Abstract
The bacterial heat shock response is characterized by the elevated expression of a number of chaperone complexes and transcriptional regulators, including the DnaJ and the HrcA proteins. Genome analysis of Bifidobacterium breve UCC 2003 revealed a second copy of a dnaJ gene, named dnaJ2, which is flanked by the hrcA gene in a genetic constellation that appears to be unique to the actinobacteria. Phylogenetic analysis using 53 bacterial dnaJ sequences, including both dnaJ1 and dnaJ2 sequences, suggests that these genes have followed a different evolutionary development. Furthermore, the B. breve UCC 2003 dnaJ2 gene seems to be regulated in a manner that is different from that of the previously characterized dnaJ1 gene. The dnaJ2 gene, which was shown to be part of a 2.3-kb bicistronic operon with hrcA, was induced by osmotic shock but not significantly by heat stress. This induction pattern is unlike those of other characterized dnaJ genes and may be indicative of a unique stress adaptation strategy by this commensal microorganism.
Bifidobacteria have been shown to be the predominant species in the gastrointestinal tract (GIT) of infants and represent the third most numerous species encountered in the colon of adult humans. In the human GIT their presence has been associated with beneficial health effects (8, 13, 16, 21). Despite the generally accepted importance of bifidobacteria as probiotic components of human GIT microflora and their use in health-promoting foods, there is a paucity of information about their physiology, phylogenetic relationship, and underlying genetics (21).
We are interested in studying the stress response in bifidobacteria because probiotic bifidobacteria must resist adverse environmental conditions that are encountered not only during food preparation and storage but also in their natural environment (i.e., the GIT).
Recently, many genes induced upon exposure of bifidobacterial cultures to stressful conditions have been identified (15, 19, 20, 22-24). These include groELS, dnaK, clpB, clp, and clpP genes. The expression of the dnaK operon and clpB is induced by osmotic shock and severe heat stress but not by moderate heat stress (22, 24), suggesting an overlap between the osmotic shock and severe heat shock regulons. Interestingly, in bifidobacteria clpB and dnaK represent the first chaperone-encoding genes to be strongly induced after exposure to very high temperature (ΔT of 13 K). In contrast, maximal transcription of heat stress-induced genes such as groESL (20) and clpC (23) occurs upon moderate shock regimens (ΔT of 6 K). Notably, transcription of the latter two genes is not induced by osmotic stress (20, 23). Thus, at least two separate regulatory pathways for coping with different types and levels of stress are operating in bifidobacteria. The first pathway corresponds to the HspR regulon that protects cells from protein damage when bifidobacteria are exposed to severe heat and osmotic shocks, while a second pathway regulated by ClgR becomes active when bifidobacterial cells are exposed to moderate heat stress (23).
In Bacillus subtilis, in which the heat shock response has been investigated in great detail, the last gene of the dnaK operon was shown to encode the repressor of the dnaK and groEL operons. This genetic constellation also appears to be conserved in other Firmicutes. This gene is named hrcA (for heat regulation at CIRCE) and encodes a protein which binds to CIRCE (for controlling inverted repeat [IR] of chaperone expression)-bearing DNA fragments (28). In the main representative genus of the group actinobacteria, Streptomyces, neither hrcA nor CIRCE is associated with the dnaK operon, but in these bacteria the hrcA gene is instead associated with a second copy of the dnaJ gene, which encodes a molecular chaperone involved in assisting the posttranslation protein folding processes (7).
In this report we describe the genetic characterization of the Bifidobacterium breve UCC 2003 hrcA-dnaJ2 genes, which appear to represent a third pattern of controlled expression of chaperone-encoding genes in the genus Bifidobacterium.
Sequence analysis of B. breve UCC 2003 hrcA locus.
The B. breve UCC 2003 genome sequence (S. Leahy, J. A. Moreno Munoz, M. O'Connell-Motherway, D. Higgins, G. F. Fitzgerald, and D. van Sinderen, unpublished data) contains two different dnaJ-like genes. The dnaJ-like gene that is found downstream of the dnaK gene (22) shares the highest degree of similarity to dnaJ of B. subtilis and was therefore designated dnaJ1. The second dnaJ-like gene, designated here as dnaJ2, is located adjacent to a gene sharing a high degree of similarity to hrcA.
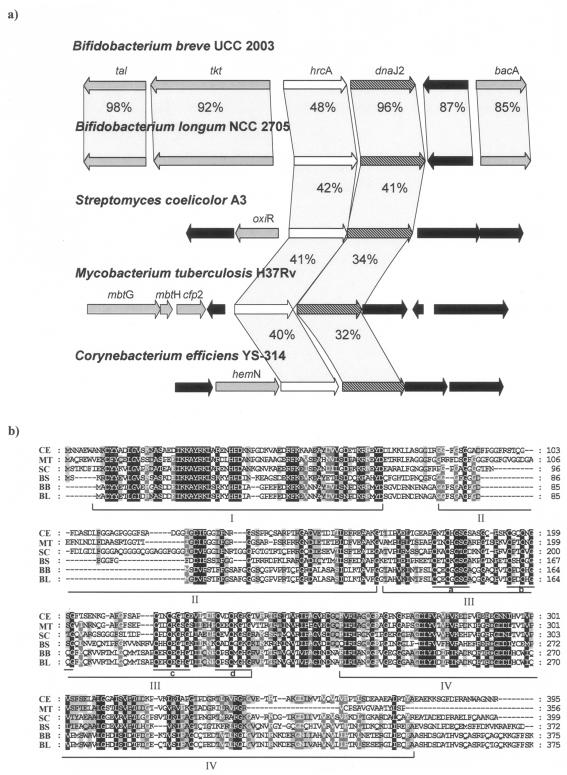
The B. breve UCC 2003 hrcA operon is preceded by the tal and tkt genes, which encode a putative transaldolase and a transketolase, respectively, and followed by a gene encoding a hypothetical protein and the bacA gene, which encodes a putative bacitracin resistance protein (Fig. 1a). Comparison of the B. breve UCC 2003 hrcA and dnaJ2 products with proteins deposited in the publicly available databases showed significant sequence similarity with the HrcA transcriptional repressor and DnaJ chaperones from other high-G+C gram-positive bacteria (e.g., Mycobacterium and Streptomyces).
FIG. 1.
(a) Comparison of the hrcA locus in B. breve UCC 2003 with corresponding loci in various other actinobacteria. Each arrow indicates an open reading frame. The length of arrows is proportional to the length of the predicted open reading frames. Corresponding genes are marked with the same color or pattern. The putative function of the proteins is indicated above each arrow (see text for explanation). Black arrows indicate hypothetical open reading frames. The amino acid identity with respect to B. breve UCC 2003 protein sequences is indicated as a percentage. (b) Alignment of the amino acid sequence for DnaJ2 proteins from gram-positive bacteria. CE, Corynebacterium efficiens YS-314; MT, Mycobacterium tuberculosis H37Rv; SC, Streptomyces coelicolor A3; BL, B. longum NCC 2705; BB, B. breve UCC 2003; BS, B. subtilis. The various conserved amino acid regions are indicated and explained in the text.
The B. breve UCC 2003 hrcA gene encodes a putative protein of 365 amino acids, with a predicted molecular mass of 38.6 kDa and a sequence 42% identical to that of Streptomyces albus G, which is the only actinobacterial hrcA gene characterized to date (7). The predicted B. breve UCC 2003 HrcA protein contains a C-terminal helix-turn-helix domain which has been described as being highly conserved among HrcA homologs.
Furthermore, the deduced B. breve HrcA sequence contained the functionally unassigned motif TIRNDMAALE, which is highly similar to the consensus TIRN(EDY)MA(DAQV)LE derived from HrcA proteins of Bacillus subtilis (27), Staphylococcus aureus (12), and Clostridium acetobutylicum (11).
The alignment of B. breve DnaJ2 sequence with those of other representative species allowed the identification of four distinct regions, which are typical of all members of the DnaJ family (Fig. 1b). The N-terminal 70 to 80 amino acids are referred to as the J domain, which is postulated to interact with the DnaK protein to stimulate its ATPase activity (26). The next region, which is rich in glycine and phenylalanine, is postulated to act as a flexible hinge necessary for the activation of the substrate binding properties of the DnaK protein (26). A third region found in the middle of the protein is characterized by four conserved repeats (marked a to d in Fig. 1b) containing the CxxCxGxG motif. This consensus is similar to the “Zink finger” motif and is believed to be involved in disulfide isomerase activity (4). The fourth domain comprises a region at the C-terminal end of the protein for which little structural or functional information is available (3).
Distribution of hrcA and dnaJ genes across bacterial genomes.
In order to assess the distribution of the hrcA-dnaJ operon across bacteria we surveyed 65 sets of available bacterial genomic data, including those for actinobacteria, Firmicutes, fusobacteria, chloroflexi, chlorobia, and cyanobacteria for the presence of the hrcA and dnaJ genes as well as their genomic locations (Table 1). The genomes of members of the group actinobacteria contained two copies of dnaJ genes, where dnaJ2 is always flanked by the hrcA gene, whereas dnaJ1 is located within the dnaK operon. Notably, only the Streptomyces avermitilis genome contains a third copy of the dnaJ gene (dnaJ3), which is placed within a second copy of the dnaK operon. In the available genomes of Firmicutes a single copy of the dnaJ gene is present, which is always located within the dnaK operon. Proteobacteria possess the same genetic constellation as Firmicutes except for certain genera (Ralstonia, Sinorhizobium, and Thermosynechoccus), in which the hrcA gene is not flanked by a DnaJ-encoding gene.
TABLE 1.
Distribution of hrcA and hrcA-dnaJ2 or hrcA-dnaK operon genetic constellation in various bacteria
| Group | Organism | Database no. | hrcA | hrcA-dnaJ2 | hrcA-dnaK operon |
|---|---|---|---|---|---|
| Actinobacteria | Bifidobacterium longum DJO10A | ZP_00120370.1 | + | + | − |
| Actinobacteria | Bifidobacterium longum NCC2705 | NP_695900.1 | + | + | − |
| Actinobacteria | Brevibacterium linens BL2 | ZP_00381522.1 | + | + | − |
| Actinobacteria | Corynebacterium diphtheriae | NP_940058.1 | + | + | − |
| Actinobacteria | Corynebacterium efficiens YS-314 | NP_738799.1 | + | + | − |
| Actinobacteria | Corynebacterium glutamicum ATCC 13032 | CAF20632.1 | + | + | − |
| Actinobacteria | Kineococcus radiotolerans SRS30216 | ZP_00198394.1 | + | + | − |
| Actinobacteria | Leifsonia xyli subsp. xyli strain CTCB07 | YP_062381.1 | + | + | − |
| Actinobacteria | Mycobacterium avium subsp. paratuberculosis strain k10 | Q73XZ5 | + | + | − |
| Actinobacteria | Mycobacterium tuberculosis H37Rv | NC_000962 | + | + | − |
| Actinobacteria | Mycobacterium tuberculosis CDC1551 | NP_216889.1 | + | + | − |
| Actinobacteria | Nocardia farcinica IFM 10152 | YP_117634.1 | + | + | − |
| Actinobacteria | Streptomyces avermitilis MA-4680 | NC_003155 | + | + | − |
| Actinobacteria | Streptomyces coelicolor A3(2) | CAB66232.1 | + | + | − |
| Actinobacteria | Propionibacterium acnes KPA171202 | YP_055626.1 | + | + | − |
| Actinobacteria | Thermobifida fusca | ZP_00293589.1 | + | + | − |
| Firmicutes | Acholeplasma laidlawii | AAM43820.1 | + | − | + |
| Firmicutes | Bacillus anthracis strain Sterne | YP_021184.1 | + | − | + |
| Firmicutes | Bacillus cereus ATCC 10987 | NP_980687.1 | + | − | + |
| Firmicutes | Bacillus clausii KSM-K16 | YP_175153.1 | + | − | + |
| Firmicutes | Bacillus licheniformis DSM 13 | YP_079885.1 | + | − | + |
| Firmicutes | Bacillus sphaericus | CAA76661.1 | + | − | + |
| Firmicutes | Bacillus subtilis | NP_390424.1 | + | − | + |
| Firmicutes | Clostridium tetani E88 | AAO36533.1 | + | − | + |
| Firmicutes | Clostridium thermocellum ATCC 27405 | ZP_00314236.1 | + | − | + |
| Firmicutes | Enterococcus faecalis V583 | NP_815028.1 | + | − | + |
| Firmicutes | Exiguobacterium sp. strain 255-15 | ZP_00182778.2 | + | − | + |
| Firmicutes | Geobacillus kaustophilus HTA426 | YP_148356.1 | + | − | + |
| Firmicutes | Lactobacillus acidophilus | YP_194110.1 | + | − | + |
| Firmicutes | Lactobacillus delbrueckii subsp. bulgaricus ATCC BAA-365 | ZP_00387602.1 | + | − | + |
| Firmicutes | Lactobacillus gasseri | ZP_00046571.1 | + | − | + |
| Firmicutes | Lactobacillus johnsonii NCC 533 | NP_965280.1 | + | − | + |
| Firmicutes | Lactobacillus plantarum WCFS1 | CAD64400.1 | + | − | + |
| Firmicutes | Lactobacillus sakei | CAA06939.1 | + | − | + |
| Firmicutes | Listeria monocytogenes | NP_464997.1 | + | − | + |
| Firmicutes | Moorella thermoacetica ATCC 39073 | ZP_00330048.1 | + | − | + |
| Firmicutes | Oceanobacillus iheyensis HTE831 | NP_692888.1 | + | − | + |
| Firmicutes | Pediococcus pentosaceus ATCC 25745 | ZP_00323327.1 | + | − | + |
| Firmicutes | Staphylococcus aureus subsp. aureus MRSA252 | YP_041051.1 | + | − | + |
| Firmicutes | Staphylococcus epidermidis ATCC 12228 | NP_764821.1 | + | − | + |
| Firmicutes | Streptococcus agalactiae 2603V/R | NP_687131.1 | + | − | + |
| Firmicutes | Streptococcus mutans UA159 | AAN57865.1 | + | − | + |
| Firmicutes | Streptococcus pneumoniae TIGR4 | ZP_00403006.1 | + | − | + |
| Firmicutes | Streptococcus pyogenes MGAS10394 | YP_060809.1 | + | − | + |
| Firmicutes | Streptococcus suis 89/1591 | ZP_00332240.1 | + | − | + |
| Firmicutes | Streptococcus thermophilus LMG 18311 | AAV61733.1 | + | − | + |
| Firmicutes | Tetragenococcus halophilus | Q93R26 | + | − | + |
| Firmicutes | Thermoanaerobacter tengcongensis MB4 | NP_622605.1 | + | − | + |
| Proteobacteria | Xanthomonas axonopodis pv. citri strain 306 | AAM36389.1 | + | − | + |
| Proteobacteria | Xanthomonas campestris pv. campestris strain ATCC 33913 | NP_636844.1 | + | − | + |
| Proteobacteria | Xanthomonas oryzae pv. oryzae KACC10331 | YP_200668.1 | + | − | + |
| Proteobacteria | Xylella fastidiosa Dixon | ZP_00039265.2 | + | − | + |
| Proteobacteria | Xylella fastidiosa Temecula1 | NP_779567.1 | + | − | + |
| Proteobacteria | Methylococcus capsulatus strain Bath | YP_114292.1 | + | − | + |
| Proteobacteria | Geobacter sulfurreducens PCA | NP_951093.1 | + | − | + |
| Proteobacteria | Anaeromyxobacter dehalogenans 2CP-C | ZP_00398344.1 | + | − | + |
| Proteobacteria | Sinorhizobium meliloti 1021 | CAC41570.1 | + | − | − |
| Proteobacteria | Ralstonia solanacearum GMI1000 | CAD16341.1 | + | − | − |
| Cyanobacteria | Thermosynechococcus elongatus BP-1 | NP_681550.1 | + | − | − |
| Cyanobacteria | Synechocystis. sp. strain PCC 6803 | NC_000911 | + | − | − |
| Spirochaetes | Leptospira interrogans | AAC35414.1 | + | − | + |
| Spirochaetes | Leptospira interrogans serovar Copenhageni strain Fiocruz L1-130 | YP_000507.1 | + | − | + |
| Fusobacteria | Fusobacterium nucleatum subsp. nucleatum ATCC 25586 | AAL94322.1 | + | − | + |
| Chloroflexi | Dehalococcoides ethenogenes 195 | YP_182107.1 | + | − | + |
| Chlorobia | Chlorobium tepidum TLS | NP_662369.1 | + | − | + |
Notably, other bacteria like the cyanobacterium Synechocystis sp. possess two DnaJ homologs, although neither of these is located in the vicinity of the hrcA gene.
The hrcA gene is uniformly present in all gram-positive bacteria studied to date (Table 1). However, the chromosomal location of the hrcA gene differs in gram-positive bacteria with low G+C content, where it is associated with the dnaK operon, in contrast to the hrcA position in high-G+C genomes, where is associated with dnaJ2. A hrcA homolog is also present in Synechocystis species and Leptospira interrogans (Table 1), both of which, according to 16S rRNA phylogeny (1), diverged at an early stage from the common ancestor of all gram-positive bacteria and Proteobacteria.
Hence, we can conclude that the hrcA-dnaJ2 genetic constellation is unique and typical of the members of the group actinobacteria.
Phylogenetic analysis based on DnaJ sequences.
The DnaJ sequence fulfills all prerequisites to be a suitable phylogenetic marker, such as very high genetic stability and a wide distribution. It has been applied to infer phylogeny in the genera Deinococcus-Thermus (2), Mycobacterium (10), and Legionella (9). In order to investigate the phylogeny of high-G+C and low-G+C gram-positive bacteria using DnaJ sequences, various DnaJ sequences were retrieved, including DnaJ1 and DnaJ2, from databases. A total of 53 sequences were thus obtained, including those from actinobacteria, Firmicutes, Proteobacteria, and cyanobacteria. Phylogeny calculations, including distance calculations and generation of phylogenetic trees, were performed using the PHYLIP package, employing default parameters (e.g., Dayhoff PAM matrix) and with the ProtDist module (5).
Moreover, since our main target was to study the phylogeny of bifidobacteria, a fragment of the dnaJ2 gene from 17 different bifidobacterial species was obtained and subsequently sequenced using a PCR-based strategy employing two primers, DNAJ2-uni (5′-CTCGGCCAGATGATGAC-3′) and DNAJ2-rev (5′-CTGAGCTT[C/G]GTGGGAATC-3′), based on two conserved regions (positions 541 to 558 and positions 1009 to 1027) from the B. breve UCC 2003 dnaJ2 gene (Fig. 1b). The reaction mixture (50 μl) contained 25 ng DNA template, which was derived from the protocol described in a previous study (17); 20 mM Tris-HCl; 50 mM KCl; 200 μM of each deoxynucleoside triphosphate; 50 pmol of each primer; 1.5 mM MgCl2; and 1 U Taq DNA polymerase (Gibco BRL, United Kingdom). Each PCR cycling profile consisted of an initial denaturation step of 5 min at 95°C, followed by amplification for 30 cycles as follows: denaturation (30 s at 95°C), annealing (30 s at 52°C), and extension (1 min at 72°C). The resulting amplicons were separated on a 1.5% agarose gel, followed by an ethidium bromide staining. PCR fragments were purified using the PCR purification spin kit (QIAGEN, West Sussex, United Kingdom) and were subsequently sequenced.
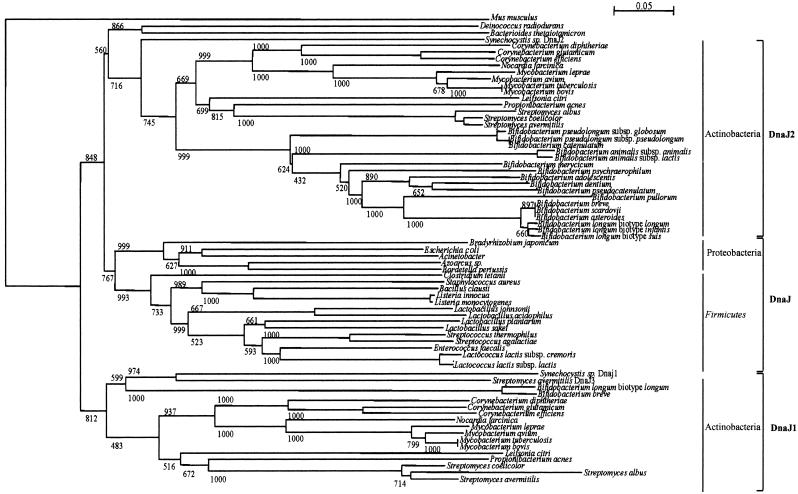
The phylogenetic tree based on DnaJ sequences shows low bootstrap scores and long branch lengths for only a very few nodes and species. In view of this, their usefulness for phylogenetic studies is limited. However, despite this caveat it should be noted that the two DnaJ homologs of actinobacteria did not form a monophyletic group in this tree, but instead the DnaJ2 proteins of actinobacteria branched together with the DnaJ sequences of Firmicutes and Proteobacteria, while the DnaJ1 sequences of actinobacteria formed a separate phylogenetic branch (Fig. 2).
FIG. 2.
Phylogenetic tree obtained using DnaJ homologs from various bacteria. The bar scale indicates phylogenetic distances. Bootstrap values are reported for a total of 1,000 replicates. The dnaJ1 and dnaJ2 gene sequences are indicated. The principal bacterial groups are indicated.
This suggests that the DnaJ of Firmicutes and Proteobacteria and DnaJ2 of actinobacteria originated from a common ancestor, which possessed two copies of DnaJ-encoding genes, and that a differential loss of gene copies did occur in Firmicutes-Proteobacteria and actinobacteria, resulting in the current dnaJ gene distribution. Of note, the close phylogenetic relationship between DnaJ of Firmicutes and DnaJ2 of actinobacteria is further illustrated by a high degree of synteny observed in the hrcA locus of these bacteria, in which the dnaJ2 genes of actinobacteria and the dnaJ genes of Firmicutes are located near the hrcA gene.
Finally, the overall similarity of DnaJ proteins roughly resembles that exhibited by HrcA sequences (1) and provides an alternative hierarchical order among gram-positive bacteria.
Conservation of dnaJ2 gene across Bifidobacterium genomes.
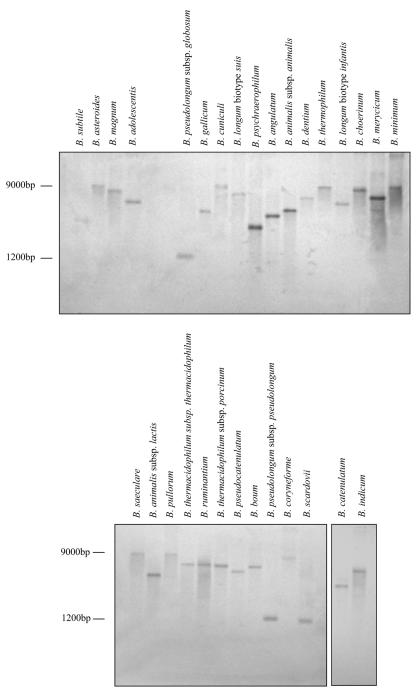
Due to the apparent high variability of dnaJ2 sequences within the genus Bifidobacterium we were able to identify this gene by PCR amplification in only 17 species of the 30 currently described species in the genus Bifidobacterium (21). Thus, in order to determine whether other bifidobacteria also contain dnaJ2 homologs, the amplified dnaJ2 DNA was hybridized to genomic DNA digested with EcoRI or HindIII of 30 different bifidobacterial species according to the method of Sambrook et al. (14). All investigated bifidobacteria yielded one single band of different sizes in all strains used (Fig. 3 and data not shown). This clearly suggests that a dnaJ2-like gene is present in most, if not all, bifidobacterial genomes.
FIG. 3.
Southern hybridization of EcoRI-digested genomic DNAs of Bifidobacterium species using the dnaJ2 gene fragment as probe. The sizes of two markers are indicated.
Kinetics of hrcA transcription following heat-osmotic stresses.
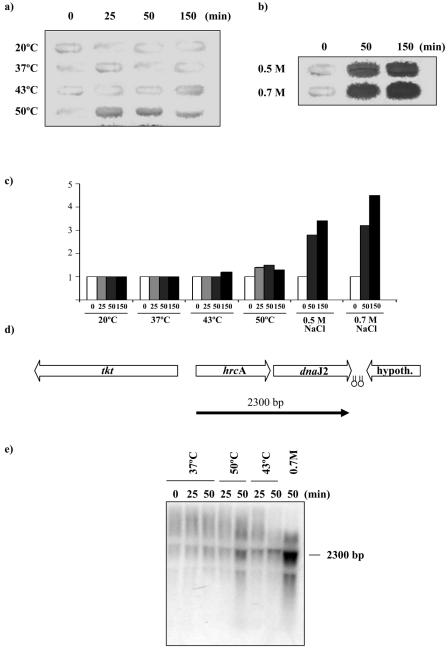
Expression of the hrcA locus in S. albus G (7) is induced by heat stress. To determine if the induction of the hrcA gene occurs upon exposure to stressful conditions in B. breve UCC 2003, slot blot hybridization was used to analyze total RNA isolated from B. breve cultures following exposure for up to 150 min to temperatures ranging from 20°C to 50°C and to NaCl concentrations of 0.5 M and 0.7 M (Fig. 4a and b). RNA isolation and slot blot hybridizations were carried out according to the method of Ventura et al. (19, 24).
FIG. 4.
Transcriptional regulation of the B. breve UCC 2003 hrcA operon. (a and b) Slot blot hybridization using RNA extracted from cells incubated up to 150 min in a range of temperatures or NaCl concentrations. (c) Quantitative representation of the induction levels of the hrcA mRNA transcripts. The numbers below each bar show the time in minutes for which each temperature or osmotic shock was applied. (d) Position of the transcripts with respect to the hrcA locus map. The estimated size of the mRNA is indicated in base pairs. Hairpins indicate possible rho-independent terminators. (e) Northern blot analysis of B. breve UCC 2003 hrcA locus performed using total mRNA isolated from cultures grown at 37°C, 43°C, or 50°C and under hyperosmotic conditions using an NaCl concentration of 0.7 M for the time indicated above each lane.
Based on the strength of the hybridization signal, the strongest expression of the hrcA gene occurred upon osmotic shock, whereas exposure to high temperatures (43°C or 50°C) or low temperature (20°C) did not appear to significantly increase the level of hrcA transcription (Fig. 4a and b). Densitometric analysis of Northern slot blots revealed that the levels of hrcA mRNA were increased at least 4.5-fold in cells that were subjected to osmotic stress for 150 min compared to unstressed cells (Fig. 4c). Furthermore, an amount of total RNA identical to that used in the induction experiment was slot blotted and hybridized using the 16S rRNA gene as a probe, in a control experiment according to the protocol described previously (23). The strength of the hybridization signal was of a similar intensity for all the slots, thus confirming the uniformity of RNA samples employed in the induction experiments (data not shown).
Transcription analysis of the hrcA locus.
Transcription of the hrcA B. breve locus was investigated by Northern blotting with two different probes encompassing hrcA and dnaJ2 genes, respectively. Total mRNA was isolated from B. breve UCC 2003 grown at 37°C, following heat shock at 43°C and 50°C or following osmotic shock at 0.7 M NaCl. Northern blot hybridizations were carried out according to the method of Ventura et al. (18). Northern blot analysis revealed a major hybridization signal corresponding to a 2.3-kb mRNA (Fig. 4d and e). This transcription pattern was also revealed for dnaJ2, where the size of the dnaJ2-hybridizing band was the same as that of the hrcA-hybridizing band, which suggests that the two genes are transcribed as a single bicistronic mRNA (data not shown). No hrcA mRNA was detected at 37°C, whereas hrcA transcripts accumulated after osmotic upshift, with maximum mRNA levels being observed about 150 min after the NaCl concentration had been increased to 0.7 M. Analysis of the nucleotide sequence of the hrcA locus revealed that the dnaJ2 gene was flanked at its 3′ end by two inverted repeats (ΔG of −25.6 and −12.2 kcal) that may function as rho-independent transcriptional terminator structures (Fig. 4d).
Determination of the transcription initiation site.
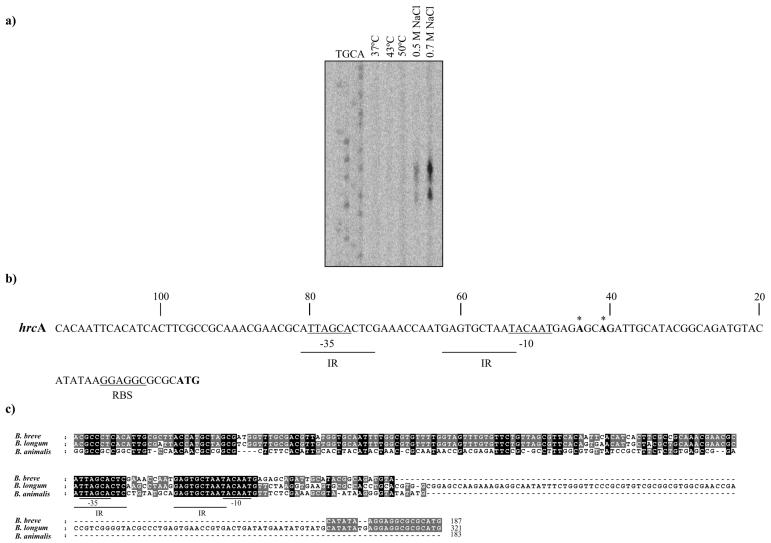
The transcription start site of the hrcA locus was determined by primer extension analysis using mRNA isolated from cultures grown at 37°C or heat shocked at 43°C or at 50°C for 150 min or following osmotic shock at 0.5 M and 0.7 M NaCl for 150 min. The 5′ end of the hrcA RNA transcript was determined following a protocol described in a previous study (18) and employing the synthetic oligonucleotide hrcA-prom (5′-CATTGCGAATGGTGGCAGAG-3′). Two transcription start sites were identified 41 bp and 44 bp upstream of the hrcA translation initiation codon using mRNA extracted from cultures subjected to osmotic shock (Fig. 5). Analysis of the putative promoter region of the hrcA reveals a potential sigma-70-promoter-like sequence resembling the consensus −10 and −35 hexamers. The DNA spacing between the −10 and −35 regions is exceptionally long, being 21 bp rather than the canonical 17 (25). The −35 hexamer of the hrcA promoter is located within a palindromic sequence (IR).
FIG. 5.
Primer extension analysis of the putative promoter sequences for the hrcA gene using an oligonucleotide targeting the 5′ ends of the hrcA gene (a). The experiment was performed using cells grown at either 37°C, 43°C, or 50°C or 0.5 M of NaCl or 0.7 M of NaCl. (b) Putative promoter sequences for the hrcA gene of B. breve UCC 2003. Underlined sequences denote the putative −10 and −35 hexamers; boldface type with an asterisk denotes the transcription start point, and boldface type without an asterisk indicates the start codon. IR indicates putative regulator sequences. RBS, ribosome binding site. (c) Comparison of the promoter sequences of three hrcA homologs from different bifidobacterial species. Shaded nucleotides indicate that the base is present in more than two of the inspected sequences.
The predicted promoter regions of hrcA genes from two bifidobacterial strains (B. breve UCC 2003 and Bifidobacterium longum NCC 2705) were aligned in an attempt to identify putative regulatory elements (Fig. 5c). For completeness, we determined by inverse PCR the putative promoter region of the hrcA gene from a more distantly related Bifidobacterium species (Bifidobacterium animalis subsp. animalis) following a protocol described previously (14, 23). The alignment of these promoter sequences revealed that the putative −10 region as well as the −35 box was highly conserved in these three bifidobacterial sequences. Moreover, the IR sequence was conserved among all three bifidobacteria examined (Fig. 5c). Interestingly, this inverted repeat resembled a regulatory structure termed CIRCE (TTAGCACTC N9 GAGTGCTAA) (28), which has been demonstrated to be the operator sequence of the heat shock protein regulator (HrcA) in both low- and high-G+C gram-positive bacteria (6, 28). A preliminary screening of the B. longum NCC 2705 and B. breve UCC 2003 genome sequence for the consensus sequences (ATTAGCACTC N9 GAGTGCTAAT) revealed the presence of this IR in the promoter region of a classical stress-induced gene such as groES (21).
Conclusions.
We identified and characterized a second dnaJ homolog which is organized with the hrcA gene in an apparently bicistronic operon in the B. breve UCC 2003 genome. Phylogenetic analysis using DnaJ proteins from low- and high-G+C gram-positive bacteria revealed a close and specific relationship between the actinobacterial dnaJ2 gene and the dnaJ gene of Firmicutes and Proteobacteria. This may indicate that the dnaJ1 gene of actinobacteria has evolved from an ancient form which existed before the evolutionary split of actinobacteria from Firmicutes or that a differential loss of dnaJ gene copies occurred during the evolution of actinobacteria and Firmicutes.
The presence of two dnaJ genes may be an indication that the encoded proteins have different physiological functions either alone or in association with DnaK, a theory that is consistent with the differential expression patterns of the two dnaJ genes. The presence of the two bifidobacterial dnaJ genes where both are activated under osmotic stress but only one (dnaJ1) is activated by high temperature may be a reflection of the fact that these bacteria normally grow in habitats like the GIT of animals where no significant temperature changes occur, whereas the osmotic conditions of the GIT are greatly variable due to the diet composition. This is corroborated by the fact that the number of chaperone-encoding genes involved in heat shock protection in actinobacteria that live in isothermal niches (e.g., the GIT of mammals) is smaller than that found in actinobacteria which are found in environments exposed to higher thermal fluctuations (e.g., soil or plant).
HrcA represents the third identified bifidobacterial transcriptional regulator, which would tune the expression of stress-induced genes. In fact, bifidobacterial genes (e.g., clpB and dnaK genes) which are highly induced upon exposure to severe heat shocks or osmotic stresses have been shown to be regulated by the transcriptional repressor protein HspR (24). Bifidobacterial genes (e.g., clpP and clpC genes) which are strongly induced upon moderate heat treatments were shown to be regulated by the transcriptional activator ClgR in conjunction with a so-far unidentified cofactor (23; unpublished data). The expression pattern of hrcA appears to be unique. In fact, bifidobacterial hrcA may represent an exception to the rule since, unlike other organisms, the hrcA and dnaJ2 genes are significantly induced upon osmotic stress rather than heat stress. Although the expression of the B. breve hrcA gene was shown to be osmoregulated, it will remain to be determined whether its activity may still be thermoregulated. Thus, additional work will be necessary in order to verify this hypothesis and to exploit the CIRCE regulon.
Nucleotide sequence accession numbers.
The nucleotide sequence data regarding the hrcA locus of B. breve UCC 2003 have been deposited in GenBank under accession number DQ144724. Furthermore, the GenBank accession numbers for dnaJ2 gene sequences generated in this study are as follows: B. animalis subsp. lactis DSM 10140, DQ144709; B. animalis subsp. animalis ATCC 25527, DQ144713; B. suis LMG 10738, DQ144710; B. psychraerophilum LMG 21775, DQ144711; B. pseudolongum subsp. pseudolongum LMG 11595, DQ144712; B. dentium LMG 11585, DQ144714; B. adolescentis LMG 11579, DQ144715; B. catenulatum LMG 11043, DQ144716; B. infantis LMG 8811, DQ144717; B. pseudolongum subsp. globosum LMG 11614, DQ144718; B. merycicum LMG 11341, DQ144719; B. scardovii LMG 11571, DQ144720; B. pseudocatenulatum LMG 11593, DQ144721; B. asteroides LMG 10735, DQ144722; B. pullorum LMG 21816, DQ144723.
Acknowledgments
This work was financially supported by Enterprise Ireland (grant BR/1998/202), by the Higher Education Authority Programme for Research in Third Level Institutions, by the Science Foundation Ireland Alimentary Pharmabiotic Centre located at University College Cork, and by the Marie Curie Development Host Fellowship (HPMD-2000-00027) to M.V.
REFERENCES
- 1.Ahmad, S., A. Selvapandiyan, and R. K. Bhatnagar. 1999. A protein-based phylogenetic tree for Gram-positive bacteria derived from hrcA, a unique heat-shock regulatory gene. Int. J. Syst. Bacteriol. 49:1387-1394. [DOI] [PubMed] [Google Scholar]
- 2.Bustard, K., and R. S. Gupta. 1997. The sequences of heat shock protein 40 (DnaJ) homologs provide evidence for close evolutionary relationship between the Deinococcus-Thermus group and cyanobacteria. J. Mol. Evol. 45:193-205. [DOI] [PubMed] [Google Scholar]
- 3.Caplan, A. J., D. M. Cyr, and M. G. Douglas. 1993. Eukaryotic homologs of Escherichia coli DnaJ: a diverse protein family that functions with Hsp70 stress proteins. Mol. Biol. Cell 4:555-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Crouy-Chanel, A., M. Kohiyama, and G. Richarme. 1995. A novel function of Escherichia coli chaperone DnaJ. J. Biol. Chem. 270:22669-22672. [DOI] [PubMed] [Google Scholar]
- 5.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 6.Georgopoulos, C., K. Liberek, M. Zylicz, and D. Ang. 1994. Properties of the heat shock proteins of Escherichia coli and the autoregulation of the heat shock response, p. 209-249. In R. I. Morimoto, A. Tissieres, and C. Georgopoulos (ed.), The biology of heat shock proteins and molecular chaperones. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 7.Grandvalet, C., G. Rapoport, and P. Mazodier. 1998. hrcA, encoding the repressor of the groEL genes in Streptomyces albus G, is associated with a second dnaJ gene. J. Bacteriol. 180:5129-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lievin, V., I. Pfeiffer, S. Hudault, F. Rochat, D. Brassart, J. R. Neeser, and A. L. Servin. 2000. Bifidobacterium strains from resident infant human gastrointestinal microflora exert antimicrobial activity. Gut 47:646-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu, H., X. Huang, Y. Kawamura, and T. Ezaki. 2003. Use of the dnaJ gene for the detection and identification of all Legionella pneumophila serogroups and description of the primers used to detect 16S rDNA gene sequences of major members of the genus Legionella. Microbiol. Immunol. 47:859-869. [DOI] [PubMed] [Google Scholar]
- 10.Morita, Y., S. Maruyama, H. Kabeya, A. Nagai, K. Kozawa, M. Kato, T. Nakajima, T. Mikami, Y. Katsube, and H. Kimura. 2004. Genetic diversity of the dnaJ gene in the Mycobacterium avium complex. J. Med. Microbiol. 53:813-817. [DOI] [PubMed] [Google Scholar]
- 11.Narberhaus, F., K. Giebeler, and H. Bahl. 1992. Molecular characterization of the dnaK gene region of Clostridium acetobutylicum, including grpE, dnaJ, and a new heat shock gene. J. Bacteriol. 174:3290-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohta, T., K. Saito, M. Kuroda, K. Honda, H. Hirata, and H. Hayashi. 1994. Molecular cloning of two new heat shock genes related to the Hsp70 genes in Staphylococcus aureus. J. Bacteriol. 176:4779-4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ouwehand, A. C., S. Salminen, and E. Isolauri. 2002. Probiotics: an overview of beneficial effects. Antonie Leeuwenhoek 82:279-289. [PubMed] [Google Scholar]
- 14.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Schmidt, G., and R. Zink. 2000. Basic features of stress response in three species of bifidobacteria: B. longum, B. adolescentis, and B. breve. Int. J. Food Microbiol. 55:41-45. [DOI] [PubMed] [Google Scholar]
- 16.Tannock, G. W. 1994. The acquisition of the normal microflora of the gastrointestinal tract, p. 1-16. In S. A. Gibson (ed.), Human health: the contribution of microorganisms. Springer, London, United Kingdom.
- 17.Ventura, M., R. Reniero, and R. Zink. 2001. Specific identification and targeted characterization of Bifidobacterium lactis from different environmental isolates by a combined multiplex-PCR approach. Appl. Environ. Microbiol. 67:2760-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ventura, M., C. Canchaya, V. Meylan, T. R. Klaenhammer, and R. Zink. 2003. Analysis, characterization, and loci of the tuf genes in Lactobacillus and Bifidobacterium and their direct application for species identification. Appl. Environ. Microbiol. 69:6908-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ventura, M., C. Canchaya, D. van Sinderen, G. F. Fitzgerald, and R. Zink. 2004. Bifidobacterium lactis DSM 10140: identification of the atp (atpBEFHAGDC) operon and analysis of its genetic structure, characteristics, and phylogeny. Appl. Environ. Microbiol. 70:3110-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ventura, M., C. Canchaya, R. Zink. G. F. Fitzgerald, and D. van Sinderen. 2004. Characterization of the groEL and groES loci in Bifidobacterium breve UCC 2003: genetic, transcriptional, and phylogenetic analysis. Appl. Environ. Microbiol. 70:6197-6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ventura, M., D. van Sinderen, G. F. Fitzgerald, and R. Zink. 2004. Insights into the taxonomy, genetics and physiology of bifidobacteria. Antonie Leeuwenhoek 86:205-223. [DOI] [PubMed] [Google Scholar]
- 22.Ventura, M., R. Zink. G. F. Fitzgerald, and D. van Sinderen. 2005. Gene structure and transcriptional organization of the dnaK operon of Bifidobacterium breve UCC 2003 and its application in bifidobacterial tracing. Appl. Environ. Microbiol. 71:487-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ventura, M., G. F. Fitzgerald, and D. van Sinderen. 2005. Genetic and transcriptional organization of the clpC locus in Bifidobacterium breve UCC 2003. Appl. Environ. Microbiol. 71:6282-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ventura, M., J. G. Kenny, Z. Zhang, G. F. Fitzgerald, and D. van Sinderen. 2005. The clpB gene of Bifidobacterium breve UCC 2003: transcriptional analysis and first insights into stress induction. Microbiology 151:2861-2872. [DOI] [PubMed] [Google Scholar]
- 25.Wagner, R. 2000. Transcription regulation in prokaryotes, p. 16-22. Oxford University Press, London, United Kingdom.
- 26.Wall, D., M. Zylic, and C. Georgopoulos. 1994. The NH2-terminal 108 amino acids of the Escherichia coli DnaJ protein stimulate the ATPase activity of DnaK and are sufficient for lambda replication. J. Biol. Chem. 269:5446-5451. [PubMed] [Google Scholar]
- 27.Wetzstein, M., U. Volker, J. Dedio, S. Lobau, S. Zuber, M. Schiesswohl, C. Herget, M. Hecker, and W. Schumann. 1992. Cloning, sequencing, and molecular analysis of the dnaK locus from Bacillus subtilis. J. Bacteriol. 174:3300-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuber, U., and W. Schumann. 1994. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. J. Bacteriol. 176:1353-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]