Abstract
We characterized the copper resistance genes in strain XvP26 of Xanthomonas campestris pv. vesicatoria, which was originally isolated from a pepper plant in Taiwan. The copper resistance genes were localized to a 7,652-bp region which, based on pulsed-field gel electrophoresis and Southern hybridization, was determined to be located on the chromosome. These genes hybridized only weakly, as determined by Southern analysis, to other copper resistance genes in Xanthomonas and Pseudomonas strains. We identified five open reading frames (ORFs) whose products exhibited high levels of amino acid sequence identity to the products of previously reported copper genes. Mutations in ORF1, ORF3, and ORF4 removed copper resistance, whereas mutations in ORF5 resulted in an intermediate copper resistance phenotype and insertions in ORF2 had no effect on resistance conferred to a copper-sensitive recipient in transconjugant tests. Based on sequence analysis, ORF1 was determined to have high levels of identity with the CopR (66%) and PcoR (63%) genes in Pseudomonas syringae pv. tomato and Escherichia coli, respectively. ORF2 and ORF5 had high levels of identity with the PcoS gene in E. coli and the gene encoding a putative copper-containing oxidoreductase signal peptide protein in Sinorhizobium meliloti, respectively. ORF3 and ORF4 exhibited 23% identity to the gene encoding a cation efflux system membrane protein, CzcC, and 62% identity to the gene encoding a putative copper-containing oxidoreductase protein, respectively. The latter two ORFs were determined to be induced following exposure to low concentrations of copper, while addition of Co, Cd, or Zn resulted in no significant induction. PCR analysis of 51 pepper and 34 tomato copper-resistant X. campestris pv. vesicatoria strains collected from several regions in Taiwan between 1987 and 2000 and nine copper-resistant strains from the United States and South America showed that successful amplification of DNA was obtained only for strain XvP26. The organization of this set of copper resistance genes appears to be uncommon, and the set appears to occur rarely in X. campestris pv. vesicatoria.
Copper compounds have been used for several decades for the control of bacterial and fungal plant pathogens; however, copper-resistant bacterial pathogens have become prevalent (1, 7, 8, 17, 26, 29, 43) and have reduced the efficacy of copper-based bactericides. Most of the genes associated with copper resistance from plant-pathogenic bacteria (7, 9, 15, 16, 41, 46) and some other bacteria, including Escherichia coli (44) and Mycobacterium scrofulaceum (20), are plasmid encoded. Chromosomal genes for copper resistance have been cloned from the plant pathogen Xanthomonas arboricola pv. juglandis (26).
Copper resistance genes in Xanthomonas campestris pv. vesicatoria were located on 188- to 200-kb self-transmissible plasmids in strains from Florida and Oklahoma (9, 41) and on a 100-kb non-self-transmissible plasmid in a strain from California(17). Although these copper resistance genes were subcloned into a cosmid, they have not been studied further. The copper resistance genes from X. arboricola pv. juglandis have the same general copABCD structure as the genes from Pseudomonas syringae (30, 31), but there are some differences in gene size and DNA sequence. The copRS regulatory genes, which are present in P. syringae pv. tomato (30), have not been found in Xanthomonas (26, 47), although copper induction has been demonstrated in X. campestris pv. vesicatoria (3).
Chromosome-encoded copper resistance was discovered in astrain of X. campestris pv. vesicatoria (strain XvP26) from Taiwan that contained only one small plasmid (15 kb) (6, 13). Since copper resistance was associated with large plasmids in this pathogen previously (25, 41), we explored the possibility that the copper resistance in this strain was similar to that associated with the chromosome of X. arboricola pv. juglandis. Furthermore, since a probe containing copper resistance genes previously isolated from a plasmid in X. campestris pv. vesicatoria did not hybridize to total genomic DNA of XvP26 under low-stringency conditions, we investigated the possibility that there is a different set of genes for resistance. The objective of this work was to compare the copper resistance genes from X.campestris pv. vesicatoria strain XvP26 with copper resistance genes from other plant-pathogenic bacteria. In addition, below we present information on the presence of putative regulatory genes associated with the copper resistance genes and evidence for copper-inducible expression of two of the genes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study and their sources are listed in Table 1. Nutrient broth cultures (BD Difco, Becton, Dickinson and Company, Sparks, MD) were grown for 24 h on a rotary shaker (150 rpm) at 28°C. Strains of E. coli were cultivated on Luria-Bertani (LB) medium at 37°C (32). A pLAFR3 cosmid library of XvP26 was maintained in E. coli DH5α on LB medium containing tetracycline. All other strains were stored in sterile tap water at room temperature or in 30% glycerol at −70°C or both. Triparental matings (19, 21) were performed on nutrient-yeast extract-glycerol agar (45). Antimicrobial agents were added to media to maintain selection for resistance markers at the following final concentrations: tetracycline, 12.5 μg/ml; kanamycin, 25 μg/ml; spectinomycin, 50 μg/ml; ampicillin, 100μg/ml; nalidixic acid, 50 μg/ml; and rifamycin SV, 75 μg/ml.
TABLE 1.
Bacterial strains and plasmids used for molecular transformation and conjugation
| Strain(s) or plasmid(s) | Relevant characteristics | Source or referencea |
|---|---|---|
| Xanthomonas campestris pv. vesicatoria strains | ||
| XvP26 | Cur Rifr | RES |
| 82-8 | Cus Rifr | RES |
| 91-118 | Cus Rifr | RES |
| ME5, 12, 16, 23, and 25 | XvP26 marker exchange mutants for ORF1, ORF2, ORF3, ORF4, and ORF5 | This studyb |
| Escherichia coli strains | ||
| DH5α | F− φ80d lacZΔM15 recA1 | Bethesda Research Labs |
| C2110 | NalrpolA | 10 |
| Plasmids | ||
| pLAFR3 | Tetrrlx+ RK2 replicon | 42 |
| pCOP35 | Cur clone from P syringae pv. tomato | DAC |
| pXjCu99 | Cur clone from X. arboricola pv. juglandis | MNS |
| pRK2073 | ColEI replicon, Tra+ Mob+ Spr, helper plasmid | 45 |
| pXvCu | Cur clone from X. campestris pv. vesicatoria 75-3 | RES |
| pXv26Cu | 27.9-kb Cur cosmid clone | This study |
| pXv26Cu-1 | 10.9-kb Cur EcoRI/HindIII subclone | This study |
| pXv26Cu-2 | 7.6-kb Cur EcoRI/XbaI subclone | This studyb |
| pORF1 (BOX+25) and pORF1 (BOX+90) | ORF1 pLAFR6 subclones | This studyc |
| pBCuSX | 6.2-kb SmaI/XbaI pBluescript subclone | This studyd |
| pL3uidA | pLAFR3 constitutive lacZ-uidA fusion | B. Staskawicz |
| pLAFR6 | pLAFR1 with trp terminators | B. Staskawicz |
| pL6uidA | pLAFR6-uidA negative control | B. Staskawicz |
| pUFR051 | pLAFR3 with pUC19 polylinker | 18 |
| pBluescript II KS(+/−) | Phagemid sequencing vector, Apr | Stratagene |
| pBSGUSK and pBSGUSX | pBluescript uidA(EcoRI) cassette vectors | This study |
| pORF1uidA, pORF3uidA, pORF4uidA, and pΔKXORF3uidA | GUS reporter constructs in pLAFR6 | This studye |
| pHoKmGUS | Kmr Apr Tn3-uidA fusion | B. Staskawicz |
| pSShe | CmrtnpA | 40 |
Copper resistance screening of Xanthomonas strains was performed either by streaking a loopful from a fresh tap water suspension of bacteria onto plates of nutrient agar containing copper sulfate at concentrations up to 200 μg/ml or by incubation in CYE broth (3, 48) containing various amounts of copper and subsequent streaking onto plates containing nutrient agar without copper.
General DNA manipulations.
Miniscale preparations of E. coli plasmid DNA were obtained by alkaline lysis as described by Sambrook et al. (38). Previously cloned DNA fragments containing copper resistance genes from different plant-pathogenic bacteria were isolated by digestion with appropriate restriction enzymes by using the conditions specified by the manufacturer (Promega Corporation, Madison, WI) for use in Southern hybridization. Restricted DNAs were separated in 0.7% agarose gels (Seakem GTG; FMC Bioproducts, Rockland, ME) in Tris-acetate-EDTA buffer by electrophoresis at 5 V cm−1. The gels were stained with ethidium bromide (10 μg/ml) for 30 min and then photographed over a UV transilluminator with type 55 Polaroid film.
Isolation of copper resistance clones.
A pLAFR3 cosmid (42) library of DNA from strain XvP26 was created and maintained in E. coli DH5α. Triparental matings were carried out by mixing mid-exponential-phase cells of X. campestris pv. vesicatoria strain 82-8 as the recipient with cosmid donors and with HB101(pRK2073) as the conjugational helper on nutrient-yeast extract-glycerol agar. The ratio of recipient to donor to helper was 2:1:1 (vol/vol/vol). After 24 h of incubation at 28°C, the mating mixtures were resuspended in sterile tap water, and aliquots were spread onto nutrient agar plates containing rifamycin, tetracycline, and copper sulfate (20 μg/ml) for selection of transconjugants and induction of copper resistance. Transconjugant colonies were subsequently transferred onto nutrient agar amended with copper sulfate (200 μg/ml) to detect clones carrying copper resistance genes.
Mapping of restriction endonuclease sites and subcloning of the DNA insert from a cosmid carrying the copper resistance gene cluster were performed by restriction digestion of the original clone with various enzymes and purification of fragments from an agarose gel by using the Wizard PCR Preps DNA purification system (Promega). Ligation of fragments into pUFR051 (18) was performed with T4 DNA ligase used according to the manufacturer's instructions. Ligation products were transformed into competent cells of E. coli DH5α produced by the calcium chloride procedure as described by Sambrook et al. (38).
Insertion mutagenesis was performed using Tn3-uidA as previously described (11). Individual insertion derivatives were analyzed by restriction enzyme digestion and mobilized into X. campestris pv. vesicatoria strain 91-118 for copper sensitivity testing. Selected insertion derivatives were used to generate marker exchange gene replacement mutants in the wild-type strain X. campestris pv. vesicatoria XvP26 as described by Bonas et al. (10) for use in complementation experiments.
In addition, pLAFR6 clones containing open reading frame 1 (ORF1) were constructed for use in complementation assays. PCR products generated using custom oligonucleotide primers (Fig. 1) BOX+25 (5′-CACCCTGTCACTCAGATGTTCC-3′), BOX+90 (5′-GAAGTGACCATGGTTTCCTCTGC-3′), and ORF1STOP (5′-GTCGCTGCACCTTTATTCCTCC-3′) were first cloned into the pGEM-T Easy vector (Promega) for sequencing and subsequently excised from the vector using EcoRI for subcloning into pLAFR6.
FIG. 1.
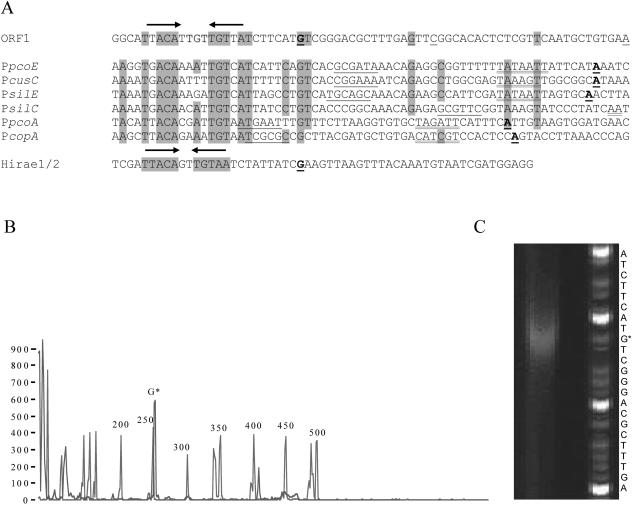
Nucleotide sequence of ORF1 (boldface type) and the upstream region, showing the locations of the copper box homology (bases enclosed in boxes) and the divergent translational start of ORF3 [(←CAT)]. Oligonucleotide primers used for subcloning and primer extension are underlined. Bases added to primers for generation of new restriction sites are indicated by lowercase superscript letters. The position of the Tn3 insertion in ORF1 marker exchange mutant ME5 is indicated by an inverted solid triangle.
DNA sequencing.
For sequence analysis, DNA fragments were cloned into the phagemid vector pBluescript II/KS (Stratagene, La Jolla, CA) using appropriate enzymes, and sequencing was initiated using the standard flanking vector T3 and T7 primers. A primer-walking strategy with custom-designed oligonucleotides was used to complete the sequencing. DNA sequencing was performed by the DNA Sequencing Core Laboratory of the Interdisciplinary Center for Biotechnology Research (University of Florida, Gainesville). The exact location of Tn3-uidA insertions was determined by sequencing using an oligonucleotide complementary to a sequence in the N terminus of the β-glucuronidase (GUS) gene (5′-GATTTCACGGGTTGGGGTTTCTACAGG-3′). DNA sequence analysis was performed with the Sequaid II program (Kansas State University, Manhattan). Database searches were performed using TBLAST (2).
β-Glucuronidase reporter assays.
β-Glucuronidase activities expressed in several X. campestris pv. vesicatoria strain backgrounds were measured by using CYE broth cultures containing 5 μM CuSO4 · 5H2O, 3CdSO4 · 8H2O, CoCl2 · 6H2O, or ZnSO4 · 7H2O to test the inducibility of ORF1, ORF3, and ORF4. In-frame fusions of uidA with ORF3 and ORF4 were constructed by excision of the uidA gene from pBSGUS clones and insertion into the pBluescript subclone pBCuSX using available restriction enzyme sites. Correct fusions were confirmed by sequencing before the fused genes were cut from the clones and moved into the vector pLAFR6 for conjugation into X. campestris pv. vesicatoria for GUS assay experiments. An ORF1-uidA fusion was obtained by cloning a 665-bp KpnI/EcoRI ORF1 PCR fragment upstream of the uidA gene in pLAFR6. Custom primers (Fig. 1) ORF1KPN (5′-CCAATCTGGCATGAGTGGTACCCAATCCC-3′) and ORF1ECO (5′-CCGAATTCATGACGTCCAGGATCACCAGGTC-3′) were used to obtain ORF1 DNA.
To assess the possible role of ORF1 and ORF2 in the regulation of expression of ORF3, a derivative reporter clone of pORF3uidA (pΔKXORF3uidA) was constructed by deletion of the 3.5-kb KpnI/XbaI fragment containing ORF1 and ORF2 and subsequent replacement of the 480-bp ORF3 upstream region with a KpnI/XbaI fragment generated by PCR using custom primers (Fig. 1) ORF1KPN and ORF1XBA (5′-CCTCTAGAGTGTGCCGAACTCAAAGCGTCC-3′).
Cells from 18-h plate cultures of transconjugants grown on CYE media containing tetracycline were suspended in fresh CYE broth, and the concentrations were standardized to an optical density at 600 nm of 0.3 for induction assays. One-milliliter portions of suspensions with or without copper were incubated at 28°C for 3 h to allow induction. The cells were then pelleted from the suspensions by centrifugation and washed once in sterile deionized water to remove the residual copper. The cell pellets were resuspended in 250 μl of MUG extraction buffer (24) and incubated at 37°C. The reactions in aliquots (25 μl) were terminated at 30-min intervals by addition of 225 μl of 0.2 M Na2CO3. Fluorescence was measured using a Cytofluor II fluorescence multiwell plate reader (PerSeptive Biosystems, Framingham, MA) calibrated with known concentrations of methyl-umbelliferone (MU) (Sigma Chemical Company) in MicroFluor B flat-bottom microtiter plates (Dynatech Laboratories, Inc., Chantilly, VA). The results were expressed as the number of units (nanomoles of MU produced per minute) normalized for CFU of bacteria in the reactions. Transconjugants carrying pL6uidA and transconjugants carrying pL3uidA were included in assays as controls for background and constitutive expression, respectively.
Primer extension analysis.
The concentration of log-phase bacteria (strain XvP26) was standardized to an optical density at 600 nm of 0.3 in fresh CYE broth with or without copper sulfate (10 μM), and the bacteria were incubated at 28°C for 1 h. Total RNA was extracted using an RNeasy Protect bacterial mini kit and an RNase-Free DNase set (QIAGEN, Valencia, CA) according to the manufacturer's instructions. Primer extension and GeneScan analysis were performed by previously described protocols (28), with some modifications, using oligonucleotides labeled at the 5′ end with 6-carboxyfluorescein. Custom oligonucleotides PE1 (5′-GACTGCAAGACCTGCCAGCCATTCAACC-3′) and PE2 (5′-GTCAGGAACAGGACGGGCATCTCAAGG-3′) (Fig. 1) were heated at 65°C for 90 min in 1× First-Strand buffer containing 10 μg of total RNA and allowed to cool at room temperature for 5 min before they were quenched on ice for 5 min. Extension was performed at 55°C for 1 h using SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA), and the preparations were heated to 70°C for 15 min before RNase A treatment. To determine the 5′ end of mRNA, the same oligonucleotides (unlabeled) were used as primers in sequencing reactions.
PCR and Southern hybridization analysis of strains.
Custom oligonucleotide primers JB8 (5′-GTGTGCCGAACTCAAAGCGTCC-3′) and JB18 (5′-GACCTGCTTGGCATACTTGAGTGC-3′), which were designed to specifically amplify portions of ORF1, ORF3, and the intergenic region between these two ORFs, were used to screen a collection of 51 pepper and 34 tomato strains of X.campestris pv. vesicatoria isolated in Taiwan between 1987 and 2000. Also, nine copper-resistant strains from the United States and South America were tested for the presence of this unique region of DNA.
DNAs from a subset of 15 representative strains tested by PCR were also examined in hybridization tests using labeled DNA of pXv26Cu-2 as a probe. For Southern hybridization, genomic DNA was isolated using the cetyltrimethylammonium bromide method (4) and digested with restriction enzymes under the conditions recommended by the manufacturer. DNA fragments were resolved by agarose gel electrophoresis and transferred to Nytran membranes (Schleicher & Schuell, Keene, NH). The DNA for probes was labeled with digoxigenin-11 dUTP, and hybridization and detection reactions were carried out using a Genius nonradioactive kit (Boehringer Mannheim, Indianapolis, IN).
Nucleotide sequence accession number.
Nucleotide and/or amino acid sequence data for the copper resistance gene cluster have been deposited in the GenBank database under accession no. AY248746.
RESULTS
Cloning and subcloning of copper resistance genes from X.campestris pv. vesicatoria strain XvP26.
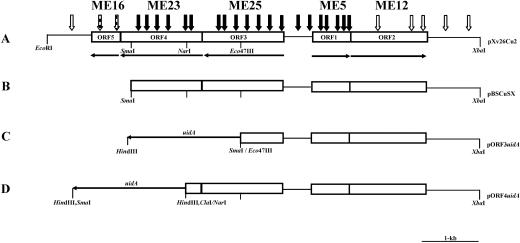
Two of the 1,100 cosmid clones tested conferred copper resistance to strain 82-8 transconjugants. A restriction enzyme map of one clone (pXv26Cu), which contained approximately 27.9 kb of insert DNA, was generated. To further localize the copper gene cluster, different fragments of the insert were first subcloned into pLAFR3. One subclone, which contained a 10.9-kb EcoRI/HindIII fragment, conferred copper resistance to strain 82-8 transconjugants on media containing 200 μg/ml of copper sulfate. The 10.9-kb fragment (pXv26Cu-1) was further subcloned as a 7,652-bp EcoRI/XbaI fragment into pUFR051 (pXv26Cu-2) and maintained copper resistance (Fig. 2).
FIG. 2.
(A) Restriction map of pXv26Cu-2, a 7,652-bp region of DNA that confers resistance to copper in XvP26. The open arrows, arrows with bars, and solid arrows indicate transposon insertions that result in transconjugants in 91-118 (copper-sensitive strain) that are resistant to copper, have intermediate sensitivity to copper, and are sensitive to copper, respectively. (B) Subclone of pXv26Cu-2 in pBluescript used to construct fusion clones shown in panels C and D. (C and D) Reporter constructs for measuring GUS activity for copper induction of ORF3 and ORF4, respectively. Construction of additional reporter clones pΔKXORF3uidA and pORF1uidA (not shown) is described in detail in Materials and Methods.
Comparison of the copper resistance genes from XvP26 withcopper resistance genes from other plant-pathogenic bacteria. The probes generated from clones containing copper resistance genes from X. campestris pv. vesicatoria strain 75-3(pXvCu), Pseudomonas syringae pv. tomato(pCop35), and X. arboricola pv. juglandis(pXjCu99) did not hybridize to total DNA digests of strain XvP26, as determined by Southern hybridization analysis using high-stringency conditions (data not shown). When the 10.9-kb fragment containing the copper resistance genes from pXv26Cu-1 was used as a probe, it exhibited only weak hybridization in low-stringency conditions with cloned plasmid-borne copper resistance genes from X. campestris pv. vesicatoria strain 75-3 and P. syringae pv. tomato and chromosome-borne copper resistance genes from X. arboricola pv. juglandis, as determined by Southern hybridization (data not shown).
Sequence analysis of the copper resistance genes.
pXv26Cu-1was subcloned by digesting the fragment with EcoRI and XbaI to produce a 7.6-kb fragment, designated pXv26Cu-2, which conferred copper resistance in 82-8 and 91-118 transconjugants. The sequence of the 7,652-bp DNA insert of pXv26Cu-2 was obtained using a combination of pBluescript subclones, a Tn3 reverse primer, and custom oligonucleotide primers. Five major open reading frames were identified (Fig. 2A), and ORF1 and ORF2 were divergently transcribed from the other three ORFs. ORF1, ORF2, ORF3, ORF4, and ORF5 were 681, 1,341, 1,428, 1,410, and 495 bp long, respectively.
ORF1 and ORF2 were selected using the alternate GTG start codon. ORF1 encodes a predicted 227-amino-acid polypeptide with the deduced amino acid sequence of a gene product exhibiting 66 and 63% identity to CopR from P. syringae(33) and to PcoR from E. coli (12), respectively. Using primer extension, the probable transcriptional start of ORF1 was identified 57 bases upstream (G) of the predicted GTG start codon and was associated with a conserved palindrome previously identified as a copper box within copper-responsive promoters (33, 34, 36) (Fig. 3C). The selected transcriptional site for ORF1 relative to the copper box aligned most closely with that of the copper-responsive repressor CopY of Enterococcus hirae (Fig. 3A). In several repeat extension reactions the presence of this strong transcript signal was sometimes accompanied by additional weak transcripts originating at a position either 42 (G), 39 (C), or 13 (A) residues upstream of the GTG codon (Fig. 3A). Two additional peaks appearing at 106 and 407 bases in the GeneScan blue (6-carboxyfluorescein) channel (Fig. 3B) were eliminated as possible ORF1 transcript sites based on subclone complementation tests. No significant differences were found in comparisons of transcript levels from total RNA obtained from uninduced and copper-induced cells (data not shown).
FIG. 3.
(A) Comparison of the ORF1 sequence with copper- and silver-inducible promoters preceded by the palindromic copper box. The upstream regions of E. coli promoters PpcoE, PcusC, and PpcoA, P. syringae promoter PcopA, and Salmonella enterica promoters PsilE and PsilS (obtained from reference 34) are aligned to highlight the copper boxes (indicated by arrows) that are found upstream of the copper- or silver-inducible promoters. Predicted −35 and −10 hexamers are indicated by single and double underlining, respectively. Transcriptional start sites that have been determined are indicated by boldface underlining. The positions of weak additional transcriptional start sites in ORF1 are indicated for the nonboldface underlined nucleotides. For comparative purposes the conserved copper box for copY of E. hirae (obtained from reference 36) is also shown (Hirae1/2). (B and C) Primer extension electropherogram (B) and image of a sequencing gel (C) used to analyze the primer extension product generated using primer PE2. Peaks corresponding to several of the GeneScan-500 ROX internal size standards (red in the actual analysis) are indicated next to the ORF1 primer extension product labeled with G* near the standard peak at 250 (blue in the actual analysis). The sequence shown is the sequence of the coding strand of ORF1.
ORF2 encodes a 447-amino-acid polypeptide that exhibits 49 and 37% identity with the putative CopS of Ralstonia metallidurans (GenBank accession number AJ278983) and PcoS of E. coli (12), respectively. ORF3 encodes a predicted 476-amino-acid polypeptide that exhibits 23% identity with a cation efflux system membrane protein, CzcC, of Alcaligenes eutrophus (35). The ORF4 product is 470 amino acids long and exhibits 64% identity with a putative copper-containing oxidoreductase protein of Sinorhizobium meliloti (5, 14). ORF5 encodes a predicted 165-amino-acid polypeptide that exhibits 47% and 51% identity with a putative copper-containing oxidoreductase signal peptide protein in S. meliloti (14) and a putative CopC protein in Aeromonas veronii (22), respectively.
Mutation and complementation analyses.
Transposon mutagenesis of pXv26Cu-2 resulted in inserts that were randomly and extensively distributed throughout the sequence based on physical mapping (Fig. 2A). Insertions in ORF1, ORF3, or ORF4 resulted in a complete loss of resistance to copper in the transconjugants screened. Insertions in ORF5 resulted in intermediate resistance to copper, and insertions in ORF2 had no noticeable effect on resistance to copper.
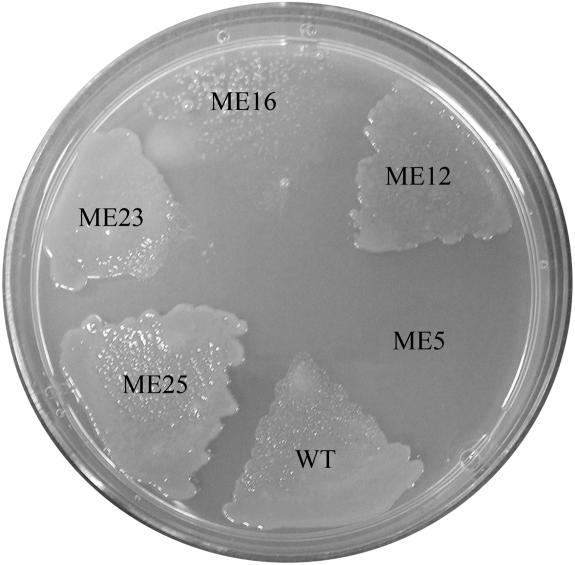
Marker exchange mutants generated in parental strain XvP26 were tested for resistance to copper. On solid media containing copper sulfate at a concentration of 120 μg/ml, only the ORF1 mutant (strain ME5) failed to grow (Fig. 4). Resistance to copper was restored in ME5 by transcomplementation with clone pORF1(BOX+90), but not by transcomplementation with clone pORF1(BOX+25). The growth of mutant ME16 (ORF5) was reduced compared to that of the wild-type strain, whereas ME12, ME25, and ME23 (representing ORF2, ORF3, and ORF4, respectively) were not affected. When the CYE broth method was used, ME25 (ORF3 mutant) appeared to be more like the wild type than ME12, ME23, and ME16 (data not shown).
FIG. 4.
Growth of wild-type strain XvP26 (WT) and marker exchange mutants on nutrient agar containing copper sulfate (120 μg/ml).
Induction of copper genes.
Upon exposure to a low level of copper, GUS activity for ORF3 and ORF4 was induced, and the levels were similar in the 91-118, ME12, and ME25 backgrounds (Table 2). The activities of both genes were considerably lower in the XvP26, ME5, ME23, and ME16 backgrounds. Co, Cd, and Zn had no significant induction effect on either gene in either the XvP26 or 91-118 background (data not shown).
TABLE 2.
Effect of copper on induction of ORF3 and ORF4 in 91-118, XvP26, and XvP26 marker exchange transconjugant backgrounds
| Recipient | β-Glucuronidase activity (U)a
|
|||||
|---|---|---|---|---|---|---|
| pORF3uidA
|
pΔKXORF3uidA
|
pORF4uidA
|
||||
| Without Cu | With Cu | Without Cu | With Cu | Without Cu | With Cu | |
| 91-118 | 41 ± 2 | 245 ± 11 | 0 | 0 | 160 ± 6 | 416 ± 10 |
| XvP26 | 14 ± 1 | 31 ± 1 | 216 ± 6 | 220 ± 7 | 31 ± 2 | 73 ± 4 |
| ME5 | 44 ± 2 | 133 ± 2 | 0 | 0 | 105 ± 1 | 268 ± 1 |
| ME12 | 116 ± 2 | 233 ± 5 | 72 ± 1 | 73 ± 2 | 116 ± 5 | 249 ± 9 |
| ME25 | 100 ± 3 | 226 ± 8 | 278 ± 10 | 319 ± 2 | 105 ± 1 | 331 ± 2 |
| ME23 | 70 ± 2 | 172 ± 7 | 0 | 43 ± 2 | 36 ± 1 | 197 ± 4 |
| ME16 | 59 ± 1 | 243 ± 8 | 0 | 56 ± 1 | 59 ± 3 | 331 ± 1 |
β-Glucuronidase activity is expressed in units (nanomoles of MU produced per minute)/1010 CFU after 3 h of induction in CYE broth with or without copper.
No GUS activity was detected when the pΔKXORF3uidA reporter clone (lacking ORF1 and ORF2) was used in the 91-118 and mutant ME5 backgrounds (Table 2). This reporter indicated that there were various levels of constitutive expression in the XvP26, ME12, and ME25 strains, with no significant induction by addition of copper. Strains ME23 and ME16 both exhibited copper-inducible expression of GUS activity for this reporter.
No GUS activity was found for pORF1uidA in the 91-118 background in our assay. A low level of constitutive activity was found for this reporter construct in XvP26 (8 ± 0.3 U) compared to the pL3uidA constitutive control (454 ± 3 U), and no significant increase was evident for either construct following addition of copper (8 ± 0.7 and 464 ± 14 U, respectively).
PCR analysis of strains.
No PCR product was amplified from the Taiwan, United States, or South American strains of X. campestris pv. vesicatoria whether they were resistant to copper or not. Only DNA from XvP26 was amplified using primers JB8 and JB18 or hybridized with the pXv26Cu-2 probe (data not shown).
DISCUSSION
We identified the determinants for copper resistance on a 7,652-bp XbaI/EcoRI chromosomal fragment in X. campestris pv. vesicatoria strain XvP26 from Taiwan. Although metal resistance is commonly associated with plasmids in bacteria (39), gene clusters associated with the chromosome that result in copper resistance have been identified in phytopathogenic Pseudomonas and Xanthomonas spp. (26, 27). Previously, copper resistance in X. campestris pv. vesicatoria was associated only with plasmids (9, 23, 41). The size of the 5.5-kb region that is functional in copper resistance in XvP26 is similar to the sizes of P syringae pv. tomato (8), X. arboricola pv. juglandis (26), and X. campestris pv. vesicatoria (46) regions, which are 4.5, 4.9, and 6.0 kb long, respectively.
As a result of sequencing of the 7.6-kb fragment, five ORFs were identified in the cluster of genes associated with copper resistance. Three of these five ORFs were required for a high level of resistance in transconjugant screening. Insertional mutation of ORF2, which exhibits homology to the CopS gene, had no observable effect on copper resistance when it was tested in the 91-118 transconjugant background. ORF5, which exhibits homology to the gene encoding a putative copper-containing oxidoreductase signal peptide protein in S. meliloti (14), was not essential for resistance to copper in plate assays, but there was delayed growth of transconjugants when it was knocked out. These results were similar to those reported for PcoE in E. coli, which has been reported to not be strictly required for copper resistance but reduced the time needed for E. coli strains to recover from copper stress (33). Francki et al. (22) observed that copper tolerance was slightly reduced by one mutation in A. veronii, similar to ORF4. A similar situation was observed when copper resistance was reduced to an intermediate level in X. campestris pv. juglandis when two ORFs whose products exhibit high levels of amino acid identity to CopC and CopD of P. syringae pv. tomato were deleted (26).
Two of the five ORFs, ORF3 and ORF4, were determined to be inducible when cultures were exposed to small amounts of copper. Although copper induction has previously been shown to occur in xanthomonads, the regulatory genes were not located (26, 46). The ORF1 product exhibits significant amino acid identity to CopR of P. syringae pv. tomato, and ORF1 contains a putative copper box motif, which previously was shown to be essential for copper-inducible activity at the PpcoA and PpcoE promoters in E. coli (37). It is unusual to find a copper box upstream of a putative copR gene in X. campestris pv. vesicatoria, considering that based on in vitro DNase I footprinting, CopR binds to the copper box upstream of PcopA (33). Munson et al. (34) provided strong proof that copper boxes upstream of PpcoA and PpcoE are the binding sites for CusR, a CopR homolog. Further confounding with regard to the copper boxes was the finding that neither ORF3 nor ORF4 contained a conserved copper box. Clearly, ORF1 is essential for copper resistance and plays a role in regulation of the system in strain XvP26.
In insertion mutant studies ORF2 (homologous to the CopS gene) was determined to be unnecessary for functional copper resistance in the 91-118 transconjugant background. One plausible explanation for this is that some region(s) outside the fragment may play a regulatory role in expression of copper resistance genes to complete the two-component signal transduction system in X. campestris pv. vesicatoria. Lee et al. (26) speculated that regulation of the copper resistance in X. arboricola pv. juglandis may be associated with a different region of the chromosome. This may also be the case in X. campestris pv. vesicatoria since ORF2, which exhibits homology to CopS, has no effect on copper resistance when it is knocked out. However, in our expression studies mutation of ORF2 resulted in lower basal expression levels and loss of the copper inducibility of ORF3 in the trans reporter, indicating that this gene has a regulatory role. There may be another gene somewhere else in the genome that functions like the CopS gene. This has been shown in E. coli, in which the pcoRS and cusRS genes encode homologous copper-responsive regulatory systems, although they cannot substitute for each other (34). Further detailed analysis, beyond the scope of this study, is needed to fully understand regulation of this system in strain XvP26.
PCR amplification analysis of strains from Taiwan collected over several years and strains from a number of regions revealed that none of the strains contained the uniquely oriented copper gene cluster found in XvP26. This could indicate that there was a rare introduction of this strain into Taiwan or chromosomal transfer from another organism or organisms. Horizontal transfer of the copper resistance gene in XvP26 has been demonstrated among strains of X. campestris pv. vesicatoria (6). The latter hypothesis may be plausible given the inability to detect the copper resistance genes in a representative collection of strains from Taiwan.
Acknowledgments
We gratefully acknowledge Akdeniz University for financial support of H.B. during this work.
REFERENCES
- 1.Adaskaveg, J. E., and R. B. Hine. 1985. Copper tolerance and zinc sensitivity of Mexican strains of Xanthomonas campestris pv. vesicatoria, causal agent of bacterial spot of pepper. Plant Dis. 69:993-996. [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen, G. L., O. Menkissoglou, and S. E. Lindow. 1991. Occurrence and properties of copper-tolerant strains of Pseudomonas syringae isolated from fruit trees in California. Phytopathology 81:648-656. [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1992. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 5.Barnett, M. J., R. F. Fisher, T. Jones, C. Komp, A. P. Abola, F. Barloy-Hubler, L. Bowser, D. Capela, F. Galibert, J. Gouzy, M. Gurjal, A. Hong, L. Huizar, R. W. Hyman, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, C. Palm, M. C. Peck, R. Surzycki, D. H. Wells, K.-C. Yeh, R. W. Davis, N. A. Federspiel, and S. R. Long. 2001. Nucleotide sequence and predicted functions of the entire Sinorhizobium meliloti pSymA megaplasmid. Proc. Natl. Acad. Sci. USA 98:9883-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basim, H., R. E. Stall, G. V. Minsavage, and J. B. Jones. 1999. Chromosomal gene transfer by conjugation in the plant pathogen Xanthomonas axonopodis pv. vesicatoria. Phytopathology 89:1044-1049. [DOI] [PubMed] [Google Scholar]
- 7.Bender, C. L., and D. A. Cooksey. 1986. Indigenous plasmids in Pseudomonas syringae pv. tomato: conjugative transfer and role in copper resistance. J. Bacteriol. 165:534-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bender, C. L., and D. A. Cooksey. 1987. Molecular cloning of copper resistance genes from Pseudomonas syringae pv. tomato. J. Bacteriol. 169:470-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bender, C. L., D. K. Malvick, K. E. Conway, S. George, and D. A. Cooksey. 1990. Characterization of pXv10A, a copper resistance plasmid in Xanthomonas campestris pv. vesicatoria. Appl. Environ. Microbiol. 56:170-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonas, U., R. Schulte, S. Fenselau, G. V. Minsavage, B. J. Staskawicz, and R. E. Stall. 1991. Isolation of a gene cluster from Xanthomonas campestris pv. vesicatoria that determines pathogenicity and the hypersensitive response on pepper and tomato. Mol. Plant-Microbe Interact. 4:81-88. [Google Scholar]
- 11.Bonas, U., R. E. Stall, and B. J. Staskawicz. 1989. Genetic and structural characterization of the avirulence gene avrBs3 from Xanthomonas campestris pv. vesicatoria. Mol. Gen. Genet. 218:127-136. [DOI] [PubMed] [Google Scholar]
- 12.Brown, N. L., S. R. Barrett, J. Camakaris, B. T. Lee, and D. A. Rouch. 1995. Molecular genetics and transport analysis of the copper-resistance determinant (pco) from Escherichia coli plasmid pRJ1004 J. Mol. Microbiol. 17:1153-1166. [DOI] [PubMed] [Google Scholar]
- 13.Canteros, B. I., G. V. Minsavage, J. B. Jones, and R. E. Stall. 1995. Diversity of plasmids in Xanthomonas campestris pv. vesicatoria. Phytopathology 85:1482-1486. [Google Scholar]
- 14.Capela, D., F. Barloy-Hubler, J. Gouzy, G. Bothe, F. Ampe, J. Batut, P. Boistard, A. Becker, M. Boutry, E. Cadieu, S. Dreano, S. Gloux, T. Godrie, A. Goffeau, D. Kahn, E. Kiss, V. Lelaure, D. Masuy, T. Pohl, D. Portetelle, A. Puehler, B. Purnelle, U. Ramsperger, C. Renard, P. Thebault, M. Vandenbol, S. Weidner, and F. Galibert. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672.11474104 [Google Scholar]
- 15.Cooksey, D. A. 1987. Characterization of a copper resistance plasmid conserved in copper-resistant strains of Pseudomonas syringae pv. tomato. Appl. Environ. Microbiol. 53:454-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooksey, D. A. 1990. Plasmid-determined copper resistance in Pseudomonas syringae from impatiens. Appl. Environ. Microbiol. 56:13-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooksey, D. A., H. R. Azad, J. S. Cha, and C. K. Lim. 1990. Copper resistance gene homologs in pathogenic and saprophytic bacterial species from tomato. Appl. Environ. Microbiol. 130:2447-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeFeyter, R., C. I. Kado, and D. W. Gabriel. 1990. Small, stable shuttle vectors for use in Xanthomonas. Gene 88:65-72. [DOI] [PubMed] [Google Scholar]
- 19.Ditta, G., S. Stanfield, D. Corbin, and D. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erardi, F. X., M. L. Failla, and J. O. Falkinham III. 1987. Plasmid-encoded copper resistance and precipitation by Mycobacterium scrofulaceum. Appl. Environ. Microbiol. 53:1951-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Figurski, D., and D. R. Helinski. 1979. Replication of an origin containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Francki, K. T., B. J. Chang, B. J. Mee, P. J. Collignon, V. Susai, and P. K. Keese. 2000. Identification of genes associated with copper tolerance in an adhesion-defective mutant of Aeromonas veronii biovar sobria. FEMS Immunol. Med. Microbiol. 29:115-121. [DOI] [PubMed] [Google Scholar]
- 23.Garde, S., and C. L. Bender. 1991. DNA probes for detection of copper resistance genes in Xanthomonas campestris pv. vesicatoria. Appl. Environ. Microbiol. 57:2435-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jefferson, R. A. 1987. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5:387-405. [Google Scholar]
- 25.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, Y., M. Hendson, N. J. Ponopoulos, and M. N. Schroth. 1994. Molecular cloning, chromosomal mapping, and sequence analysis of copper resistance genes from Xanthomonas campestris pv. juglandis: homology with small blue copper proteins and multicopper oxidase. J. Bacteriol. 176:173-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim, C. K., and D. A. Cooksey. 1993. Characterization of chromosomal homologs of the plasmid-borne copper resistance operon of Pseudomonas syringae. J Bacteriol. 175:4492-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lloyd, A. L., B. J. Marshall, and B. J. Mee. 2005. Identifying cloned Helicobacter pylori promoters by primer extension using FAM-labelled primer and GeneScan analysis. J. Microbiol. Methods 60:291-298. [DOI] [PubMed] [Google Scholar]
- 29.Marco, G. M., and R. E. Stall. 1983. Control of bacterial spot of pepper initiated by strains of Xanthomonas campestris pv. vesicatoria that differ in sensitivity to copper. Plant Dis. 67:779-781. [Google Scholar]
- 30.Mellano, M. A., and D. A. Cooksey. 1988. Nucleotide sequence and organization of copper resistance genes from Pseudomonas syringae pv. tomato. J. Bacteriol. 170:2879-2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mellano, M. A., and D. A. Cooksey. 1988. Induction of the copper resistance operon from Pseudomonas syringae. J. Bacteriol. 170:4399-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 33.Mills, S. D., C. A. Jasalavich, and D. A. Cooksey. 1993. A two-component regulatory system required for copper-inducible expression of the copper resistance operon of Pseudomonas syringae. J. Bacteriol. 175:1656-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munson, G. P., D. L. Lam, F. W. Outten, and T. V. O'Halloran. 2000. Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J. Bacteriol. 182:5864-5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nies, D. H., A. Nies, L. Chu, and S. Silver. 1989. Expression and nucleotide sequence of a plasmid-determined divalent cation efflux system from Alcaligenes eutrophus. Proc. Natl. Acad. Sci. USA 86:7351-7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Portmann, R., D. Magnani, J. V. Stoyanov, A. Schmechel, G. Multhaup, and M. Solioz. 2004. Interaction kinetics of the copper-responsive CopY repressor with the cop promoter of Enterococcus hirae. J. Biol. Inorg. Chem. 9:396-402. [DOI] [PubMed] [Google Scholar]
- 37.Rouch, D. A., and N. L. Brown. 1997. Copper-inducible transcriptional regulation at two promoters in the Escherichia coli copper resistance determinant pco. Microbiology 143:1191-1202. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 39.Silver, S., and L. T. Phung. 1996. Bacterial heavy metals: new surprises. Annu. Rev. Microbiol. 50:753-789. [DOI] [PubMed] [Google Scholar]
- 40.Stachel, S. E., G. An, C. Flores, and E. W. Nester. 1985. A Tn3 lacZ transposon for the random generation of β-galactosidase gene fusions: application to the analysis of gene expression in Agrobacterium. EMBO J. 4:891-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stall, R. E., D. C. Loschke, and J. B. Jones. 1986. Linkage of copper resistance and avirulence loci on a self-transmissible plasmid in Xanthomonascampestris pv. vesicatoria. Phytopathology 76:240-243. [Google Scholar]
- 42.Staskawicz, B., D. Dahlbeck, N. Keen, and C. Napoli. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sundin, G. W., A. L. Jones, and D. W. Fulbright. 1989. Copper resistance in Pseudomonas syringae pv. syringae from cherry orchards and its associated transfer in vitro and in planta with a plasmid. Phytopathology 79:861-865. [Google Scholar]
- 44.Tetaz, T. J., and R. K. Luke. 1983. Plasmid-controlled resistance to copper in Escherichia coli. J. Bacteriol. 154:1263-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turner, P., C. Barber, and M. Daniels. 1984. Behaviour of the transposons Tn5 and Tn7 in Xanthomonas campestris pv. campestris. Mol. Gen. Genet. 195:101-107. [Google Scholar]
- 46.Voloudakis, A. E., C. L. Bender, and D. A. Cooksey. 1993. Similarity between copper resistance genes from Xanthomonas campestris and Pseudomonas syringae. Appl. Environ. Microbiol. 59:1627-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Voloudakis, A. E., T. M. Reignier, and D. A. Cooksey. 2005. Regulation of resistance to copper in Xanthomonas axonopodis pv. vesicatoria. Appl. Environ. Microbiol. 71:782-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zevenhuizen, L. P. T. M., J. Dolfing, E. J. Eshuis, and J. Scholtern- Koerselman. 1979. Inhibitory effects of copper on bacteria related to the free ion concentration. Microb. Ecol. 5:139-164. [DOI] [PubMed] [Google Scholar]