Abstract
Fresh feces, manure slurry (from earthen lagoons and/or concrete pits), and drinking and surface water samples were collected from 28 pig farms in the Midwestern United States. All samples were tested for hepatitis E virus (HEV) RNA by reverse transcription-PCR. Seven of 28 farms had fecal samples that contained HEV. Of 22 farms where pit samples were accessible, 15 contained HEV, and of 8 farms that had lagoons, 3 contained HEV. The highest virus titers were 10 and 103 genome equivalents per 60 ml of manure slurry in lagoon and pit samples, respectively. None of the water samples tested HEV positive. To determine the infectivity of the HEV found in the positive farm 19 lagoon (designated L19) or farm 12 pit (designated P12) samples, pigs were inoculated either intravenously (n = 3) or orally (n = 3) with the L19 or P12 manure slurry. Four pigs inoculated intravenously with prototype swine HEV served as positive controls. All positive-control pigs shed HEV in feces and 3 of 4 developed anti-HEV antibodies. Two pigs in the intravenously inoculated P12 group shed HEV in feces, and one of the pigs seroconverted to anti-HEV antibodies. None of the pigs in the negative-control, L19 oral, L19 intravenous, or P12 oral group shed HEV in feces. The findings indicate that HEV found in pig manure slurry was infectious when inoculated intravenously. Pit manure slurry is a potential source of HEV infection and for contamination of the environment. Contamination of drinking or surface water with HEV was not found on or near the pig farms.
Hepatitis E virus (HEV) is the etiologic agent of an acute self-limiting non-A, non-B hepatitis in humans (30). The virus is a nonenveloped positive-sense single-stranded RNA virus and was recently given taxonomic status in the genus of Hepevirus, a member of the family Hepeviridae (9). Human infection with HEV typically manifests as acute icteric hepatitis. The overall mortality is low, except for pregnant women, who may suffer severe and fatal hepatitis with mortality rates up to 25% (18, 19). HEV infection is distributed worldwide and is endemic to parts of Africa and Asia (18, 32).
Human HEV outbreaks in regions of endemicity are reportedly associated with flooding and heavy rains and spread through the fecal-oral route of transmission (7, 8). HEV infection in industrialized countries of nonendemicity is sporadic and is often reported in human patients with a history of traveling to countries of endemicity; however, there are increasing cases of sporadic human HEV infection in countries of nonendemicity such as Japan, Greece, Italy, and the United States in patients without a history of traveling to regions of endemicity. The human HEV discovered in industrialized countries of nonendemicity appears to be closely related to the swine HEV found in the same countries (4, 8, 28, 29, 31).
Pigs are a natural host for swine HEV (13, 25, 26). The fecal-oral route is thought to be the primary mode of HEV transmission (8, 36). Pigs infected with HEV shed the virus in feces in large amounts for 3 to 4 weeks, whereas viremia is transient and persists for 1 to 2 weeks (10, 15, 17). Current pig-raising practices allow repeated exposure of pigs to feces and thus to potentially high doses of HEV (17, 36).
Food-borne or blood transfusion routes have been implicated as likely means of origin of sporadic cases of human HEV infection in industrialized countries of nonendemicity (20, 21, 33, 37). Genomic comparison of HEV from human cases of sporadic acute or fulminant hepatitis E in Japan revealed a high homology with swine HEV isolates found in pig livers in nearby grocery stores, indicating that swine HEV in inadequately cooked pig liver may be transmitted orally to humans (37). Contamination of drinking or surface water in the vicinity of pig production facilities is possible and of concern (32). The status of HEV in pig manure slurry in storage facilities such as concrete pits and earthen lagoons remains to be investigated. If HEV is indeed present in stored pig manure slurry, it needs to be determined if the virus is infectious to pigs or other animals. The objectives of the present study were: (i) to determine whether HEV is present in pig feces, stored manure slurry, on-site drinking water supplies, and surface water near swine production facilities and (ii) to determine if HEV found in pig manure slurry and/or water samples is infectious to naive pigs.
MATERIALS AND METHODS
Fecal sample collection and processing.
In the late summer and autumn of 2002, 28 pig farms located in the Midwestern United States (Iowa) were visited and sampled. Clinical evidence suggests that pigs in the United States most likely become infected with swine HEV between 8 and 16 weeks of age (22, 25). Because pig farms with various production styles were sampled, pigs of different age groups ranging from newborns to adults were present on the 28 farms (Table 1). Approximately 50 g of fresh feces from 5 live pigs ranging from 2 to 6 months of age (Table 1) were collected from the floor of the pig barns, pooled, and prepared as a 10% (wt/vol) fecal suspension in diethyl pyrocarbonate (DEPC) water-based phosphate-buffered saline (PBS).
TABLE 1.
Detection of HEV RNA in pooled pig feces and environmental samples collected from 28 pig farms
| Farm identification no. | Type of farma | Feces
|
Pit HEV RNA (virus titer)b | Lagoon HEV RNA (virus titer)b | |
|---|---|---|---|---|---|
| Mean age of pigs sampled (wk) | HEV RNA | ||||
| 1 | Farrow to finish | 20 | −c | + (103) | NAd |
| 2 | Farrow | 24 | − | − | − |
| 3 | Farrow to finish | 16 | − | − | − |
| 4 | Farrow to finish | 16 | − | − | NA |
| 5 | Grow to finish | 24 | − | + (10) | NA |
| 6 | Grow to finish | 20-24 | − | NA | − |
| 7 | Grow to finish | 16 | − | + (10) | NA |
| 8 | Grow to finish | 12-16 | − | − | NA |
| 9 | Grow to finish | 16-20 | + | + (10) | NA |
| 10 | Wean to finish | 12 | − | − | + (10) |
| 11 | Farrow to finish | 12 | − | + (10) | NA |
| 12 | Grow to finish | 20-24 | + | + (103) | NA |
| 13 | Farrow to finish | 20-24 | − | + (10) | NA |
| 14 | Grow to finish | 12-20 | − | + (10) | NA |
| 15 | Grow to finish | 8-20 | − | + (102) | NA |
| 16 | Grow to finish | 8 | − | + (10) | NA |
| 17 | Farrow to finish | 20-24 | − | NA | + (10) |
| 18 | Wean to finish | 20 | + | + (10) | NA |
| 19 | Farrow to finish | 20 | + | NA | + (10) |
| 20 | Wean to finish | 20-24 | − | − | NA |
| 21 | Farrow to finish | 16-20 | + | + (10) | NA |
| 22 | Grow to finish | 20 | + | + (10) | NA |
| 23 | Farrow to finish | 12-16 | − | NA | NA |
| 24 | Grow to finish | 16-20 | − | NA | NA |
| 25 | Grow to finish | 16-20 | + | + (102) | − |
| 26 | Grow to finish | 16-20 | − | + (102) | NA |
| 27 | Grow to finish | 16-20 | − | − | NA |
| 28 | Wean to finish | 16 | − | NA | − |
| Total | 7/28 | 15/22 | 3/8 | ||
Differences in the types of farms are due to the age groups of pigs present on the farm. Farrow (adults and newborn to 3-week-old piglets), farrow to finish (adults, newborns, and pigs up to approximately 24 weeks of age), grow to finish (pigs from approximately 10 weeks to 24 weeks of age), wean to finish (pigs from weaning [2 weeks] to market age, approximately 24 weeks).
HEV genome titer is given as genome equivalents per 60 ml of manure slurry sample, determined by semi-qualitative RT-PCR.
−, negative for HEV RNA by qualitative RT-PCR; +, positive for HEV RNA by qualitative RT-PCR.
NA, sample not available at the farm.
Manure slurry sample collection and processing.
Samples of manure slurry in pits (22/28 farms had accessible pits) and lagoons (8/28 farms had lagoons) were randomly obtained from each manure slurry storage facility and collected in a 50-ml sterile centrifuge tube. Ten manure slurry samples were collected from the concrete pit under the slats of each finishing facility. The samples were taken from 10 randomly selected sites in each pit. Each sample was collected from a depth between 30 and 60 cm below the surface and directly below the pig pens. For the lagoon manure slurry samples, 10 samples were randomly collected at a depth of 120 and 200 cm under the surface crust.
All samples were transported on ice to the laboratory. The 10 tubes of each manure slurry sample from each storage facility (pit and/or lagoon) were pooled together in two aliquots of 250 ml. All fecal suspensions and manure slurry samples were kept at −80°C until tested by reverse transcription (RT)-PCR assay. Portions (60 ml) of the pooled manure slurry samples were subjected to ultracentrifuge protocols to concentrate virus particles (27). The manure slurry samples were clarified at 1,100 × g for 10 min at 4°C. The supernatant was ultracentrifuged at 229,600 × g for 1 h at 4°C to pellet the virus particles. The virus pellet was then diluted with 10 ml of 0.25 M glycine buffer (pH 9.4) on ice for 30 min. The resuspended virus pellet was recentrifuged at 12,000 × g for 15 min at 4°C to discard solid materials. The supernatant of the virus pellet was saved and ultracentrifuged at 229,600 × g for 1 h at 4°C to produce a final virus concentrate. The final virus concentrate was resuspended with 400 μl of DEPC water-based PBS. Two aliquots of 200 μl each were prepared for qualitative nested RT-PCR and for semiquantitative nested RT-PCR and kept at −80°C until tested. The manure slurry concentrate samples that were positive for HEV RNA by the qualitative RT-PCR had their titers for HEV further determined by the semiquantitative RT-PCR assay.
Water sample collection and processing.
On each farm visited, 10 liters of on-site drinking water and 10 liters of water from the nearest upstream and downstream surface water (creeks, streams, or rivers) within a 0.4- to 6.4-kilometer radius of the pig farms were collected and concentrated. All water samples were collected in a sterile 10-liter container. Drinking water samples were directly obtained from a hydrant or faucet in or near the pig barns. Surface water samples were directly collected from the water source by submerging the container below the water surface. The water samples were transported in a cooler with ice packs to the laboratory and were processed for virus concentration upon arrival within a day of collection.
The concentration protocols were modified from those of Abbaszadegan et al. (1) and the guidelines for detection of enteric viruses recommended by U.S. Environmental Protection Agency (3). Water samples (10 liters) were run through a sterile unit of a filter housing (CUNO, Inc., Meriden, CT) and cartridge (Virosorb MDS1; CUNO, Inc., Meriden, CT). All flowthrough water was discarded. Once the filtering process was complete, 1 liter of autoclaved 0.25 M glycine buffer (pH 9.4) was used to elute the entrapped virus particles in the filter cartridge. The elution process was performed twice for 15 min. The first viral eluate was used for the second elution process. The final virus eluate was neutralized at pH 7 to 7.4 with 1 N HCl (pH 1.5), mixed vigorously, and kept on ice before proceeding to the virus concentration process by organic flocculation (3). It has been suggested that HEV is relatively stable under acid and mildly alkaline conditions (30), and previously, it has been confirmed that acidic pH enhances the structure and stability of the HEV capsid protein (38). Therefore, the neutralized virus eluate was adjusted to lower its pH to 3.5 with 1 N HCl and 12 g of autoclaved beef extract powder and stirred vigorously for 15 min. The pH-adjusted virus eluate was then centrifuged at 3,000 × g for 10 min at 4°C. The supernatant of the virus eluate was discarded, and the resulting pellet was resuspended with 50 ml of DEPC water-based PBS. The suspended virus pellet was made into 2 aliquots of 25 ml. One aliquot was subjected to the centrifugal concentration process to adjust the eluate volume for qualitative RT-PCR testing and the other was kept at −80°C for an archival purpose. The virus eluate for RT-PCR was subjected to 1,100 × g for 20 min in a centrifugal filter unit (Centricon; Millipore Corporation, Billerica, MA) to concentrate the eluate volume for the RNA extraction steps.
To determine the sensitivity of the water concentration used in the present study, 10 liters of tap water was spiked with 1 ml of a swine HEV infectious fecal pool (10% wt/vol) with an HEV titer of 102 genome equivalents (GE) per ml which resulted in a final concentration of 10−2 GE per ml water. The swine HEV-spiked water was processed through the protocols described above.
Nested HEV RT-PCR.
Hepatitis E virus is heterogenic in U.S. swine populations (13). To account for this, two different nested HEV RT-PCR approaches were used (10, 13). Both PCR assays amplify portions of the ORF2 gene. The universal nested RT-PCR assay designed to detect genetically variable strains of HEV (13) was used to test the concentrated manure slurry (collected either from concrete pit or earthen lagoon facilities) and concentrated water samples (collected either from surface water sources or on-site drinking water), since the extent of genetic variation among the HEV isolates was unknown. The U.S. prototype nested RT-PCR assay designed to specifically detect the prototype U.S. strain of swine HEV (10) was used in the bioassay due to 1-log-higher sensitivity (13).
RNA isolation was similar for all samples and assays. The modified spin column method (QIAamp; QIAGEN, Chatsworth, CA) was used to extract total RNA from 140 μl of concentrated manure slurry and water samples or fecal suspensions (2). The RNA extract (11.5 μl) was then immediately used for the reverse transcription reaction for HEV cDNA synthesis.
For the universal nested RT-PCR, the first-round PCR primers were forward primer 3156N (5′-AATTATGCC[T]CAGTAC[T]CGG[A]GTTG-3′) and reverse primer 3157N(5′-CCCTTA[G]TCC[T]TGCTGA[C]GCATTCTC-3′). The second-round primers were the second forward primer 3158N (5′-GTT[A]ATGCTT[C]TGCATA[T]CATGGCT-3′) and the second reverse primer 3159N (5′-AGCCGACGAAATCAATTCTGTC-3′) (13). For the PCR parameters used in the universal RT-PCRs, we strictly followed the protocol published by Huang et al. (13). The second-round amplified PCR product of 348 bp in length was visualized after 2% agarose gel electrophoresis.
For the U.S. prototype nested RT-PCR assay, the first-round PCR primers were forward primer F1 (5′-AGCTCCTGTACCTGATGTTGACTC-3′) and reverse primer R1 (5′-CTACAGAGCGCCAGCCTTGATTGC-3′). The second-round PCR primers were forward primer F2 (5′-GCTCACGTCATCTGTCGCTGCTGG-3′) and reverse primer R2 (5′-GGGCTGAACCAAAATCCTGACATC-3′) (10). To synthesize HEV cDNA, portions (11.5 μl) of RNA extract were amplified in a reverse transcription reaction with R1 reverse primer and Superscript II reverse transcriptase (Invitrogen) at 42°C for 1 h. The cDNA (10 μl) was used as a template in a 100-μl PCR mixture. The first-round PCR was initiated with the activation of AmpliTaq Gold DNA polymerase (Applied Biosystems) at 95°C for 9 min, followed by repeated 39 cycles of denaturation, annealing, and extension at 94°C for 1 min, 52°C for 1 min, and 72°C for 1.5 min, respectively, and finally, an incubation at 72°C for 7 min. Similar PCR parameters were applied to the second-round PCR. The second-round amplified PCR product of 266 bp in length was visualized after 2% agarose gel electrophoresis.
Semiquantitative nested RT-PCR for titration of HEV.
To determine the virus titer in samples that tested positive for HEV RNA, serial 10-fold dilutions in DEPC water-based PBS, ranging from 10−1 to 10−6 dilution, were made and subjected to the universal RT-PCR. The virus titer was reported as GE at the highest dilution found positive by the RT-PCR (23).
HEV genomic sequencing and phylogenetic analysis.
PCR products amplified from pooled feces and manure slurry were purified using the glass milk procedure with a GENECLEAN kit (Bio 101, Inc., Carlsbad, CA) and then directly sequenced at the Virginia Tech DNA Sequencing Facility. Sequences of the PCR products were determined for both DNA strands. The primer sequences of the universal RT-PCR were excluded from the sequence and phylogenetic analysis. The resulting partial 222-bp sequences of positive feces and manure slurry were analyzed and compared with the corresponding regions of prototype swine HEV and some known strains of swine HEV available in the GenBank database by the MacVector computer program (Oxford Molecular, Inc., Campbell, CA). Phylogenetic analysis was conducted with the aid of the PAUP program (David Swofford, Smithsonian Institute, Washington, D.C., distributed by Sinauer Associates, Inc., Sunderland, MA). A heuristic search with 1,000 replicates was used to produce a phylogenetic tree. The geographic origins and the GenBank accession numbers of the nucleotide sequences of the HEV strains, representing four major genotypes (8) and including the recently characterized avian HEV (11), used in the phylogenetic and sequence analyses are as follows: human HEV (hHEV) Sar-55 (Pakistan, accession no. M80851, genotype 1), hHEV Mexican (Mexico, accession no. M74506, genotype 2), prototype swine HEV (United States, accession no. AF082843, genotype 3), hHEV U.S.-2 (United States, accession no. AF060669, genotype 3), hHEV T1 (China, accession no. AJ272108, genotype 4), and avian HEV (United States, accession no. AY043166).
Virus inocula.
The swine HEV inocula used for inoculation of the positive-control pigs were prepared from an infectious stock of feces collected from pigs intravenously infected with the prototype U.S. strain of swine HEV (25). The inocula contained an HEV titer of 104.5 50% pig infectious doses per ml as determined by intravenous inoculation, approximately equivalent to 106 GE per ml (24). The inocula were kept at −80°C until used.
Swine bioassay.
To determine if HEV RNA found in manure slurry samples represented infectious virus particles, the HEV-positive pit and lagoon samples with the highest titer of HEV measured by semiquantitative RT-PCR were prepared as inocula for naive pigs in a swine bioassay. Nineteen 6-week-old, specific-pathogen-free pigs were randomly separated into 6 groups of 3 to 4 pigs. All pigs were anti-HEV seronegative by enzyme-linked immunosorbent assay (ELISA) and free of HEV in feces by swine HEV-specific nested RT-PCR. Three pigs served as uninoculated controls. Four positive-control pigs were inoculated intravenously with the prototype U.S. swine strain (24). Four groups of three pigs each were inoculated with 25 ml of either an HEV-positive pit or lagoon sample via the intravenous or oral route. The selected lagoon sample was collected from farm no. 19 and designated L19, and the selected pit sample was collected from farm no. 12 and designated P12.
For preparation of the inocula, portions (150 ml) of the manure slurry collected from L19 or P12 were dissolved in sterile DEPC water-based PBS to produce a manure slurry suspension. The suspensions were clarified at 1,100 × g for 10 min at 4°C. Each pig was given 25 ml of the supernatant by intravenous (i.v.) inoculation. Due to volume limitations, the intravenous inoculation was performed for 5 consecutive days using 5 ml of the inocula per pig per day. The accumulative HEV titer each pig received by intravenous inoculation was approximately 10 and 103 GE for L19 and P12, respectively. For oral inoculation, 25 ml of pooled pig manure slurry from L19 or P12 was given to naive pigs all at one time by oral gavage. The HEV titer each pig received for oral inoculation was approximately 10 and 103 GE for L19 and P12, respectively. Serum samples and fecal swabs were collected weekly for anti-HEV serum antibodies and detection of HEV RNA. The bioassay was terminated on 56 days postinoculation (dpi).
Detection of anti-HEV immunoglobulin G antibodies by ELISA.
Serum samples were tested by an ELISA using a purified 55-kDa truncated recombinant putative capsid protein of human HEV Sar-55 strain as the antigen (24, 25). Each serum sample was tested in duplicate, and mean optical density values above 0.3 were regarded as positive samples. Preimmune and hyperimmune sera were used as negative and positive controls, respectively.
RESULTS
Detection of HEV in feces, manure slurry, and water samples.
HEV RNA was detected in pooled fresh feces from 7 of the 28 farms. Fifteen of 22 farms were found to have HEV-positive manure slurry samples collected from concrete storage pits. Three of 8 lagoons were positive for HEV RNA (Table 1). The highest virus titer in manure slurry samples that tested positive for HEV RNA was approximately 10 and 103 GE in L19 and P12, respectively (Table 1).
The swine HEV-spiked water was concentrated to determine the efficacy of the concentration process and was tested by swine HEV-specific RT-PCR assay and found to be positive for swine HEV RNA. However, none of the concentrated water samples collected from on-site drinking water supplies or the nearest surface water sources (creeks, streams, or rivers) were positive for HEV RNA (data not shown).
Genetic variation of HEV isolates in pooled feces and pit or lagoon manure slurry samples.
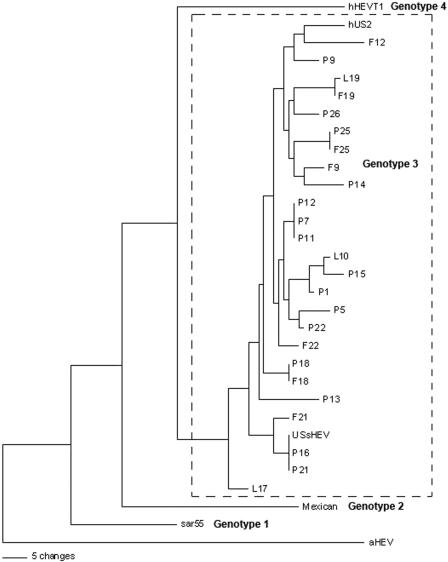
Of 7 pooled fecal samples positive for HEV RNA, nucleotide sequence analyses of the amplicons revealed that the 7 HEV isolates shared 88 to 94% identity with each other and 88 to 96% identity with the prototype U.S. swine HEV. A phylogenetic tree was built based on 222-bp-long sequences. Phylogenetic analysis indicates that HEVs amplified from feces or manure slurry samples collected from either pit or lagoon facilities are segregated within genotype 3, which is where the prototype swine HEV and the US-2 strain of human HEV are clustered (Fig. 1). There were two farms (farm no. 18 and 19) where the HEVs found in pig feces and the pit or lagoon manure slurry samples were more closely related to each other than to the HEVs from different farms. On one farm (F18 and P18), the HEVs from feces and pit manure slurry were identical (Fig. 1).
FIG. 1.
A phylogenetic tree based on analysis of a 222-bp nucleotide sequence of a portion of the ORF2 gene of HEV. Representative HEV strains of 4 major genotypes (sar55, human HEV Pakistan strain [genotype 1]; Mexican, human HEV Mexican strain [genotype 2]; USsHEV, prototype swine HEV strain [genotype 3]; hUS2, human HEV U.S. strain [genotype 3]; hHEVT1, Chinese human HEV T1 strain [genotype 4]) and a recently classified avian HEV strain were included in the analysis. F, P, and L represent HEV sequences amplified from pooled fecal, pit, and lagoon samples, respectively, which are presented in Table 1. The number following the sample initial refers to the farm identification listed in Table 1. A scale bar reflecting the numbers of character state changes is provided for comparison of genetic distance.
Determination of infectivity of HEV found in manure slurry samples.
The swine bioassay results are summarized in Table 2. All positive-control pigs shed HEV in feces from 14 to 49 dpi, and 3 of 4 positive-control pigs developed anti-HEV antibodies by 56 dpi. Uninoculated control pigs remained HEV-free in feces and negative for anti-HEV immunoglobulin G antibodies by the termination of the study at 56 dpi. Two pigs inoculated intravenously with the P12 inocula became infected with swine HEV. The presence of HEV RNA in feces was detected in one pig in the P12-inoculated group by 14 dpi and in the other pig by 21 dpi. Both pigs continued to shed HEV in feces through 28 dpi. The pig that shed HEV in feces from 14 to 28 dpi seroconverted to HEV, whereas the other pig remained seronegative on termination of the study at 56 dpi (Table 2). One of 3 pigs inoculated intravenously with manure slurry from L19 developed anti-HEV antibodies by 56 dpi (Table 2). The amplicon of HEV RNA from feces collected at 21 dpi from a pig (designated pig I) inoculated intravenously with P12 was 100% identical to the amplicon of HEV RNA from the P12 intravenous inoculum that the pig received. The amplicon of HEV RNA from feces collected at 21 dpi from the other pig (designated pig II) in the P12 intravenous group was 93% identical to the P12 intravenous inoculum that the pig received.
TABLE 2.
Swine bioassay results: detection of HEV RNA in feces and anti-HEV serum antibodies in pigs inoculated with manure slurry samples
| Inoculum (titer as genome equivalent) | Route | No. of HEV RNA-positive pigs at each dpi
|
anti-HEV (56 dpi)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0a | 7 | 14 | 21 | 28 | 35 | 42 | 49 | 56 | Incidenceb | ELISA ODc | ||
| None (uninoculated) | 0/3b | 0/3 | 0/3 | NTd | NT | NT | NT | NT | 0/3 | 0/3 | ||
| L19 (10) | Oral | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | NT | NT | 0/3 | 0/3 | |
| L19 (10) | i.v. | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | NT | NT | 0/3 | 1/3 | 0.414 |
| P12 (103) | Oral | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | NT | 0/3 | 0/3 | |
| P12 (103) | i.v. | 0/3 | 0/3 | 1/3 | 2/3 | 2/3 | 0/3 | 0/3 | NT | 0/3 | 1/3 | 0.306 |
| Positive control (106) | i.v. | 0/4 | 3/4 | 4/4 | 4/4 | 4/4 | 4/4 | 4/4 | 4/4 | 0/4 | 3/4 | 0.425-1.269 |
Fecal samples were collected at 0 to 56 days post inoculation.
Number of pigs positive/number of pigs tested.
Optical density (OD) of the positive samples; cutoff = 0.3.
NT, not tested.
DISCUSSION
Fifteen of 22 farms (68%) were found to have HEV-positive manure slurry samples. The concentration protocols used in this study for detection of HEV in manure slurry offer a reliable method to detect small amounts of HEV RNA in large volumes of manure slurry. In contrast, only 6 of the 15 farms (40%) also had detectable HEV RNA in pooled fresh fecal samples. This might be due to the low numbers (5 pigs) of individual pigs sampled at each time point or due to RT-PCR inhibitors that might have been present in the feces. An internal control for determination of the presence of RT-PCR inhibitors could have been used to further confirm this but was not part of our protocol at that time.
In the present study, all manure slurry samples were collected during the late summer and autumn months of a year when the ambient temperatures were quite warm compared to those in the winter months in the Midwestern United States. The high ambient temperatures at the time the manure slurry samples were collected could have enhanced the biodegradation of organic waste in the manure slurry storage facilities and decreased the amounts of HEV nucleic acids detectable by RT-PCR and the viability of the virus assayed. In contrast, low temperatures during winter months may preserve the infectivity of viruses in environmental samples for longer periods (5).
The HEV isolates from the pooled feces and manure slurry were genetically diverse but all clustered in genotype 3 with the prototype U.S. swine HEV. Nucleotide sequence comparison showed that HEVs isolated from the pooled feces and pit and/or lagoon samples of the same farm are genetically more closely related to each other than those from different farms. However, the HEV isolates from the pooled feces and manure samples of farm no. 9 and 12 were more genetically distinct than those of the other farms (Fig. 1). This may indicate multiple strains of HEV isolates circulating on one farm. It is common to have growing pigs from more than one breeding source present on a farm at the same time or over time. Wild rats likely reside on pig farms. Therefore, it is also possible that rats are an alternative source for contamination of pig manure slurry storage facilities, since a high prevalence of anti-HEV antibodies has been reported in wild rats (12, 14). Interestingly, all HEVs found in the manure slurry samples tested in the present study are phylogenetically clustered in genotype 3, which is genetically distant from the rat HEV reported in Nepal, which clusters in genotype 1 (12). Wild deer may also gain access to manure slurry in lagoons in the Midwestern United States. Since evidence of food-borne zoonotic transmission of HEV from deer to humans has been reported in Japan (33), it is reasonable to speculate that wild deer may be another reservoir for HEV. To date, the status of anti-HEV antibodies in wild deer has not been reported in the United States. It remains to be determined whether HEV exists in the U.S. wild deer populations and if deer may contribute to contamination of the environment with HEV and subsequent HEV transmission to humans, pigs, and other animals.
The swine bioassay indicates that HEV found in pig manure slurry was infectious to naive pigs when inoculated intravenously but not when inoculated orally. Intravenous inoculation is commonly done in HEV studies, and it has been shown previously that HEV shed from pigs inoculated intravenously was infectious to naive contact pigs (16, 23). Experimental studies indicate that fecal-oral transmission of HEV in pigs may occur; however, it likely requires a higher dose and repeated exposure for successful transmission (16, 17). The results of this study are consistent with the previous findings that experimental oral inoculation is less efficient and requires a higher virus load to induce HEV infection compared to intravenous inoculation (8, 17), since none of the 6 pigs inoculated orally became infected, whereas 2 of 6 pigs inoculated intravenously shed detectable amounts of HEV in feces. However, it needs to be noted that, although all pigs received the same total amount of inoculum, oral inoculation was only done once. Intravenous inoculation was done on 5 consecutive days to reduce the risk of adverse reactions from giving large volumes of fecal material i.v. It is possible that the frequency of inoculation (1 inoculation versus 5 inoculations) influenced the results.
It has been demonstrated that HEV titers are typically higher in feces than in blood (16). Because of this knowledge, and the additional cost of testing, viremia was not test for or confirmed in the present study. One pig inoculated intravenously with the L19 inoculum seroconverted to HEV by 56 dpi without detectable shedding of the virus in feces (Table 2). A possible explanation for this discrepancy may be that the pig did become infected and had brief localized replication of HEV in the intestines (35) and that viremia and virus shedding in feces may have been intermittent and undetected by weekly RT-PCR testing. An alternative explanation is that the virus in the L19 intravenous inoculum was able to induce an immune response in that particular pig; however, as the swine bioassay results indicate, the virus titer of 10 GE failed to induce HEV infection. It has been demonstrated that the effective titer to induce HEV infection, replication, and virus shedding in feces in pigs was 102 GE when inoculated intravenously (17).
One pig from the positive control group and one pig from the P12 intravenously inoculated (pig I) group had detectable shedding of HEV in feces, yet they remained HEV seronegative at the end of the study (Table 2). It has been previously demonstrated that a small proportion of pigs experimentally inoculated intravenously with swine HEV may have a delayed response to the infection and that development of anti-HEV antibodies may occur after the 56-day duration of the experiment (17, 25).
Partial nucleotide sequences of the PCR amplicon amplified from the P12 inocula and from 1 of the pigs (pig I) inoculated intravenously with the P12 inocula were identical and were different from the amplicon amplified from the positive-control pigs inoculated intravenously with the prototype swine HEV. This confirms that the virus found in the P12 inocula was infectious, replicated in pig I, and subsequently shed in the feces of that pig. The lack of complete nucleotide identity between the virus found in feces from pig II and the P12 inocula (and pig I) may reflect the genetic variability of different HEVs (HEV quasispecies) found in the manure slurry, since the amplicon from feces collected from pig II was different from that in the P12 manure slurry or the prototype swine HEV.
The swine bioassay findings are in accordance with a report of analysis in raw sewage from an urban area in Spain where HEV present in the biological waste facilities remained infectious to experimentally inoculated rhesus monkeys (27). Subsequently, the authors reported the presence of HEV in raw sewage from pig slaughterhouses. However, an animal bioassay was not performed to determine the infectivity of the HEV found in the raw pig sewage (28). HEV has recently been found in raw human sewage collected from industrialized countries, including the United States. According to partial nucleotide sequence analysis, the HEV found in the human sewage was more homologous to the prototype U.S. swine HEV than to the US-1 and US-2 strains of human HEV (98.4% versus 91.0%) (6).
HEV was not found in the drinking or surface water samples tested with the concentration methods used in the present study. The methods used in the current study were able to detect HEV in spiked water at a titer of 10−2 GE per ml water. The dilution of the concentrated water samples with sterile PBS and the extraction protocols using the microspin column technique have been reported to minimize the inhibition of PCR by potential inhibitors that have contaminated samples prior to PCR (2, 34). It is, therefore, possible that either the water samples tested in the study lacked HEV particles or the level of contamination by HEV was below the detection ability of the concentration methods applied in the study. It also needs to be noted that sampling was done only at one time point, thus not accounting for any seasonal effects, such as recent rainfall, that may increase the amount of fecal material in water. Colder weather may also better preserve virus. To conclude that HEV is not present in water samples, more samples collected over time and during various seasonal conditions will need to be tested.
Acknowledgments
We thank Robert H. Purcell and Suzanne U. Emerson of the National Institutes of Health, Bethesda, Maryland, for providing the Sar-55 antigen and the swine HEV inocula. We are grateful to Dennis Winter and Doug Quam for access to pig farms.
The study was supported by a grant from the National Pork Board Pork Checkoff Dollars (United States).
REFERENCES
- 1.Abbaszadegan, M., P. Stewart, and M. LeChevallier. 1999. A strategy for detection of viruses in groundwater by PCR. Appl. Environ. Microbiol. 65:444-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aggarwal, R., and K. McCaustland. 1998. Hepatitis E virus RNA detection in serum and feces specimens with the use of microspin columns. J. Virol. Methods 74:209-213. [DOI] [PubMed] [Google Scholar]
- 3.Anonymous. 1995. Detection of enteric viruses, p. 9/87-9/99. In Standard methods. U.S. Environmental Protection Agency, Government Printing Office, Cincinnati, Ohio.
- 4.Banks, M., R. Bendall, S. Grierson, G. Heath, J. Mitchell, and H. Dalton. 2004. Human and porcine hepatitis E virus strains, United Kingdom. Emerg. Infect. Dis. 10:953-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bøtner, A. 1991. Survival of Aujesky's disease virus in slurry at various temperatures. Vet. Microbiol. 29:225-235. [DOI] [PubMed] [Google Scholar]
- 6.Clemente-Casares, P., S. Pina, M. Buti, R. Jardi, M. Martin, S. Bofill-Mas, and R. Girones. 2003. Hepatitis E virus epidemiology in industrialized countries. Emerg. Infect. Dis. 9:448-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corwin, A. L., N. T. K. Tien, K. Bounlu, J. Winarno, M. P. Putri, K. Laras, R. P. Larasati, N. Sukri, T. Endy, H. A. Sulaiman, and K. C. Hyams. 1999. The unique riverine ecology of hepatitis E virus transmission in South-East Asia. Trans. R. Soc. Trop. Med. Hyg. 93:255-260. [DOI] [PubMed] [Google Scholar]
- 8.Emerson, S. U., and R. H. Purcell. 2003. Hepatitis E virus. Rev. Med. Virol. 13:145-154. [DOI] [PubMed] [Google Scholar]
- 9.Emerson, S. U., D. Anderson, A. Arankalle, X.-J. Meng, M. Purdy, G. G. Schlauder, and S. A. Tsarev. 2004. Hepevirus, p. 851-855. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, London, United Kingdom.
- 10.Halbur, P. G., C. Kasorndorkbua, C. Gilbert, D. Guenette, M. B. Potters, R. H. Purcell, S. U. Emerson, T. E. Toth, and X. J. Meng. 2001. Comparative pathogenesis of infection of pigs with hepatitis E viruses recovered from a pig and a human. J. Clin. Microbiol. 39:918-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haqshenas, G., H. L. Shivaprasad, P. R. Woolcock, D. H. Read, and X. J. Meng. 2001. Genetic identification and characterization of a novel virus related to human hepatitis E virus from chickens with hepatitis-splenomegaly syndrome in the United States. J. Gen. Virol. 82:2449-2462. [DOI] [PubMed] [Google Scholar]
- 12.He, J., B. L. Innis, M. P. Shrestha, E. T. Clayson, R. M. Scott, K. J. Linthicum, G. G. Musser, S. C. Gigliotti, L. N. Binn, R. A. Kuschner, and D. W. Vaughn. 2002. Evidence that rodents are a reservoir of hepatitis E virus for humans in Nepal. J. Clin. Microbiol. 40:4493-4498. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Huang, F. F., G. Haqshenas, D. K. Guenette, P. G. Halbur, S. K. Schommer, F. W. Pierson, T. E. Toth, and X. J. Meng. 2002. Detection by reverse transcription-PCR and genetic characterization of field isolates of swine hepatitis E virus from pigs in different geographic regions of the United States. J. Clin. Microbiol. 40:1326-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kabrane-Lazizi, Y., J. B. Fine, J. Elm, G. E. Glass, H. Higa, A. Diwan, C. J. Gibbs, Jr., X.-J. Meng, S. U. Emerson, and R. H. Purcell. 1999. Evidence for widespread infection of wild rats with hepatitis E virus in the United States. Am. J. Trop. Med. Hyg. 61:331-335. [DOI] [PubMed] [Google Scholar]
- 15.Kasorndorkbua, C., B. J. Thacker, P. G. Halbur, D. K. Guenette, R. M. Buitenwerf, R. L. Royer, and X.-J. Meng. 2003. Experimental infection of pregnant gilts with swine hepatitis E virus. Can. J. Vet. Res. 67:303-306. [PMC free article] [PubMed] [Google Scholar]
- 16.Kasorndorkbua, C., D. K. Guenette, F. F. Huang, P. J. Thomas, X.-J. Meng, P. G. Halbur. 2004. Routes of transmission of swine hepatitis E virus in pigs. J. Clin. Microbiol. 42:5047-5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasorndorkbua, C., P. G. Halbur, P. J. Thomas, D. K. Guenette, T. E. Toth, and X. J. Meng. 2002. Use of a swine bioassay and a RT-PCR assay to assess the risk of transmission of swine hepatitis E virus in pigs. J. Virol. Methods 101:71-78. [DOI] [PubMed] [Google Scholar]
- 18.Krawczynski, K., K. McCaustland, E. Mast, P. O. Yarbough, M. Purdy, M. O. Favorov, and J. Spellbring. 1996. Elements of pathogenesis of HEV infection in man and experimentally infected primates, p. 317-328. In Y. Buisson, P. Coursaget, M. Kane (ed.), Enterically transmitted hepatitis viruses. La Simarre, Tours, France.
- 19.Kumar, A., M. Beniwal, P. Kar, J. B. Sharma, and N. S. Murthy. 2004. Hepatitis E in pregnancy. Int. J. Gynaecol. Obstet. 85:240-244. [DOI] [PubMed] [Google Scholar]
- 20.Matsubayashi, K., Y. Nagaoka, H. Sakata, S. Sato, K. Fukai, T. Kato, K. Takahashi, S. Mishiro, M. Imai, N. Takeda, and H. Ikeda. 2004. Transfusion-transmitted hepatitis E caused by apparently indigenous hepatitis E virus strain in Hokkaido, Japan. Transfusion 44:934-940. [DOI] [PubMed] [Google Scholar]
- 21.Matsuda, H., K. Okada, K. Takahashi, and S. Mishiro. 2003. Severe hepatitis E virus infection after ingestion of uncooked liver from a wild boar. J. Infect. Dis. 188:944. [DOI] [PubMed] [Google Scholar]
- 22.Meng, X. J., B. Wiseman, F. Elvinger, D. K. Guenette, T. E. Toth, R. E. Engle, S. U. Emerson, and R. H. Purcell. 2002. Prevalence of antibodies to hepatitis E virus in veterinarians working with swine and in normal blood donors in the United States and other countries. J. Clin. Microbiol. 40:117-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meng, X.-J., P. G. Halbur, J. S. Haynes, T. S. Tsareva, J. D. Bruna, R. L. Royer, R. H. Purcell, and S. U. Emerson. 1998. Experimental infection of pigs with the newly identified swine hepatitis E virus (swine HEV), but not with human strains of HEV. Arch. Virol. 143:1405-1415. [DOI] [PubMed] [Google Scholar]
- 24.Meng, X.-J., P. G. Halbur, M. S. Shapiro, S. Govindarajan, J. D. Bruna, I. K. Mushahwar, R. H. Purcell, and S. U. Emerson. 1998. Genetic and experimental evidence for cross-species infection by swine hepatitis E virus. J. Virol. 72:9714-9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng, X.-J., R. H. Purcell, P. G. Halbur, J. R. Lehman, D. M. Webb, T. S. Tsareva, J. S. Haynes, B. J. Thacker, and S. U. Emerson. 1997. A novel virus in swine is closely related to the human hepatitis E virus. Proc. Natl. Acad. Sci. USA 94:9860-9865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng, X. J., S. Dea, R. E. Engle, R. Friendship, Y. S. Lyoo, T. Sirinarumitr, K. Urairong, D. Wang, D. Wong, D. Yoo, Y. Zhang, R. H. Purcell, and S. U. Emerson. 1999. Prevalence of antibodies to the hepatitis E virus (HEV) in pigs from countries where hepatitis E is common or rare in the human population. J. Med. Virol. 59:297-302. [PubMed] [Google Scholar]
- 27.Pina, S., J. Jofre, S. U. Emerson, R. H. Purcell, and R. Girones. 1998. Characterization of a strain of infectious hepatitis E virus isolated from sewage in an area where hepatitis E is not endemic. Appl. Environ. Microbiol. 64:4485-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pina, S., M. Buti, M. Cotrina, J. Piella, and R. Girones. 2000. HEV identified in serum from humans with acute hepatitis and in sewage of animal origin in Spain. J. Hepatol. 33:826-833. [DOI] [PubMed] [Google Scholar]
- 29.Piper-Jenks, N., H. W. Horowitz, and E. Schwartz. 2000. Risk of hepatitis E infection to travelers. J. Travel Med. 7:194-199. [DOI] [PubMed] [Google Scholar]
- 30.Purcell, R. H., and S. U. Emerson. 2001. Hepatitis E virus, p. 3051-3061. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 31.Schlauder, G. E., and I. K. Mushahwar. 2001. Genetic heterogeneity of hepatitis E virus. J. Med. Virol. 65:282-292. [DOI] [PubMed] [Google Scholar]
- 32.Smith, J. L. 2001. A review of hepatitis virus. J. Food Prot. 64:572-586. [DOI] [PubMed] [Google Scholar]
- 33.Tei, S., N. Kitajima, K. Takahashi, and S. Mishiro. 2003. Zoonotic transmission of hepatitis E virus from deer to human beings. Lancet 362:371-373. [DOI] [PubMed] [Google Scholar]
- 34.Wilde, J., J. Eiden, and R. Yolken. 1990. Removal of inhibitory substances from human fecal specimens for detection of group A rotaviruses by reverse transcriptase and polymerase chain reactions. J. Clin. Microbiol. 28:1300-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams, T. P. E., C. Kasorndorkbua, P. G. Halbur, G. Haqshenas, D. K. Guenette, T. E. Toth, and X. J. Meng. 2001. Evidence of extrahepatic sites of replication of the hepatitis E virus in a swine model. J. Clin. Microbiol. 39:3040-3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu, J.-C., C.-M. Chen, T.-Y. Chiang, W.-H. Tsai, W.-J. Jeng, I.-J. Sheen, C.-C. Lin, and X.-J. Meng. 2002. Spread of hepatitis E virus among different-aged pigs: two-year survey in Taiwan. J. Med. Virol. 66:488-492. [DOI] [PubMed] [Google Scholar]
- 37.Yazaki, Y., H. Mizuo, M. Takahashi, T. Nishizawa, N. Sasaki, Y. Gotanda, and H. Okamoto. 2003. Sporadic acute or fulminant hepatitis E in Hokkaido, Japan, may be food-borne, as suggested by the presence of hepatitis E virus in pig liver as food. J. Gen. Virol. 84:2351-2357. [DOI] [PubMed] [Google Scholar]
- 38.Zafrullah, M., Z. Khursheed, S. Yadav, D. Sahgal, S. Jameel, and F. Ahmad. 2004. Acidic pH enhances structure and structural stability of the capsid protein of hepatitis E virus. Biochem. Biophys. Res. Commun. 313:67-73. [DOI] [PubMed] [Google Scholar]