Abstract
The Ras family of GTPase proteins has been shown to control morphogenesis in many organisms, including several species of pathogenic fungi. In a previous study, we identified a gene encoding a fungus-specific Ras subfamily homolog, rasB, in Aspergillus fumigatus. Here we report that deletion of A. fumigatus rasB caused decreased germination and growth rates on solid media but had no effect on total biomass accumulation after 24 h of growth in liquid culture. The ΔrasB mutant had an irregular hyphal morphology characterized by increased branching. Expression of rasBΔ113-135, a mutant transgene lacking the conserved rasB internal amino acid insertion, did not complement the deletion phenotype of delayed growth and germination rates and abnormal hyphal morphology. Virulence of the rasB deletion strain was diminished; mice infected with this strain exhibited ∼65% survival compared to ∼10% with wild-type and reconstituted strains. These data support the hypothesis that rasB homologs, which are highly conserved among fungi that undergo hyphal growth, control signaling modules important to the directional growth of fungal hyphae.
Aspergillus fumigatus is the predominant mold pathogen of immunosuppressed patient populations (12). This organism inhabits soil and organic debris where it produces conidia, the infective propagules that are disseminated by aerosolization (12). In order for A. fumigatus conidia to cause invasive aspergillosis, inhaled conidia must undergo processes that are essential to the establishment and progression of disease and to growth mechanisms common to all filamentous fungi. First, the conidia must complete germination, a process that involves isotropic growth, mitosis, and emergence of the initial germ tube. In order to continue growth and consequently invade tissue, the nascent germling must elongate by a process termed apical extension. The result of this growth process is long, tube-like hyphae, the characteristic morphology of filamentous fungi. The molecular mechanisms that control germination and apical extension and their contribution to virulence in A. fumigatus remain unknown.
Homologs of the Ras family of GTPase proteins have been shown to contribute to morphology and virulence in several pathogenic fungi (14). Control of morphogenetic pathways by Ras proteins can be achieved by several different mechanisms. For example, the RSR1 homologs of Saccharomyces cerevisiae (11), Candida albicans (23), and Ashbya gossypii (3) are hypothesized to control components of the polarisome directly by effecting their recruitment and/or stabilization at the site of polarization. Control of a wider range of developmental processes by Ras homlogs can be seen in both Aspergillus nidulans (6, 18, 21) and A. fumigatus (7). The rasA gene product from both of these filamentous fungi has been implicated in controlling events in germination, including mitosis, as well as in completion of the asexual developmental cycle. Although the mechanism is unknown, the Neurospora crassa NC-ras2 gene, a rasB homolog, has also been shown to regulate apical growth of hyphae, cell wall biosynthesis, and conidia formation (10). Among the previously studied pathogenic fungi, Ras subfamily activity has been shown to be essential for wild-type virulence in Cryptococcus neoformans (RAS1) (2), C. albicans (CaRAS1 and CaRSR1) (13, 23), and A. fumigatus (rhbA) (20).
Here we report that a strain lacking the A. fumigatus rasB gene is viable but reveals a profound growth defect that is more severe than previously reported for a strain expressing a dominant-negative (DN) rasB allele (7). The aberrant growth processes, caused by the ΔrasB mutation, lead to decreased virulence in a mouse model of invasive aspergillosis.
MATERIALS AND METHODS
A. fumigatus strains and growth conditions.
All strains were maintained on and harvested from Aspergillus minimal medium (AMM) agar plates, modified to contain 10 mM ammonium tartrate as the nitrogen source (5). Submerged cultures for determination of biomass and germination rates were grown in YG (1% yeast extract-2% glucose) medium as indicated. Determination of total biomass was performed by inoculating 104 conidia into preweighed, sterile 50-ml centrifuge tubes containing 5 ml of YG medium. After 24 and 48 h of growth at 37°C with shaking at 250 rpm, each tube was frozen in an ethanol-dry-ice bath and then lyophilized for 24 h before a final dry weight was recorded. Germination experiments were performed by inoculating coverslip cultures with 105 conidia/ml in YG medium. The numbers of swollen conidia initiating a germ tube were enumerated at 15-min intervals. For growth rate analysis, 104 conidia were spotted in the center of AMM agar plates, and the change in colony diameter was monitored over 48 h. All experiments were performed in triplicate.
Construction of the rasB deletion strain.
In order to delete rasB, a deletion construct was designed to replace the entire rasB coding region with a hygromycin resistance cassette by homologous recombination. PCR Primers were designed to amplify a 3-kb region 5′ of the rasB predicted ATG start codon (5′-AAGACTGAGAATACTACC-3′ and 5′-AATAGCTCTAGACGCACCCGTAGGTCACC-3′) and a 3-kb region 3′ of the predicted rasB stop codon (5′-TTATCGTCTAGAGAACACATTAGCATTCGC-3′ and 5′-ACCCTAGCATGCCGAACAGGACCGTCGTGGC-3′). These PCR fragments were separately cloned into the pGEM T-Easy cloning vector (Promega) to serve as flanking regions for the recombination event. The 3′ flanking arm was then added to the cloned 5′ arm as an XbaI/SpeI fragment. An SalI/XbaI hygromycin resistance cassette from pAN7-1 was then cloned into the center of the 5′ and 3′ flanking regions, completing the deletion construct. A complementation vector was designed to undergo a single crossover integration adjacent to the newly disrupted gene locus. This vector was built by subcloning a 3-kb PCR fragment of genomic sequence containing 1 kb of rasB promoter, the entire rasB genomic sequence, and 1 kb of downstream flanking sequence into pGEM T-Easy. A phleomycin resistance cassette was subcloned into this vector as a KpnI/AgeI fragment from pBCphleo into the KpnI/AgeI restriction sites found 3′ of the rasB coding sequence. Both vectors were linearized at a unique XmnI site in the backbone and used for protoplast transformation, as previously described (7). In order to confirm that rasB deletion and reconstitution were achieved, Southern blotting was performed on BamHI-digested genomic DNA, using a fragment of the rasB promoter sequence as the probe.
Deletion of the rasB internal amino acid insertion.
In order to create the rasBΔ113-135 mutant, an overlap PCR technique was used to delete the nucleotides encoding the internal amino acid insertion from cloned rasB cDNA. The overlap PCR involved three amplification steps. In step 1, the area 5′ of the internal insertion from the rasB start codon to position 336 was amplified with primers 5′-ATCGTTCATATGTCGGGGAAAATGACGTTG-3′ (NdeI site in bold) and 5′-TGGGCTGACTCCTTGACC-3′. Step 2 involved amplification of the area 3′ of the insertion from position 406 to the rasB stop codon, with primers 5′-GGTCAAGGAGTCAGCCCACGTCCCCGTGATGCTAGTCG-3′ and 5′-ATTATCGCGGCCGCATGGATGTTCGTGGACGC-3′ (NotI in bold). In step 3, the PCR products from steps 1 and 2 were pooled and fused through PCR employing the primers 5′-ATCGTTCATATGTCGGGGAAAATGACGTTG-3′ and 5′-ATTATCGCGGCCGCATGGATGTTCGTGGACGC-3′. Fusion of products 1 and 2 relied on the overlapping region, shown above as underlined sequence. The rasBΔ113-135 cDNA (now a NdeI/NotI fragment) was cloned into TOPO 2.1 (Invitrogen), using T/A cloning, and sequenced (University of Cincinnati DNA Core Facility). Upon alignment with the wild-type rasB cDNA sequence, the sequence of the selected clone revealed that only the nucleotides encoding the insertion were deleted and that no other mutations were introduced. The rasBΔ113-135 cDNA was then cloned under the control of the rasB endogenous promoter (1 kb upstream of the rasB ATG) in a vector carrying the phleomycin resistance cassette. This new expression vector was then linearized by XmnI digestion and transformed into ΔrasB strain protoplasts. Ectopic integration of a single copy of rasBΔ113-135 was confirmed by Southern blot analysis, and expression was confirmed by reverse transcription-PCR (data not shown).
Microscopy procedures and image analysis.
Specimens grown in liquid culture for microscopic analysis were grown for the times indicated and mounted in 50% glycerol. Microscopy was performed on an Olympus AX80 microscope equipped with a digital camera. Digital images were analyzed using Magnafire 2.1 and Photoshop 6.0 software. Bright-field microscopy was performed on an Olympus BH-2 microscope.
Murine model of invasive aspergillosis.
Strain CF-1 mice (Charles River) were immunosuppressed with a single dose of cyclophosphamide (150 mg/kg of body weight injected intraperitoneally on day −3) followed by a single dose of triamcinolone acetonide (40 mg/kg of body weight injected subcutaneously in the nape of the neck on day −1). The mice were anesthetized with 3.5% isofluorane and inoculated intranasally with 105 conidia in 20 μl of sterile saline on day 0. Mortality was monitored for the next 14 days, and mice that appeared moribund were sacrificed by CO2 euthanasia. The lungs, kidneys, and brain of all mice were plated onto Inhibitory Mold Agar (Becton Dickinson), and the genotypes of isolates were confirmed by genomic Southern blot analysis. Statistical significance was assessed by Kruskall-Wallis analysis, and pairwise analysis was performed post hoc by Dunn's procedure using SigmaStat software.
Analysis of fungal burden by QPCR.
To quantitate fungal burden, strain CF-1 mice were immunosuppressed with a single dose of triamcinolone acetonide (40 mg/kg of body weight injected subcutaneously in the nape of the neck on day −1) and then given a sublethal dose (5 × 104 conidia) of each strain by intranasal inoculation. On days 0, 2, 4, and 6 postinoculation, three mice from each group were sacrificed, and 100 mg of tissue was removed from the upper right lung and processed following a modified FastDNA prep kit (Biogene) protocol. In short, fresh lung tissue was suspended in 1 ml of buffer CLS-TC (a cell lysis solution used for animal tissues and bacteria) containing 20 μg/ml proteinase K. Samples were subjected to mechanical disruption in a Mini-BeadBeater (Biospec Products)for 30 sec at 50 rpm. This treatment was applied three times for each sample, with cooling on ice between disruptions. The remaining DNA extraction, washing, and elution were performed according to the manufacturer's protocol. Samples were then analyzed by a modification of a previously described Molecular Beacon quantitative-PCR (QPCR) assay (4). The A. fumigatus ITS2 region and the murine GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene were used as amplification targets in the QPCR assay, and a plasmid containing the S. cerevisiae PMA1 gene was employed as an internal control.
Calculation of average lesion area.
Image analysis was performed as previously described (20). In short, the average lesion area was determined by using tissue sections of the lower left lung from mice inoculated with the isogenic set that were sacrificed at days 2, 4, and 6 postinoculation. Tissue was fixed in 10% buffered formalin and submitted for processing (University of Cincinnati Comparative Pathology facility). Images of silver-stained lesions were captured with an Olympus AX80 microscope equipped with a digital camera, and the lesion area was measured using ProImage image analysis software. The lesion border was defined as the perimeter of the region containing visual fungal mass, not including surrounding tissue damage. All lesions present in three consecutive 5-μm sections from each of three mice per strain were measured. The data were analyzed by one-way analysis of variance. Pairwise analysis was performed post hoc by using the Holm-Sidak method. Fungal burden was determined by dividing the sum of all lesion areas in each section by the total area of the section.
RESULTS
A. fumigatus rasB is nonessential.
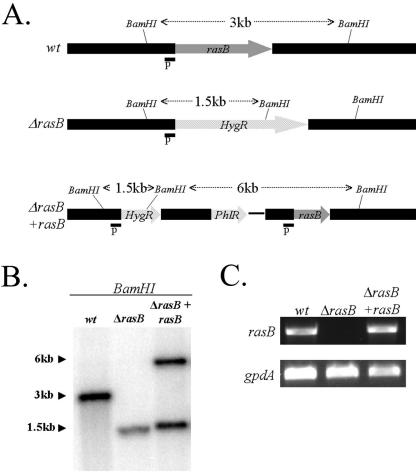
Previous studies revealed that expression of a constitutively inactive rasB mutant in a wild-type A. fumigatus background led to a decrease in germination and growth rates as well as abnormal hyphal morphology (7). However, since this phenotype relies on the balance between the dominant-negative mutant and wild-type RasB proteins, we sought to create a strain of A. fumigatus lacking rasB. Using standard Aspergillus transformation techniques, the entire rasB coding region was replaced with a deletion cassette containing the hygromycin resistance marker (Fig. 1A). In order to ensure that all phenotypes noted in the rasB deletion strain (ΔrasB) were due to the specific targeting of rasB, the ΔrasB strain was reconstituted by reintroduction of the genomic rasB sequence directly adjacent to the mutated locus (Fig. 1A). Deletion and reconstitution of the rasB coding sequence (yielding the ΔrasB+rasB strain) were verified by Southern blot analysis (Fig. 1B). This method revealed a 3-kb fragment for the wild type, a smaller 1.5-kb fragment for the ΔrasB strain, and both a 1.5-kb ΔrasB fragment plus a 6-kb fragment from the complementation construct. These variations in band sizes were expected due to the introduction of new BamHI sites from the hygromycin and phleomycin resistance cassettes. In addition, reverse transcription-PCR employing gene-specific primers confirmed the loss and reappearance of rasB messenger RNA in the ΔrasB and ΔrasB+rasB strains, respectively, (Fig. 1C).
FIG. 1.
Deletion of A. fumigatus rasB. (A) Graphical representation of the rasB genomic locus from the wild-type, ΔrasB, and reconstituted strains (ΔrasB+rasB). Probes (P) used for Southern blotting are shown as small black boxes. Relative positions of the BamHI restriction sites are given. (B) Genomic Southern blot of wild-type, ΔrasB, and ΔrasB+rasB DNA digested with BamHI. (C) Reverse transcription-PCR of the isogenic set, showing loss and recovery of rasB message RNA in the ΔrasB and reconstituted strains, respectively. The loading control is A. fumigatus gpdA.
A. fumigatus rasB regulates germination and radial growth.
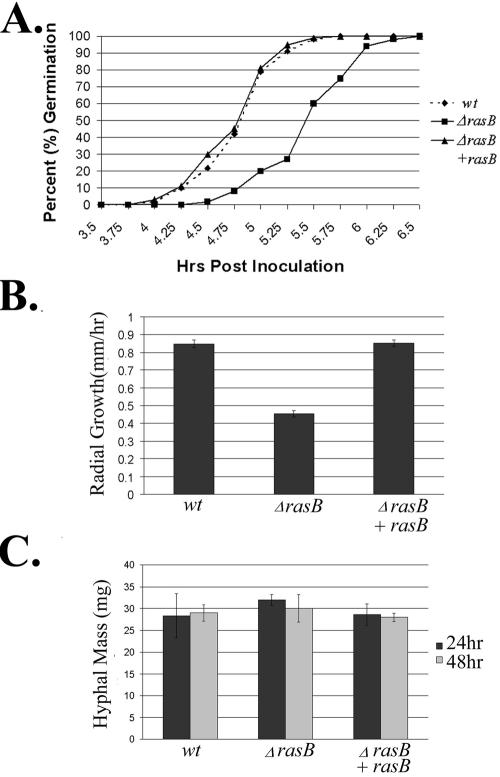
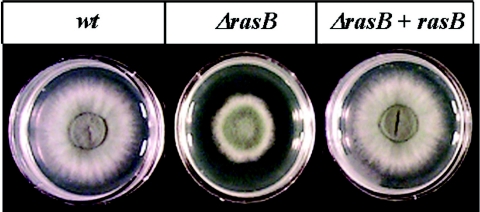
Because expression of a constitutively inactive rasB caused aberrations in growth and germination rates in a wild-type background (7), we sought to quantify both of these developmental processes in a strain completely devoid of rasB activity. In medium containing glucose as the carbon source, the ΔrasB mutant displayed delayed germination, characterized by an approximately 45-min lag in initiation. After the initial lag, the rate of germination for all strains appeared similar, with the wild-type and reconstituted strains reaching 100% germination after 5.5 h and the ΔrasB mutant reaching completion within 6.5 h (Fig. 2A). As measured by a solid agar growth assay, radial outgrowth of the ΔrasB mutant was also decreased to 50% that of the wild-type or reconstituted strains between 24 and 48 h (Fig. 2B). Due to the slower outgrowth, the ΔrasB mutant forms a denser colony with a morphology that displays a decreased peripheral growth zone and irregular borders (Fig. 3). Because the ΔrasB colony morphology is more compact, we also measured total biomass generated by each strain in submerged culture. Surprisingly, although the germination and growth rates were both delayed for the ΔrasB mutant strain, the total hyphal mass in liquid culture was not significantly different from the wild type after 24 or 48 h of growth (Fig. 2C).
FIG. 2.
Germination, radial growth, and biomass of the isogenic set. (A) Graphic representation of germination rates from the wild-type (♦), ΔrasB (▪), and ΔrasB+rasB (▴) strains. Conidia from each strain were inoculated onto coverslips immersed in YG medium at 37°C. (B) Radial growth rates. Conidia (104) from each strain were inoculated onto AMM solid agar and incubated for 48 h at 37°C. (C) Comparison of the total biomass of the isogenic set grown for 24 or 48 h at 37°C.
FIG. 3.
Colony morphology of the wild-type, ΔrasB, and reconstituted strains.
A. fumigatus rasB regulates hyphal development.
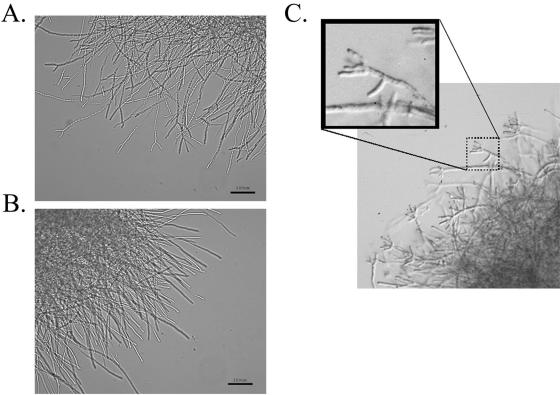
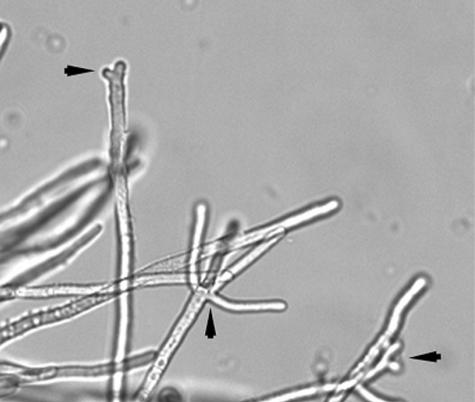
The initial steps in development of the ΔrasB strain, though temporally slowed, were morphologically similar to the wild-type strain. These steps include isotropic growth, subsequent initiation of mitosis, and formation of the initial germ tube (data not shown). However, after 12 h of submerged growth, the hyphal morphology of the ΔrasB mutant displayed a hyper-branching phenotype, characterized by increased apical branching (Fig. 4A) that was not seen in the wild type (Fig. 4B). Incubations in submerged culture for 24 h revealed the development of exacerbated apical branching (Fig. 4C). These phenotypes were not observed in the wild-type or reconstituted strains grown under identical conditions (data not shown).
FIG. 4.
Hyphal morphology of the ΔrasB mutant. (A) The ΔrasB mutant grown for 12 h at 37°C with shaking at 250 rpm. (B) Wild-type A. fumigatus grown for 12 h at 37°C and 250 rpm. (C) Enlarged image of the ΔrasB strain grown for 24 h in YG medium at 37°C and 250 rpm.
The internal amino acid insertion is essential to RasB function.
To determine the importance of the conserved amino acid insertion to the function of the rasB gene product, a rasB mutant was created in which the nucleic acid sequence encoding the insertion was deleted by overlap PCR. The mutant cDNA, rasBΔ113-135, was introduced into the ΔrasB background as a single-copy, ectopic integration with expression driven by the endogenous rasB promoter. This mutant leaves the effector domain, the GTP/GDP binding domains, and the CAAX motif intact (Fig. 5). Although expression of the rasBΔ113-135 gene was confirmed by reverse transcription-PCR, it was unable to complement the mutant phenotype in terms of germination, radial growth rates, or colony morphology (data not shown). Moreover, the strain expressing rasBΔ113-135 continued to display the apically branched phenotype that was characteristic of the ΔrasB mutant (Fig. 6).
FIG. 5.
Predicted protein sequences of the RasBΔ113-135 gene product. Sequence alignment of the predicted protein sequences for RasB (top line) and RasBΔ113-135 (bottom line). Conserved Ras protein domains are indicated above the sequences.
FIG. 6.
Phenotype of the rasBΔ113-135 mutant. Hyphal morphology of the rasBΔ113-135 strain after 12 h of growth at 37°C with shaking at 250 rpm. Arrowheads indicate sites of multiple branching.
A. fumigatus rasB is necessary for wild-type virulence.
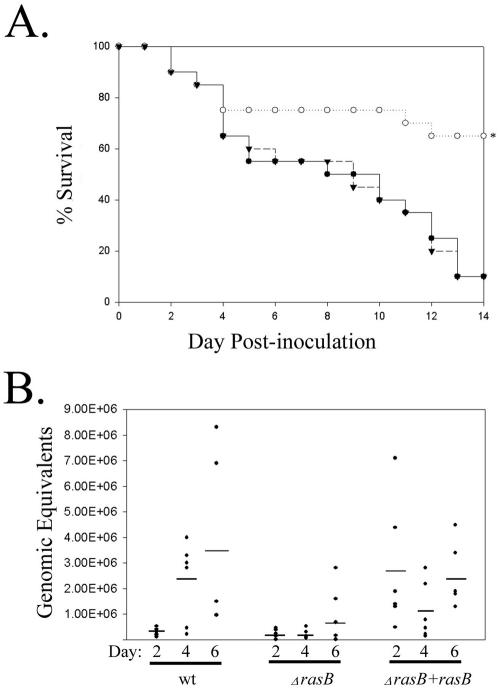
To determine the contribution of RasB activity to the virulence of A. fumigatus, freshly harvested conidia from the wild-type, ΔrasB, and the reconstituted strains were inoculated into immunocompromised mice. The mice were then monitored for mortality for 14 days postinoculation. Although early mortality was nearly identical among all groups, the remaining mice receiving the deletion mutant survived significantly longer than those in the other groups (Fig. 7A). There was no significant difference between the mortality in the wild-type and reconstituted groups, and no mortality was observed in saline controls. Notably, of the surviving mice that received the ΔrasB strain, only 30% still harbored viable fungus at the end of the experiment. In all of these mice, the presence of fungus was confined to the lungs. In contrast, all of the surviving mice that received either the wild-type or reconstituted strain harbored viable fungus in multiple organs (data not shown).
FIG. 7.
Reduced virulence and fungal burden of the ΔrasB strain. (A) Survival plot comparison of the wild-type (▪), ΔrasB (○), and ΔrasB+rasB (▾) strains employed in a murine model of invasive aspergillosis. (B) QPCR analysis of fungal burden at days 2, 4, and 6 in mice inoculated with the isogenic set. Genomic equivalents were calculated from a standard curve generated from A. fumigatus DNA and normalized to mouse GAPDH. Dots represent duplicate QPCR results for each of three mice per group per day. Bars represent the average genomic equivalents for all mice at each time point.
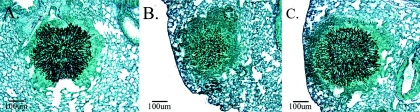
In order to determine the overall fungal burden for each isolate, a separate experiment was performed in which immunosuppressed mice were given a sublethal dose of conidia, and three mice per group were sacrificed at days 0, 2, 4, and 6 postinoculation. The fungal burden of the lung was then assessed by two methods, QPCR and morphometry. Total DNA extracted from lung tissue of mice infected with members of the isogenic set was analyzed by QPCR, targeting the ITS2 region (Fig. 7B). Although the variability in each group prevented the differences from reaching significance, the QPCR data showed a definite trend toward more fungus, as measured in genome equivalents, in the lungs of mice infected with the wild-type and reconstituted strains in comparison to ΔrasB strain-infected animals (Fig. 7B). The average lesion area produced by infection with each strain was also compared by morphometric analysis of silver-stained lung sections. In mice infected with the ΔrasB strain, the average lesion area remained low throughout the time course, whereas the average areas in the wild-type and reconstituted groups were significantly higher at days 4 and 6 postinoculation (Table 1). Tissue invasion was evident for each strain at all three time points (Fig. 8).
TABLE 1.
Average lesion area
| Strain | Avg area (RU ± SEM) by no. of days postinoculation (n)a
|
||
|---|---|---|---|
| 2 | 4 | 6 | |
| Wild type | 46 ± 9 (10) | 777 ± 105 (10) | 2,926 ± 753 (9) |
| ΔrasB | 44 ± 5 (10) | 98 ± 24 (8)b | 364 ± 44 (11)b |
| ΔrasB + rasB | 66 ± 9 (10) | 846 ± 124 (9) | 2,973 ± 714 (10) |
RU, relative units; n, number of lesions per group used to quantitate the area.
P < 0.01.
FIG. 8.
Representative day 6 lesions from the wild-type (A), ΔrasB (B) and ΔrasB+rasB (C) strains. Tissue sections were stained with Grocott silver stain.
DISCUSSION
A. fumigatus rasB is a member of the Ras subfamily of GTP-binding proteins. We previously showed that RasB homologs are restricted to fungi that grow as hyphae during some part of their normal life cycle (7). The rasB homologs form a separate group of Ras proteins that is characterized by the presence of a unique ∼20-amino-acid insertion that encodes a conserved domain of unknown function. In our previous work on rasB, a DN rasB allele was expressed in a wild-type background. In order to delineate more clearly the function of RasB, a null mutant was produced. Deletion of rasB resulted in exaggeration of the phenotypes we obtained by expressing a DN rasB allele. For example, expression of a DN rasB mutant in a wild-type background caused a 20-min lag in the initiation of germination (7). In comparison, germ tube production in the ΔrasB strain was delayed by nearly 45 min. Once germination began, however, the rates of both the DN rasB and ΔrasB strains were the same as the rate of the wild type. Radial growth on solid agar was decreased by 50% in the ΔrasB strain, resulting in a colony that displayed a decreased peripheral growth zone and scalloped borders. Interestingly, although radial growth was markedly reduced in the ΔrasB mutant, the total biomass of the wild-type, ΔrasB, and reconstituted strains did not differ after 24 or 48 h of submerged culture. These findings are similar to those reported for a Trichoderma virens strain lacking TmkA, a mitogen-activated protein kinase involved in biocontrol properties and conidiation. Deletion of TmkA in T. virens produces a mutant with a decreased radial growth rate yet no change in germination rates or rates of increase in biomass in comparison to the wild-type T. virens strain (17). These seemingly contradictory data may be due to actual production of a similar hyphal mass that is represented in a more compact colony morphology for the ΔrasB mutant. Conversely, the growth characteristics of the different strains may vary in liquid culture, lessening the effect of the decreased radial growth rate observed on solid agar.
Production of hyphae that undergo increased apical branching, or tip-splitting, was another phenotype noted in the rasB deletion strain. In results reported here, the ΔrasB strain, after 12 h of growth in submerged culture, displayed numerous apically branching hyphae. Allowing this mutant to grow for 24 h in submerged culture caused exacerbated apical branching and produced structures that resembled aberrant conidiophores. Conidiation in submerged culture for Aspergillus species is normally considered a starvation phenotype (1). We previously showed that expression of DN rasB in a wild-type background of A. fumigatus resulted in formation of conidiophores that were able to produce conidia in liquid culture (7). In contrast, no conidia were produced by the structures present in the ΔrasB mutant. Discordant results between phenotypes obtained by expressing an inactive mutant compared with phenotypes of a null mutant are not unusual. For example, the mitogen-activated protein kinases of S. cerevisiae, Kss1p and Fus1p, have inhibitory functions that are independent of their kinase activities (15). These regulatory functions were elucidated when the phenotypes of the kinase mutants were compared with those of the null mutants. In the case of RasB function in submerged conidiation, perhaps the DN rasB product continues to play a structural role that enables inappropriate activation of the conidiation pathway.
The conserved amino acid insertion, found exclusively among rasB homologs, represents an intriguing area of RasB function. Extensive searches of protein databases failed to provide information that might suggest a function for this domain. However, the possibility of regulation by phosphorylation is suggested by the presence of predicted phosphorylation sites at amino acids S113, S115, and S120 (data not shown). Our data suggest that the conserved RasB internal domain is required for RasB function because a mutant lacking this region was unable to complement the ΔrasB phenotypes of reduced growth rate, delayed germination rates, and abnormal colony and hyphal morphology. Also, comparisons of predicted RasB and RasBΔ113-135 protein structures by the Swiss-Model database identified no predicted changes introduced by the deletion of the internal insertion (data not shown). This raises the possibility that amino acids 113 to 135 are essential to the function of RasB rather than the loss of function of RasBΔ113-135 being due to changes in the protein structure. Further structure-function studies on this region will be necessary to elucidate its role in RasB function and/or regulation.
Although the ΔrasB mutant had reduced radial growth on plates, its growth rate in liquid culture was indistinguishable from the wild type. Since the overall growth rate of A. fumigatus would be expected to affect fungal burden in the host (19), we tested the virulence of this mutant in a mouse model of invasive aspergillosis. Although it may be assumed that a decreased radial growth rate would lead to less total mass and less tissue invasion, this may not always be the case. For example, the tmkA mutant of T. virens, having a wild-type biomass, displays decreased virulence compared to wild-type T. virens (17). Mice infected with the ΔrasB strain had fewer deaths and smaller average lesion size and appeared to harbor lower fungal burdens. These findings indicate that, at least in this case, growth on solid agar was a better indicator of in vivo growth potential for this isogenic set.
The reduction in virulence seen in the ΔrasB mutant appears to be the result of reduced radial growth in the absence of RasB. The identification of genes that function in polarized growth is not foreign to the filamentous fungi, and, in fact, many genes are now considered to be polarity-related genes (reviewed in references 8 and 16). For example, the Ashbya gossypii GTPase proteins AgCdc42, AgRho1 and AgRho3 play various roles in the polarized growth process. AgCdc42 has been shown to play an early role in the establishment of polarity, defined by the loss of germ tube emergence in an AgCdc42 deletion strain (16, 22). In contrast, AgRho1 and AgRho3 are involved in polarity maintenance through the stabilization of cell wall integrity and the coordination of actin assembly (16, 22). Some polarity genes also affect the normal branching patterns of filamentous fungi, as seen in the A. fumigatus ΔrasB mutant. In A. nidulans, a formin, SepA, and a septin, AspB, have been shown to localize to sites of branch emergence (reviewed in reference 16). However, deletion of either of these genes causes different aberrant branching patterns in the resultant A. nidulans mutant. Deletion of sepA causes a tip-splitting/apical-branching phenotype, whereas aspB deletion causes increased subapical branching (16).
It is enticing to speculate that the rasB homologs may also be polarity-related genes, signaling through conserved pathways that directly control multiple aspects of apical growth, including polarity establishment, maintenance, and branching. Perhaps even more interesting is that the data presented here, relating polarized growth to pathogenesis, emphasize the importance of delineating growth dynamics in the filamentous fungi. Only two Ras subfamily members among the filamentous fungi have been implicated in polarized growth: N. crassa NC-ras2, a homolog of A. fumigatus rasB, and RSR1. The RSR1 genes of S. cerevisiae, C. albicans, and Ashbya gossypii are other Ras-family homologs that are also known to play a role in apical growth. In S. cerevisiae, RSR1 is known as the bud-site selector that functions in normal budding patterns and is essential for pseudohyphal growth (11). The RSR1 of Ashbya gossypii plays a slightly different role; it appears to stabilize polarisome components at the actively growing tip in this filamentous fungus (3). The CaRSR1 gene of C. albicans is also involved in bud-site selection and is necessary for full virulence (23). In fact, recent data have shown that CaRSR1 may play a role similar to AgRSR1 during the hyphal and invasive growth of C. albicans (9). Homology searches of the available genome databases reveal that A. fumigatus does not have a typical RSR1 homolog and that homologs of RSR1 and rasB may be mutually exclusive. In fact, studies in our laboratory have revealed that heterologous expression of A. fumigatus rasB in an S. cerevisiae RSR1 deletion strain does not correct the randomized budding pattern of that mutant (data not shown). Perhaps this is not surprising since the proteins are fairly dissimilar from a sequence homology standpoint (32% homology). However, based on the RSR1 data compiled from these other organisms, rasB may be hypothesized to serve an RSR1-type function in recruiting polarisome components to the apical compartment and/or promoting their stabilization at the hyphal apex in organisms that contain a RasB homolog but no RSR1. Such a function for A. fumigatus RasB would explain the decreased germination rates, slowed radial growth, and increased tip-splitting that develop in the ΔrasB mutant. Experiments to examine the intracellular localization and interacting partners of RasB will be required to test this hypothesis.
In summary, the data presented here suggest that RasB plays a role in hyphal tip maintenance and polarized growth of A. fumigatus. These findings also relate polarized growth of a fungal organism to pathogenesis, again emphasizing the importance of delineating growth dynamics in the filamentous fungi.
Acknowledgments
We thank Jay Card for his assistance with photography.
This work was supported by National Institutes of Health grants AI41119 (J.C.R.) and AI48746 (D.S.A.).
REFERENCES
- 1.Adams, T. H., J. K. Wieser, and J. H. Yu. 1998. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62:35-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alspaugh, J. A., L. M. Cavallo, J. R. Perfect, and J. Heitman. 2000. RAS1 regulates filamentation, mating and growth at high temperature of Cryptococcus neoformans. Mol. Microbiol. 36:352-365. [DOI] [PubMed] [Google Scholar]
- 3.Bauer, Y., P. Knechtle, J. Wendland, H. Helfer, and P. Philippsen. 2004. A Ras-like GTPase is involved in hyphal growth guidance in the filamentous fungus Ashbya gossypii. Mol. Biol. Cell 15:4622-4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beauvais, A., D. Maubon, S. Park, W. Morelle, M. Tanguy, M. Huerre, D. S. Perlin, and J. P. Latgé. 2005. Two α(1 to 3) glucan synthases with different functions in Aspergillus fumigatus. Appl. Environ. Microbiol. 71:1531-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cove, D. 1966. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta 113:5-56. [DOI] [PubMed] [Google Scholar]
- 6.Fillinger, S., M.-K. Chaveroche, K. Shimizu, N. Keller, and C. d'Enfert. 2002. cAMP and Ras signaling independently control spore germination in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 44:1001-1016. [DOI] [PubMed] [Google Scholar]
- 7.Fortwendel, J. R., J. C. Panepinto, A. E. Seitz, D. S. Askew, and J. C. Rhodes. 2004. Aspergillus fumigatus rasA and rasB regulate the timing and morphology of asexual development. Fungal Genet. Biol. 41:129-139. [DOI] [PubMed] [Google Scholar]
- 8.Harris, S. D., and M. Momany. 2004. Polarity in filamentous fungi: moving beyond the yeast paradigm. Fungal Genet. Biol. 4:391-400. [DOI] [PubMed] [Google Scholar]
- 9.Hausauer, D. L., M. Gerami-Nejad, C. Kistler-Andersonn, and C. A. Gale. 2005. Hyphal guidance and invasive growth in Candida albicans require the Ras-like GTPase Rsr1p and its GTPase-activating protein Bud2p. Eukaryot. Cell 4:1273-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kana-uchi, A., C. T. Yamashiro, S. Tanabe, and T. Murayama. 1997. A ras homologue of Neurospora crassa regulates morpholgy. Mol. Gen. Genet. 254:427-432. [DOI] [PubMed] [Google Scholar]
- 11.Kawasaki, R., K. Fujimura-Kamada, H. Toi, H. Kato, and K. Tanaka. 2003. The upstream regulator, Rsr1p, and downstream effectors, Gic1p and Gic2p, of the Cdc42p small GTPase coordinately regulate initiation of budding in Saccharomyces cerevisiae. Genes Cells 8:235-250. [DOI] [PubMed] [Google Scholar]
- 12.Latgé, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leberer, E., D. Harcus, D. Dignard, L. Johnson, S. Ushinsky, D. Y. Thomas, and K. Schröppel. 2001. Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signaling pathways in the pathogenic fungus Candida albicans. Mol. Microbiol. 42:673-687. [DOI] [PubMed] [Google Scholar]
- 14.Lengeler, K. B., R. C. Davidson, C. D'Souza, T. Harashima, W. C. Shen, P. Wang, X. Pan, M. Waugh, and J. Heitman. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64:746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madhani, H. D., C. A. Styles, and G. R. Fink. 1997. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell 91:673-684. [DOI] [PubMed] [Google Scholar]
- 16.Momany, M. 2002. Polarity in filamentous fungi: establishment, maintenance, and new axes. Curr. Opin. Biol. 5:580-585. [DOI] [PubMed] [Google Scholar]
- 17.Mukherjee, P. K., J. Latha, R. Hadar, and B. A. Horwitz. 2003. TmkA, a mitogen-activated protein kinase of Trichoderma virens, is involved in biocontrol properties and repression of conidiation in the dark. Eukaryot. Cell 2:446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osherov, N., and G. May. 2000. Conidial germination in Aspergillus nidulans requires Ras signaling and protein synthesis. Genetics 155:647-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paisley, D., G. D. Robson, and D. W. Denning. 2005. Correlation between in vitro growth rate and in vivo virulence in Aspergillus fumigatus. Med. Mycol. 43:397-401. [DOI] [PubMed] [Google Scholar]
- 20.Panepinto, J. C., B. G. Oliver, J. R. Fortwendel, D. L. Smith, D. S. Askew, and J. C. Rhodes. 2003. Deletion of the Aspergillus fumigatus gene encoding the Ras-related protein RhbA reduces virulence in a model of invasive pulmonary aspergillosis. Infect. Immun. 71:2819-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Som, T., and V. S. R. Kolaparthi. 1994. Developmental decisions in Aspergillus nidulans are modulated by Ras activity. Mol. Cell. Biol. 14:5333-5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wendland, J., and P. Philippsen. 2001. Cell polarity and hyphal morphogenesis are controlled by multiple Rho-protein modules in the filamentous ascomycete Ashbya gossypii. Genetics 151:557-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yaar, L., M. Mevarech, and Y. Koltin. 1997. A Candida albicans RAS-related gene (CaRSR1) is involved in budding, cell morphogenesis and hypha development. Microbiology 143:3033-3044. [DOI] [PubMed] [Google Scholar]