIFERNDNISFL. A 96-bp gene fragment encoding the N-terminal part of C. aminovalericum NADH oxidase was cloned using an oligonucleotide designed from the N-terminal sequence. Using the 96-bp nucleotide fragment, a genome fragment of approximately 4 kb was cloned and sequenced, and three open reading frames (ORFs) were identified as orf1, noxA, and orf2 (Fig. 1A). The cloned gene for NADH oxidase was 1,347 bp long and encoded 448 amino acid residues. The primary structure of NADH oxidase deduced from noxA shows high sequence similarity to E. faecalis NADH oxidase (NCBI accession no. AAO81372; 66% identity). The proposed domains for flavin adenine dinucleotide binding and NADH binding and the site of a redox-active cysteine residue in Enterococcus faecalis NADH oxidase (Cys42) (58) are conserved in C. aminovalericum NoxA. By the genome database search for investigating the distribution of this protein in clostridia, C. aminovalericum NoxA homologues were identified in Clostridium perfringens (accession no. BAB80440; 29% identity) and Clostridium tetani (accession no. AAO35751; 28% identity). Both proteins conserved redox-active Cys42 and domains for flavin adenine dinucleotide binding and NADH binding, suggesting that they function as NADH oxidase, but both proteins have not been characterized in terms of function.
FIG. 1.
Cloning and expression of the noxA gene of C. aminovalericum. (A) Genome structure around the noxA gene. (B) Northern blot of 15 μg of C. aminovalericum total RNA probes with noxA amplified by PCR. 0, just before the start of aeration with 3% O2-97% N2 at the mid-exponential phase; 10, after 10 min of 3% O2-97% N2 aeration; 30, after 30 min of 3% O2-97% N2 aeration. The estimated sizes of the observed transcripts are indicated on the right. Ethidium bromide staining of the rRNA genes for confirmation of equal loading of RNA samples is shown below the autoradiogram.
ORF1, located 641 bp upstream from the start codon of noxA, encodes a protein of 211 amino acids, and proteins showing similarity are found in Streptococcus mutans (called hypothetical protein SMU.1470c [NCBI accession no. NP_721820]; 46% identity) and Enterococcus faecium (called uncharacterized conserved protein [accession no. ZP_00287298]; 46% identity). None of these proteins has been characterized in terms of function. ORF2, located 481 bp downstream from the terminal codon of noxA, encodes a protein of 156 amino acids, and similar proteins are found in Enterococcus faecium (called a putative transposase [accession no. AAF73101]; 78% identity) and C. acetobutylicum (called IS605/IS200-like transposase [accession no. AAK81457]; 73% identity).
Transcriptional expression of genes encoding NADH oxidase from C. aminovalericum.
We assumed that the C. aminovalericum NADH oxidase would play a central role in the oxygen consumption observed in this bacterium (34). Actually, the activity of NADH oxidase in the cell extract was activated more than sixfold after 30 min of flushing with 3% O2 (34). To determine whether the expression of the noxA gene corresponds with the elevation of enzyme activity, Northern analysis was performed (Fig. 1B). The results show that the noxA gene was monocistronically transcribed and strongly up-regulated by 10 min after the start of microoxic aeration. These results, as well as the enzyme kinetics data (34), indicate that NoxA is involved in the oxygen metabolic system of C. aminovalericum.
Examination of the C. acetobutylicum genome to identify the genes involved in oxygen metabolism and active oxygen scavenging.
For further attempts to identify the genes that are required for oxygen metabolic systems in clostridia, we used C. acetobutylicum, a well-investigated solvent producer that grows microoxically by consuming oxygen, as in the case of C. aminovalericum. The whole genome sequence of C. acetobutylicum is available in public databases (54). By using key words such as oxygen metabolism, reactive oxygen species (ROS) scavengers, and the maintenance of the redox environment of a cell, we examined the C. acetobutylicum genome database for choosing genes including C. aminovalericum NoxA homologues. The genes chosen in this study are listed in Table 1. The functions of most of these proteins in C. acetobutylicum have not been characterized, and they are all presumed to be located in cytosol. The specific primers used to amplify the target genes are listed in Table 1. Biological reduction of oxygen typically occurs by means of terminal oxidases such as cytochrome oxidase. Interestingly, cytochrome bd-type oxidases have recently been discovered in strict anaerobes such as Bacteroides species (5), Desulfovibrio gigas (42), and Moorella thermoacetica (16) as a functional terminal oxidase. In the C. acetobutylicum genome, genes encoding cytochrome bd-type oxidase or other membrane-bound-type oxidases are not found.
TABLE 1.
List of genes and PCR primers used in this study
| Gene identification | Gene name | Primer sequence (5′-3′) |
|---|---|---|
| nror gene cluster | ||
| CAC2448 | nror | F, AGATGATTTATATGAAAAGCAC |
| R, AATGTATTTATCTTCTTGTG | ||
| CAC2449 | fprA2 | F, AGTTCTAAATCCTAGTCTCC |
| R, CTCAGATGGAACAAATAAAC | ||
| CAC2450 | dsr | F, ATGAATAACGATTTATCAATTTAC |
| R, TTATATATCTGCTTTCCATAGG | ||
| CAC2451 | orf2451 | F, GAGCTTAATATAATAGTTCC |
| R, ACATTTATTTAATAGCAGCC | ||
| CAC2452 | fld1 | F, GTCGAGGAGGAATTATTATG |
| R, TCTTCCTTACTAGGTGCCTC | ||
| Glutathione peroxidase operon 1 | ||
| CAC1547 | trxA1 | F, GGCAATGAAATGATACAAGAG |
| R, TGTCCATAGAAATCCACTCC | ||
| CAC1548 | trxB1a | |
| CAC1549 | gpx3a | |
| Glutathione peroxidase operon 2 | ||
| CAC1570 | gpx1 | F, TTGGGAGGTACGATTATGTC |
| R, TCTTCCATGCTTATATCTTC | ||
| CAC1571 | gpx2a | |
| Thioredoxin-thioredoxin reductase operon | ||
| CAC3082 | trxB2 | F, AGGAGAGCCTGTAATAGTTG |
| R, TAAAGGAGCATCACAAGTGG | ||
| CAC3083 | trxA2a | |
| BCP and TSA family proteins | ||
| CAC0327 | bcp | F, TTTAGAGGTGATATTTATGG |
| R, TCCTATTGGCGTTTTCTACC | ||
| CAC3306 | tpx | F, TAAACAGTATGCTATGAAAG |
| R, GCTGCTTGTAACACTTCATC | ||
| Rubrerythrin homologue | ||
| CAC2575 | rubY | F, TATATGAAATCACTTAAAGG |
| R, TGAGGATGAAGACATGCTGG | ||
| CAC3018 | rubZ | F, GGAAGTATATTATTATGAGTG |
| R, CATTGTTCTATATTCTCATC | ||
| SOD homologue | ||
| CAC1363 | sodC | F, TGAACACAGAGAGAACGAGG |
| R, CTAACAGCTCTAATAACACC | ||
| CAC2567 | sodB | F, AATTTTAATTCACTAGCTGG |
| R, ATAGACATAAAGGGACATAC |
Genes whose expression levels were determined to be the same as that of a gene in a polycistronic unit.
O2-responsive nror gene cluster.
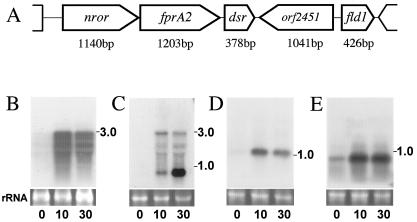
In C. acetobutylicum, the database search identified several genes that encode C. aminovalericum NoxA homologues; however, their identity scores were not high (the products of genes CAC0764, CAC1674, CAC3371, and CAC3408 show homology but less than 17% identity with NoxA). The nror gene (CAC2448) encodes a protein showing the closest similarity with C. aminovalericum NoxA (22% identity) in the C. acetobutylicum genome. This protein was formerly characterized as NADH-rubredoxin oxidoreductase (NROR) by Guedon and Petitdemange (24). Although the reactivity of C. acetobutylicum NROR with oxygen has not been determined, redox-active Cys42 is not conserved in C. acetobutylicum NROR, suggesting that NROR itself does not function as an NADH oxidase. C. acetobutylicum NROR reduces rubredoxin efficiently; however, its actual function in vivo has not been elucidated because the rd gene (accession no. CAC2778), which encodes rubredoxin, the expected electron acceptor from NROR, is located 340 kb apart from nror (24). Northern blot analysis showed that nror is transcribed tricistronically with other two genes (CAC2449, named fprA2, and CAC2450, named dsr), which encode an A-type flavoprotein homologue and a desulfoferrodoxin homologue, respectively (Fig. 2A). These genes are faintly expressed during anaerobic growth but are strongly upregulated within 10 min of the start of aeration (Fig. 2B). To our knowledge, there is no study on O2-responsive nror in bacteria. Interestingly, the dsr gene is transcribed not only tricistronically with nror and fprA2 genes but also monocistronically by its own promoter. By using the dsr region as a probe, tricistronic upregulation after 10 min was observed, and also, strong upregulation of dsr was detected after 30 min (Fig. 2C). These results suggested that two types of transcriptional regulators regulate the expression of the dsr gene. We are now analyzing the expression mechanism of the dsr gene.
FIG. 2.
(A) A gene cluster downstream of the nror gene (CAC2448) encoding NADH rubredoxin oxidoreductase. (B to E) Northern blots of 15 μg of C. acetobutylicum total RNA probed with (B) the fprA2 gene, (C) the dsr gene, (D) the orf2451 gene (CAC2451), or (E) the fld1 gene amplified by PCR. The autoradiogram probed with the PCR-amplified nror gene showed a profile similar to that when fprA2 was used as a probe (data not shown). 0, just before the start of aeration with 5% O2-95% N2 at the mid-exponential phase; 10, after 10 min of 5% O2-95% N2 aeration; 30, after 30 min of 5% O2-95% N2 aeration. The estimated sizes of the observed transcripts are indicated on the right. Ethidium bromide staining of the rRNA genes for confirmation of equal loading of RNA samples is shown below the autoradiogram.
Interestingly, fprA2 encodes a protein showing homology to an oxygen-induced A-type flavoprotein (NCBI accession no. NP_347663) (encoded by CAC1027, named fprA1; 40% identity) identified by two-dimensional electrophoresis in our previous study (35). FprA homologues are distributed predominantly in anaerobes. Desulfovibrio vulgaris FprA (accession no. Q9F0J6) and Moorella thermoacetica FprA (accession no. Q9FDN7) were recently determined to function as nitric oxide reductases, but not as oxidases, because oxidase turnover causes irreversible inactivation of the enzymes (60, 61). C. acetobutylicum FprA2 shows 28% identity with D. vulgaris FprA and M. thermoacetica FprA; however, the numbers and positions of the conserved cysteine residues, which are important for iron binding and the redox state, do not correspond. Thus, there is a possibility that oxygen-responsive C. acetobutylicum FprA1 and FprA2 are involved in the oxygen metabolic system.
C. acetobutylicum Dsr shows homology to Desulfovibrio vulgaris desulfoferrodoxin (NCBI accession no. P20418; 43% identity) and Desulfovibrio desulfuricans desulfoferrodoxin (accession no. P22076; 42% identity) (7, 52). Desulfoferrodoxin is reported to catalyze the superoxide reductase (SOR) reaction (44, 45). The SOR reaction is believed to be advantageous to anaerobic bacteria, in comparison to SOD, because SOR produces no oxygen by reducing O2− (23, 29). In C. acetobutylicum Dsr, one Cys-X-Y-Cys and one Cys-Cys motif that are conserved in Desulfovibrio proteins and are responsible for the binding of redox-active center I [Fe(SCys)4], which has a distorted tetrahedral sulfur coordination sphere and is presumably an electron transfer center (17), are not conserved. C. acetobutylicum Dsr contains a [Fe(His)4Cys] site, called center II in desulfoferrodoxin family proteins (12, 68). Lombard etal. reported that center II is the O2−-reactive center because it is rapidly oxidized by O2− (45). A BLAST search found one orthologue of C. acetobutylicum Dsr, which lacks center I, in the genome database of Treponema pallidum (named desulfoferrodoxin [accession no. AAC65791]; 36% identity). This protein has been well characterized and was reported to function as superoxide reductase (31, 46). These results suggested that C. acetobutylicum Dsr belongs to the same family of desulfoferrodoxins and functions as superoxide reductase. Based on the gene structure and the expression profile, we propose that the proteins encoded by the nror operon might function as novel multiple-enzyme complexes (NROR-FprA2-Dsr) in NAD(P)H-dependent oxygen metabolism or ROS scavenging.
There are two genes localized downstream of dsr (Fig. 2A), one of which encodes a methyltransferase homologue fused to a MerR-type transcriptional regulator homologue at the N-terminal part (CAC2451, named ORF2451). A BLAST search revealed no fusion proteins distributed among organisms. MerR-type transcriptional regulators have been found in a wide range of bacterial genera and have been shown to mediate environmental responses to stress including heavy metals, drugs, or oxidative stress (1, 6, 56, 65). The MerR-type family regulators conserve homologous N-terminal helix-turn-helix DNA-binding domains but different C-terminal domains, which enable them to bind a variety of coeffectors (inducers) (6). The N-terminal part of ORF2451 (60 amino acid residues from the N terminus) shows high similarity to the DNA-binding region of MerR-type regulatory proteins and conserves the helix-turn-helix domain (Fig. 3A). The C-terminal part (approximately 250 amino acid residues from the C terminus) shows similarity to some bacterial hypothetical proteins named menaquinone biosynthesis methyltransferase or S-adenosyl-l-methionine-dependent methyltransferase in genome projects (i.e., possible menaquinone biosynthesis methyltransferase in Pyrococcus abyssi [NCBI accession no. CAB49332]; 34% identity). These top-BLAST-hit proteins have not been characterized in terms of function. The C-terminal part of ORF2451 shows low similarity with the functionally identified Escherichia coli C-methyltransferase (named ubiE [accession no. M80749]; 13.5% identity and 50.6% similarity) (Fig. 3B). This protein is involved in the S-adenosyl-l-methionine-dependent C-methyltransferase step of both ubiquinone and menaquinone biosynthesis (40). Thus, ORF2451 may be a novel MerR-type O2-responsive transcriptional regulator with methyltransferase activity.
FIG. 3.
Amino acid sequence alignments. (A) Alignment of ORF2451 with the N-terminal DNA-binding domains of functionally characterized MerR family proteins. Proteins and their corresponding NCBI accession numbers are as follows: B. subtilis BmrR (accession no. CAB56371) (1), B. subtilis MtaN (accession no. CAB15677) (4), P. aeruginosa MerR (accession no. CAA83897) (56), and E. coli CueR (accession no. AAC73589) (57). H T H, DNA-binding helix-turn-helix motif. By comparison of ORF2451 and E. coli CueR, identical residues are asterisked, and similar residues are dotted (37% identity and 58% similarity). (B) Comparison of the amino acid sequence of the ORF2451 C-terminal region and the functionally characterized E. coli UbiE (E UbiE) (accession no. AAA67628) (40). Identical residues are asterisked, and similar residues are dotted.
The gene named fld1 (NCBI accession no. CAC2452) encodes a flavodoxin homologue. A BLAST search revealed highly homologous proteins distributed among Clostridium species. Flavodoxin is a flavin mononucleotide protein that replaces bacterial ferredoxin in hydrogen evolution and nitrogen fixation in vitro (37, 38). Flavodoxin is obtained from Clostridium pasteurianum grown in iron-deficient medium (37). It is suggested that flavodoxin replaces ferredoxin in vivo since iron-deficient cells contain very little ferredoxin. These two genes are localized in tandem, but the orf2451 gene (CAC2451) is in an opposite direction. Northern blot analysis revealed that the expression of both genes is up-regulated by 10 min after the start of aeration (Fig. 2C and D). In Escherichia coli, flavodoxin is reported to belong to the soxRS oxidative stress regulon, but the role of flavodoxin in oxidative stress is unknown (20).
Oxygen-responsive glutathione peroxidase, thioredoxin, and thioredoxin reductase.
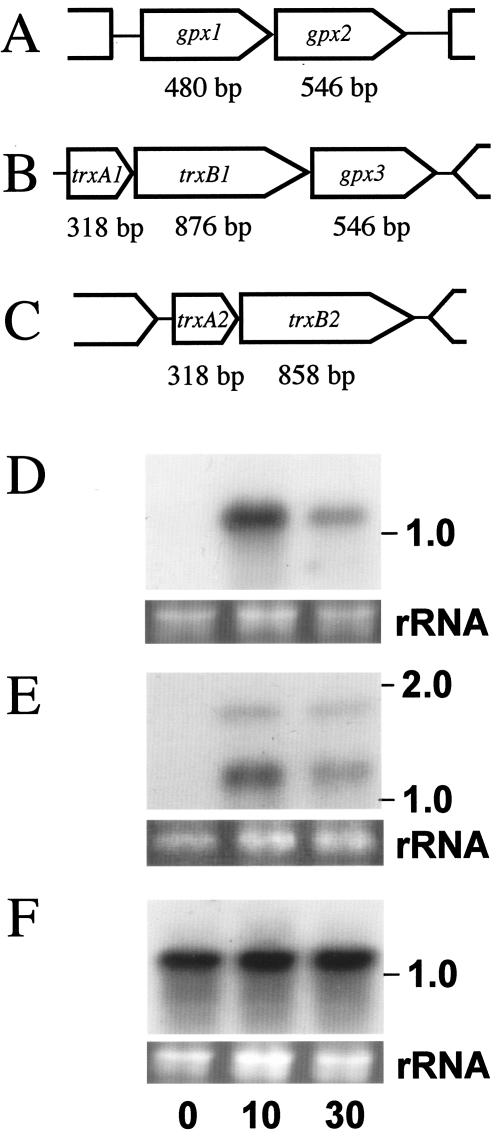
The genes that encode glutathione peroxidase homologues are located in different parts of the genome; two of them are localized tandemly as a dicistronic unit (CAC1570 [named gpx1] and CAC1571 [named gpx2]) (Fig. 4A). gpx3 (CAC1549) is in the same polycistronic unit as two other genes, trxB1 and trxA1, which encode a thioredoxin reductase homologue and a thioredoxin homologue, respectively (Fig. 4B). gpx1 encodes a protein with 159 amino acid residues, and gpx2 and gpx3 encode proteins with 181 amino acid residues. Gpx1 and Gpx2 show 51% identity, and Gpx2 and Gpx3 show 55% identity. Gpx1 shows 55% identity with the functionally identified Synechocystis Gpx1 (19). Among bacterial genome data, it is difficult to find a gene cluster in which gpx1 and gpx2 are located in tandem. Northern analysis indicated that both gpx1 and gpx2 are dicistronically transcribed and strongly upregulated within 10 min after O2 flushing (Fig. 4D). gpx3 is also transcribed tricistronically and is strongly upregulated within 10 min after O2 flushing (Fig. 4E). Gpx3 may function in a multiple-enzyme complex with thioredoxin and thioredoxin reductase for ROS scavenging.
FIG. 4.
Gene clusters of (A) two glutathione peroxidase-like proteins, (B) thioredoxin-, thioredoxin reductase- and glutathione peroxidase-like proteins, and (C) thioredoxin-thioredoxin reductase. (D to F) Northern blots of 15 μg of C. acetobutylicum total RNA probed with (D) the gpx1 gene, (E) the trxA1 gene, or (F) the trxB2 gene amplified by PCR. 0, just before the start of aeration with 5% O2-95% N2 at the mid-exponential phase; 10, after 10 min of 5% O2-95% N2 aeration; 30, after 30 min of 5% O2-95% N2 aeration. The estimated sizes of the observed transcripts are indicated on the right. Ethidium bromide staining of the rRNA genes for confirmation of equal loading of RNA samples is shown below the autoradiogram.
A gene encoding a homologue of oxygen-responsive TrxB1 is found in the C. acetobutylicum genome. The gene CAC3082 encodes a thioredoxin reductase homologue named TrxB2 which shows 59% identity with TrxB1. trxB2 is located in tandem with a gene encoding a thioredoxin homologue, named trxA2 (CAC3083) (Fig. 4C). TrxA2 shows 41% identity with the above-mentioned oxygen-responsive TrxA1. Northern blot analysis indicated that trxA2-trxB2 is dicistronically transcribed and expressed constitutively (Fig. 4F).
Rubrerythrin homologues.
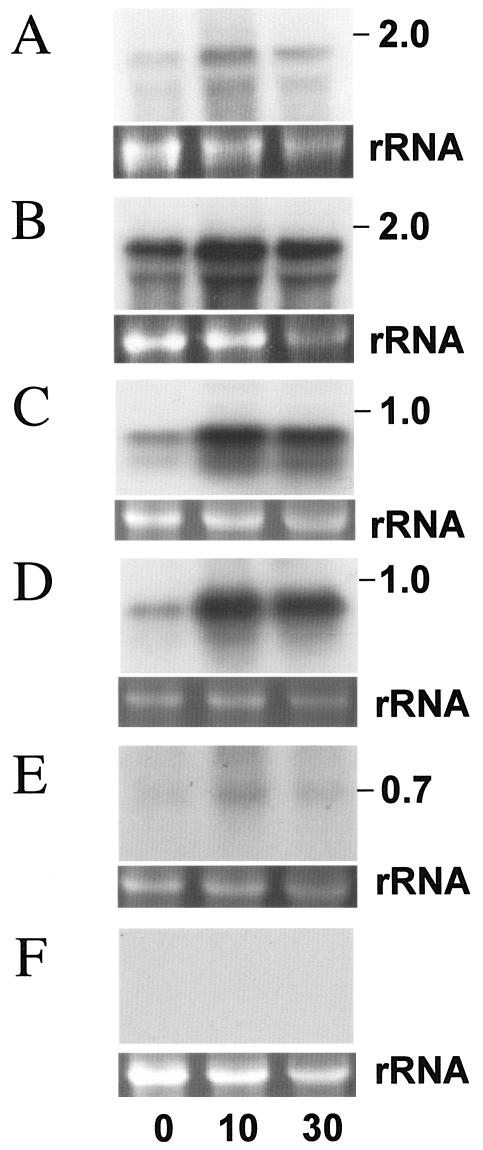
In C. acetobutylicum, tandemly linked rubrerythrins (NCBI accession no. NP_350180 and NP_350181) were identified as O2-induced proteins, and the genes encoding these proteins are upregulated within 10 min after flushing with 5% O2 (35). These genes are regulated by heat shock and H2O2 treatment (48). In the C. acetobutylicum genome, genes encoding rubrerythrin homologues are found as listed in Table 1. Unlike the two rubrerythrin homologues with conserved Fe-binding clusters at their N termini, the remaining two rubrerythrin homologues (RubY, encoded by a gene named rubY [CAC2575], and RubZ, encoded by a gene named rubZ [CAC3018]) have the conserved Fe-binding cluster in their C-terminal regions. This type of rubrerythrin is well characterized and reported to function as SOD or cytoplasmic peroxidase (13, 15, 21, 41, 47, 64). rubY was found to be moderately up-regulated within 10 min after O2 flushing, but rubZ did not show strong up-regulation within 30 min after O2 flushing (Fig. 5A and B). May et al. reported that the expression of rubY is not influenced by air stress (48). This result may be the reason that air stress (20% O2) makes the medium highly oxic, which causes the cells to stop growing.
FIG. 5.
Northern blots of 15 μg of C. acetobutylicum total RNA probed with (A) the rubY gene, (B) the rubZ gene, (C) the bcp gene, (D) the tpx gene, (E) the sodC gene, and (F) the sodB gene amplified by PCR. 0, just before the start of aeration with 5% O2-95% N2 at the mid-exponential phase; 10, after 10 min of 5% O2-95% N2 aeration; 30, after 30 min of 5% O2-95% N2 aeration. The estimated sizes of the observed transcripts are indicated on the right. Ethidium bromide staining of the rRNA genes for confirmation of equal loading of RNA samples is shown below the autoradiogram.
AhpC/TSA/BCP family proteins.
Genes encoding thiol peroxidase (Tpx) family proteins were found in the C. acetobutylicum genome, suggesting that C. acetobutylicum possesses those antioxidative peroxiredoxins. Thiol peroxidases are mainly classified into three different clusters, the alkyl hydroperoxidase component (AhpC), bacterioferritin comigratory proteins (BCP), and thiol-specific antioxidant protein (TSA) (9, 25, 30). These thiol peroxidases reduce hydroperoxides with the use of other electron donor proteins (8, 30, 53). Genes encoding a BCP homologue (CAC0327, named bcp) and a TSA homologue (CAC3306, named tpx), but no clear orthologue of AhpC, are found in the C. acetobutylicum genome. Tpx shows 38% identity with the functionally characterized E. coli Tpx (accession no. AAC45284). C. acetobutylicum Tpx has conserved Cys58 and Cys92 (corresponding to redox-active Cys61 and Cys95 in E. coli Tpx) but lacks Cys85, the deletion of which has a slight effect on the reaction rate of E. coli Tpx (3). Bcp shows 37% identity to the functionally characterized E. coli BCP (accession no. AAC75533), and the three cysteine residues (corresponding to Cys45, Cys50, and Cys99 in E. coli BCP) are well conserved (30). Northern blot analysis indicated that both genes are transcribed monocistronically and are highly expressed within 10 min after 5% O2 flushing (Fig. 5C and D). To our knowledge, there is no study on the transcriptional response of bcp to oxidative stress in bacteria. In E. coli, expression of the gene encoding Tpx did not respond to oxidative stress (9).
SOD.
Superoxide dismutase is well characterized in many organisms as a scavenger of the superoxide anions that arise from the one-electron reduction of oxygen (49). In aerobic organisms, this enzyme is essential for ROS scavenging; however, its role is not well characterized in clostridia. In C. acetobutylicum, the genes that encode an Fe/Mn-type SOD homologue (CAC2567, named sodB) and a Cu/Zn-type SOD homologue (CAC1363, named sodC) are located in different parts of the genome. SodB shows 31% identity with functionally characterized Aquifex pyrophilus Fe-SOD (accession no. 1COJA) (43). SodB does not show significant identity (24% in overlapping regions) with O2-responsive C. perfringens CPE1236. SodC shows 33% identity with the functionally characterized chloroplast-localized Cu/Zn SOD of Arabidopsis thaliana (accession no. O78310) (36). Upon Northern blot analysis, sodC was found to be up-regulated within 10 min of O2 flushing, but no expression of sodB was detected (Fig. 5E and F).
Enzyme activities for oxygen and active oxygen scavenging.
In C. acetobutylicum, many genes that encode putative functional proteins related to oxygen metabolisms and ROS scavenging are up-regulated under microoxic growth conditions. These results suggest that many enzymes are required for normal growth under microoxic conditions. Enzyme activities after 30 min of flushing with O2 are shown in Table 2.
TABLE 2.
Enzyme activities for oxygen and ROS scavenging in C. acetobutylicum after 30 min of 5% O2-95% N2 aeration
| Enzyme | Activityb (mU/mg protein)
|
Induction (fold) | |
|---|---|---|---|
| 0 min | 30 min | ||
| NADH oxidase | 35.9 ± 1.3 | 74.2 ± 0.4 | 2.1 |
| NADPH oxidase | 6.93 ± 0.34 | 14.7 ± 0.65 | 2.1 |
| NADH peroxide reductase | |||
| H2O2 | 0.20 ± 0.08 | 23.1 ± 1.30 | 116 |
| t-Butyl hydroperoxide | 0.23 ± 0.12 | 4.75 ± 0.53 | 20.7 |
| Cumene hydroperoxide | 0.09 ± 0.03 | 4.40 ± 0.17 | 48.9 |
| NADPH peroxide reductase | |||
| H2O2 | NDb (<0.1) | 10.7 ± 0.20 | >100 |
| t-Butyl hydroperoxide | 0.47 ± 0.10 | 4.24 ± 0.71 | 9.0 |
| Cumene hydroperoxide | 0.29 ± 0.18 | 3.71 ± 0.18 | 12.8 |
| Glutathione peroxidase | 0.87 ± 0.17 | 0.26 ± 0.09 | 0.3 |
| SODa | 0.75 ± 0.29 | 0.97 ± 0.35 | 1.3 |
U/mg protein.
ND, not detected.
The rate of activation of NAD(P)H oxidase activity was low compared to that of C. aminovalericum. This may be due to the lack of H2O-forming NADH oxidase in C. acetobutylicum. Interestingly, the activities of NADH- and NADPH-dependent H2O2 reductase are dramatically increased after aeration. In addition, NADH- and NADPH-dependent alkyl hydroperoxide reductase activities (determined as t-butyl hydroperoxide and cumene hydroperoxide reductase activities) are also dramatically increased. In cyanobacteria, the glutathione peroxidase homologues Slr1171 and Slr1992 do not exhibit glutathione peroxidase activities but show NADPH-dependent alkyl hydroperoxide reductase activities (19). In C. acetobutylicum, three genes encoding homologues of glutathione peroxidase that show homology to Synechocystis glutathione peroxidases are strongly up-regulated by aeration, but the corresponding glutathione peroxidase activity was not activated, suggesting that the NADPH alkyl hydroperoxide reductase activity might be carried out by these glutathione peroxidase-like proteins. In addition, genes encoding BCP and TSA are strongly up-regulated in response to O2. Both proteins act as Trx-linked peroxidases capable of reducing NAD(P)H-dependent H2O2 and alkyl hydroperoxides in vitro (3, 8, 30). Two sets of thioredoxin-thioredoxin reductase genes are found in the C. acetobutylicum genome, one of which (trxA1-trxB1-gpx3) is up-regulated within 10 min after O2 stress, while the other (trxA2-trxB2) is expressed constitutively. The actual electron transfer components to BCP and TSA remain to be identified; these two sets of thioredoxin-thioredoxin reductases are the possible electron donor proteins for these two peroxiredoxins. Furthermore, previously identified O2-responsive rubrerythrins (NCBI accession no. NP_350180 and NP_350181) (35, 48) might be involved in NAD(P)H-dependent O2, H2O2, or alkyl hydroperoxide reductase activities because the N-terminal parts of these rubrerythrins show high degrees of identity with rubredoxin, which is a small protein predicted to be involved in some electron transfer systems for oxygen and ROS metabolism (10, 11, 14, 22, 66). Proteins encoded by the O2-responsive nror gene cluster (Fig. 2A) are also considered to involve in NAD(P)H-dependent O2 or ROS reductase activities.
These results suggest that not only oxygen metabolism but also active oxygen and lipid peroxide scavenging enzymes are important for the microoxic growth of C. acetobutylicum. We are now investigating the individual functions of the proteins encoded by O2-responsive genes, the mechanisms of oxygen-responsive promoters, and the distribution of oxygen response systems among clostridia.
Acknowledgments
We thank Tohru Kodama and Junichi Nakagawa for valuable discussions. We also thank Yoshimasa Sagane, Aoi Nakai, Soushi Domae, Akinori Anzai, and Yousuke Shirai for helpful technical assistance at the Tokyo University of Agriculture.
REFERENCES
- 1.Ahmed, M., C. M. Borsch, S. S. Taylor, N. Vazquez-Laslop, and A. A. Neyfakh. 1994. A protein that activates expression of a multidrug efflux transporter upon binding the transporter substrates. J. Biol. Chem. 269:28506-28513. [PubMed] [Google Scholar]
- 2.Andreesen, J. R., H. Bahl, and G. Gottschalk. 1991. Introduction to the physiology and biochemistry of the genus Clostridium, p. 27-62. In N. P. Minton and D. J. Clark (ed.), Biotechnology handbook, vol. 3. Plenum Press, New York, N.Y. [Google Scholar]
- 3.Baker, L. M., and L. B. Poole. 2003. Catalytic mechanism of thiol peroxidase from Escherichia coli. Sulfenic acid formation and overoxidation of essential CYS61. J. Biol. Chem. 278:9203-9211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baranova, N. N., A. Danchin, and A. A. Neyfakh. 1999. Mta, a global MerR-type regulator of the Bacillus subtilis multidrug-efflux transporters. Mol. Microbiol. 31:1549-1559. [DOI] [PubMed] [Google Scholar]
- 5.Baughn, A. D., and M. H. Malamy. 2004. The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature 427:441-444. [DOI] [PubMed] [Google Scholar]
- 6.Brown, N. L., J. V. Stoyanov, S. P. Kidd, and J. L. Hobman. 2003. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27:145-163. [DOI] [PubMed] [Google Scholar]
- 7.Brumlik, M. J., G. Leroy, M. Bruschi, and G. Voordouw. 1990. The nucleotide sequence of the Desulfovibrio gigas desulforedoxin gene indicates that the Desulfovibrio vulgaris rbo gene originated from a gene fusion event. J. Bacteriol. 172:7289-7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cha, M. K., H. K. Kim, and I. H. Kim. 1995. Thioredoxin-linked “thiol peroxidase” from periplasmic space of Escherichia coli. J. Biol. Chem. 270:28635-28641. [DOI] [PubMed] [Google Scholar]
- 9.Cha, M. K., W. C. Kim, C. J. Lim, K. Kim, and I. H. Kim. 2004. Escherichia coli periplasmic thiol peroxidase acts as lipid hydroperoxide peroxidase and the principal antioxidative function during anaerobic growth. J. Biol. Chem. 279:8769-8778. [DOI] [PubMed] [Google Scholar]
- 10.Chen, L., M. Y. Liu, J. Legall, P. Fareleira, H. Santos, and A. V. Xavier. 1993. Purification and characterization of an NADH-rubredoxin oxidoreductase involved in the utilization of oxygen by Desulfovibrio gigas. Eur. J. Biochem. 216:443-448. [DOI] [PubMed] [Google Scholar]
- 11.Chen, L., M. Y. Liu, J. Legall, P. Fareleira, H. Santos, and A. V. Xavier. 1993. Rubredoxin oxidase, a new flavo-hemo-protein, is the site of oxygen reduction to water by the “strict anaerobe” Desulfovibrio gigas. Biochem. Biophys. Res. Commun. 193:100-105. [DOI] [PubMed] [Google Scholar]
- 12.Coelho, A. V., P. Matias, V. Fülop, A. Thomson, A. Gonzalez, and M. A. Carrondo. 1997. Desulfoferrodoxin structure determined by MAD phasing and refinement to 1.9-Å reveals a unique combination of a tetrahedral FeS4 centre with a square pyramidal FeSN4 centre. J. Inorg. Biochem. 2:680-689. [Google Scholar]
- 13.Coulter, E. D., N. V. Shenvi, and D. M. Kurtz, Jr. 1999. NADH peroxidase activity of rubrerythrin. Biochem. Biophys. Res. Commun. 255:317-323. [DOI] [PubMed] [Google Scholar]
- 14.Cypionka, H. 2000. Oxygen respiration by Desulfovibrio species. Annu. Rev. Microbiol. 54:827-848. [DOI] [PubMed] [Google Scholar]
- 15.Das, A., E. D. Coulter, D. M. Kurtz, Jr., and L. G. Ljungdahl. 2001. Five-gene cluster in Clostridium thermoaceticum consisting of two divergent operons encoding rubredoxin oxidoreductase-rubredoxin and rubrerythrin-type A flavoprotein-high-molecular-weight rubredoxin. J. Bacteriol. 183:1560-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das, A., R. Silaghi-Dumitrescu, L. G. Ljungdahl, and D. M. Kurtz, Jr. 2005. Cytochrome bd oxidase, oxidative stress, and dioxygen tolerance of the strictly anaerobic bacterium Moorella thermoacetica. J. Bacteriol. 187:2020-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Devreese, B., P. Tavares, J. Lampreia, N. Van Damme, J. Le Gal, J. J. Moura, J. Van Beeumen, and I. Moura. 1996. Primary structure of desulfoferrodoxin from Desulfovibrio desulfuricans ATCC 27774, a new class of non-heme iron proteins. FEBS Lett. 385:138-142. [DOI] [PubMed] [Google Scholar]
- 18.Flohe, L., and W. A. Gunzler. 1984. Assays of glutathione peroxidase. Methods Enzymol. 105:112-121. [DOI] [PubMed] [Google Scholar]
- 19.Gaber, A., M. Tamoi, T. Takeda, Y. Nakano, and S. Shigeoka. 2001. NADPH-dependent glutathione peroxidase-like proteins (Gpx-1, Gpx-2) reduce unsaturated fatty acid hydroperoxides in Synechocystis PCC 6803. FEBS Lett. 499:32-36. [DOI] [PubMed] [Google Scholar]
- 20.Gaudu, P., and B. Weiss. 2000. Flavodoxin mutants of Escherichia coli K-12. J. Bacteriol. 182:1788-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geissmann, T. A., M. Teuber, and L. Meile. 1999. Transcriptional analysis of the rubrerythrin and superoxide dismutase genes of Clostridium perfringens. J. Bacteriol. 181:7136-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomes, C. M., G. Silva, S. Oliveira, J. LeGall, M. Y. Liu, A. V. Xavier, C. Rodrigues-Pousada, and M. Teixeira. 1997. Studies on the redox centers of the terminal oxidase from Desulfovibrio gigas and evidence for its interaction with rubredoxin. J. Biol. Chem. 272:22502-22508. [DOI] [PubMed] [Google Scholar]
- 23.Grunden, A. M., F. E. Jenney, Jr., K. Ma, M. Ji, M. V. Weinberg, and M. W. Adams. 2005. In vitro reconstitution of an NADPH-dependent superoxide reduction pathway from Pyrococcus furiosus. Appl. Environ. Microbiol. 71:1522-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guedon, E., and H. Petitdemange. 2001. Identification of the gene encoding NADH-rubredoxin oxidoreductase in Clostridium acetobutylicum. Biochem. Biophys. Res. Commun. 285:496-502. [DOI] [PubMed] [Google Scholar]
- 25.Hofmann, B., H. J. Hecht, and L. Flohe. 2002. Peroxiredoxins. Biol. Chem. 383:347-364. [DOI] [PubMed] [Google Scholar]
- 26.Holdeman, L. V., E. P. Cato, and W. E. C. Moor. 1977. Anaerobe laboratory manual, 4th ed. Virginia Polytechnic Institute and State University, Blacksburg, Va.
- 27.Holland, K. T., J. S. Knapp, and J. G. Shoesmith. 1987. Anaerobes and oxygen, p. 4-12. In K. T. Holland, J. S. Knapp, and J. G. Shoesmith (ed.), Anaerobic bacteria. Chapman and Hall, New York, N.Y.
- 28.Jean, D., V. Briolat, and G. Reysset. 2004. Oxidative stress response in Clostridium perfringens. Microbiology 150:1649-1659. [DOI] [PubMed] [Google Scholar]
- 29.Jenney, F. E., Jr., M. F. Verhagen, X. Cui, and M. W. Adams. 1999. Anaerobic microbes: oxygen detoxification without superoxide dismutase. Science 286:306-309. [DOI] [PubMed] [Google Scholar]
- 30.Jeong, W., M. K. Cha, and I. H. Kim. 2000. Thioredoxin-dependent hydroperoxide peroxidase activity of bacterioferritin comigratory protein (BCP) as a new member of the thiol-specific antioxidant protein (TSA)/alkyl hydroperoxide peroxidase C (AhpC) family. J. Biol. Chem. 275:2924-2930. [DOI] [PubMed] [Google Scholar]
- 31.Jovanovic, T., C. Ascenso, K. R. Hazlett, R. Sikkink, C. Krebs, R. Litwiller, L. M. Benson, I. Moura, J. J. Moura, J. D. Radolf, B. H. Huynh, S. Naylor, and F. Rusnak. 2000. Neelaredoxin, an iron-binding protein from the syphilis spirochete, Treponema pallidum, is a superoxide reductase. J. Biol. Chem. 275:28439-28448. [DOI] [PubMed] [Google Scholar]
- 32.Karnholz, A., K. Kuse, A. Gossner, A. Schramm, and H. L. Drake. 2002. Tolerance and metabolic response of acetogenic bacteria toward oxygen. Appl. Environ. Microbiol. 68:1005-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawasaki, S., T. Nakagawa, Y. Nishiyama, Y. Benno, T. Uchimura, K. Komagata, K. Kozaki, and Y. Niimura. 1998. Effect of oxygen on the growth of Clostridium butyricum (type species of the genus Clostridium), and the distribution of enzymes for oxygen and for active oxygen species in clostridia. J. Ferment. Bioeng. 86:368-372. [Google Scholar]
- 34.Kawasaki, S., J. Ishikura, D. Chiba, T. Nishino, and Y. Niimura. 2004. Purification and characterization of an H2O-forming NADH oxidase from Clostridium aminovalericum: existence of an oxygen-detoxifying enzyme in an obligate anaerobic bacteria. Arch. Microbiol. 181:324-330. [DOI] [PubMed] [Google Scholar]
- 35.Kawasaki, S., J. Ishikura, Y. Watamura, and Y. Niimura. 2004. Identification of O2-induced peptides in an obligatory anaerobe, Clostridium acetobutylicum. FEBS Lett. 571:21-25. [DOI] [PubMed] [Google Scholar]
- 36.Kliebenstein, D. J., R. A. Monde, and R. L. Last. 1998. Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiol. 118:637-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knight, E., Jr., A. J. D'Eustachio, and R. W. Hardy. 1966. Flavodoxin: a flavoprotein with ferredoxin activity from Clostrium pasteurianum. Biochim. Biophys. Acta 113:626-628. [DOI] [PubMed] [Google Scholar]
- 38.Knight, E., Jr., and R. W. Hardy. 1966. Isolation and characteristics of flavodoxin from nitrogen-fixing Clostridium pasteurianum. J. Biol. Chem. 241:2752-2756. [PubMed] [Google Scholar]
- 39.Küsel, K., A. Karnholz, T. Trinkwalter, R. Devereux, G. Acker, and H. L. Drake. 2001. Physiological ecology of Clostridium glycolicum RD-1, an aerotolerant acetogen isolated from sea grass roots. Appl. Environ. Microbiol. 67:4734-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, P. T., A. Y. Hsu, H. T. Ha, and C. F. Clarke. 1997. A C-methyltransferase involved in both ubiquinone and menaquinone biosynthesis: isolation and identification of the Escherichia coli ubiE gene. J. Bacteriol. 179:1748-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lehmann, Y., L. Meile, and M. Teuber. 1996. Rubrerythrin from Clostridium perfringens: cloning of the gene, purification of the protein, and characterization of its superoxide dismutase function. J. Bacteriol. 178:7152-7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lemos, R. S., C. M. Gomes, M. Santana, J. LeGall, A. V. Xavier, and M. Teixeira. 2001. The ‘strict’ anaerobe Desulfovibrio gigas contains a membrane-bound oxygen-reducing respiratory chain. FEBS Lett. 496:40-43. [DOI] [PubMed] [Google Scholar]
- 43.Lim, J. H., Y. G. Yu, I. G. Choi, J. R. Ryu, B. Y. Ahn, S. H. Kim, and Y. S. Han. 1997. Cloning and expression of superoxide dismutase from Aquifex pyrophilus, a hyperthermophilic bacterium. FEBS Lett. 406:142-146. [DOI] [PubMed] [Google Scholar]
- 44.Liochev, S. I., and I. Fridovich. 1997. A mechanism for complementation of the sodA sodB defect in Escherichia coli by overproduction of the rbo gene product (desulfoferrodoxin) from Desulfoarculus baarsii. J. Biol. Chem. 272:25573-25575. [DOI] [PubMed] [Google Scholar]
- 45.Lombard, M., M. Fontecave, D. Touati, and V. Niviere. 2000. Reaction of the desulfoferrodoxin from Desulfoarculus baarsii with superoxide anion. Evidence for a superoxide reductase activity. J. Biol. Chem. 275:115-121. [DOI] [PubMed] [Google Scholar]
- 46.Lombard, M., D. Touati, M. Fontecave, and V. Niviere. 2000. Superoxide reductase as a unique defense system against superoxide stress in the microaerophile Treponema pallidum. J. Biol. Chem. 275:27021-27026. [DOI] [PubMed] [Google Scholar]
- 47.Lumppio, H. L., N. V. Shenvi, A. O. Summers, G. Voordouw, and D. M. Kurtz, Jr. 2001. Rubrerythrin and rubredoxin oxidoreductase in Desulfovibrio vulgaris: a novel oxidative stress protection system. J. Bacteriol. 183:101-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.May, A., F. Hillmann, O. Riebe, R. J. Fischer, and H. Bahl. 2004. A rubrerythrin-like oxidative stress protein of Clostridium acetobutylicum is encoded by a duplicated gene and identical to the heat shock protein Hsp21. FEMS Microbiol. Lett. 238:249-254. [DOI] [PubMed] [Google Scholar]
- 49.McCord, J. M., and I. Fridovich. 1969. Superoxide dismutase. An enzymatic function for erythrocuprein (hemocuprein). J. Biol. Chem. 244:6049-6055. [PubMed] [Google Scholar]
- 50.Mitchell, W. J. 2001. Biology and physiology, p. 49-104. In H. Bahl and P. Durre (ed.), Clostridia. Wiley-VCH, Weinheim, Germany.
- 51.Morris, J. G. 1976. Oxygen and the obligatory anaerobe. J. Appl. Bacteriol. 40:229-244. [DOI] [PubMed] [Google Scholar]
- 52.Moura, I., P. Tavares, J. J. Moura, N. Ravi, B. H. Huynh, M. Y. Liu, and J. LeGall. 1990. Purification and characterization of desulfoferrodoxin. A novel protein from Desulfovibrio desulfuricans (ATCC 27774) and from Desulfovibrio vulgaris (strain Hildenborough) that contains a distorted rubredoxin center and a mononuclear ferrous center. J. Biol. Chem. 265:21596-21602. [PubMed] [Google Scholar]
- 53.Niimura, Y., L. B. Poole, and V. Massey. 1995. Amphibacillus xylanus NADH oxidase and Salmonella typhimurium alkyl hydroperoxide reductase flavoprotein components show extremely high scavenging activity for both alkyl hydroperoxide and hydrogen peroxide in the presence of S. typhimurium alkyl hydroperoxide reductase 22-kDa protein component. J. Biol. Chem. 270:25645-25650. [DOI] [PubMed] [Google Scholar]
- 54.Nolling, J., G. Breton, M. V. Omelchenko, K. S. Makarova, Q. Zeng, R. Gibson, H. M. Lee, J. Dubois, D. Qiu, J. Hitti, Y. I. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucaille, M. J. Daly, G. N. Bennett, E. V. Koonin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O'Brien, R. W., and J. G. Morris. 1971. Oxygen and growth and metabolism of Clostridium acetobutylicum. J. Gen. Microbiol. 68:307-318. [DOI] [PubMed] [Google Scholar]
- 56.O'Halloran, T., and C. Walsh. 1987. Metalloregulatory DNA-binding protein encoded by the merR gene: isolation and characterization. Science 235:211-214. [DOI] [PubMed] [Google Scholar]
- 57.Outten, F. W., C. E. Outten, J. Hale, and T. V. O'Halloran. 2000. Transcriptional activation of an Escherichia coli copper efflux regulon by the chromosomal MerR homologue, cueR. J. Biol. Chem. 275:31024-31029. [DOI] [PubMed] [Google Scholar]
- 58.Ross, R. P., and A. Claiborne. 1992. Molecular cloning and analysis of the gene encoding the NADH oxidase from Streptococcus faecalis 10C1. Comparison with NADH peroxidase and the flavoprotein disulfide reductases. J. Mol. Biol. 227:658-671. [DOI] [PubMed] [Google Scholar]
- 59.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 60.Silaghi-Dumitrescu, R., K. Y. Ng, R. Viswanathan, and D. M. Kurtz, Jr. 2005. A flavo-diiron protein from Desulfovibrio vulgaris with oxidase and nitric oxide reductase activities. Evidence for an in vivo nitric oxide scavenging function. Biochemistry 44:3572-3579. [DOI] [PubMed] [Google Scholar]
- 61.Silaghi-Dumitrescu, R., D. M. Kurtz, Jr., L. G. Ljungdahl, and W. N. Lanzilotta. 2005. X-ray crystal structures of Moorella thermoacetica FprA. Novel diiron site structure and mechanistic insights into a scavenging nitric oxide reductase. Biochemistry 44:6492-6501. [DOI] [PubMed] [Google Scholar]
- 62.Sneath, P. H. A. 1986. Endospore-forming gram-positive rods and cocci, p.1104-1207. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology. Williams & Wilkins Co., Baltimore, Md.
- 63.Storz, G., L. A. Tartaglia, S. B. Farr, and B. N. Ames. 1990. Bacterial defenses against oxidative stress. Trends Genet. 6:363-368. [DOI] [PubMed] [Google Scholar]
- 64.Sztukowska, M., M. Bugno, J. Potempa, J. Travis, and D. M. Kurtz, Jr. 2002. Role of rubrerythrin in the oxidative stress response of Porphyromonas gingivalis. Mol. Microbiol. 44:479-488. [DOI] [PubMed] [Google Scholar]
- 65.Tsaneva, I. R., and B. Weiss. 1990. soxR, a locus governing a superoxide response regulon in Escherichia coli K-12. J. Bacteriol. 172:4197-4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weinberg, M. V., F. E. Jenney, Jr., X. Cui, and M. W. Adams. 2004. Rubrerythrin from the hyperthermophilic archaeon Pyrococcus furiosus is a rubredoxin-dependent, iron-containing peroxidase. J. Bacteriol. 186:7888-7895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woods, D. R., and D. T. Jones. 1986. Physiological responses of Bacteroides and Clostridium strains to environmental stress factors. Adv. Microb. Physiol. 28:1-64. [DOI] [PubMed] [Google Scholar]
- 68.Yeh, A. P., Y. Hu, F. E. Jenney, Jr., M. W. Adams, and D. C. Rees. 2000. Structures of the superoxide reductase from Pyrococcus furiosus in the oxidized and reduced states. Biochemistry 39:2499-2508. [DOI] [PubMed] [Google Scholar]