Abstract
RNA-binding proteins are important in many aspects of RNA processing, function, and destruction. One class of such proteins contains the RNA recognition motif (RRM), which consists of about 90 amino acid residues, including the canonical RNP1 octapeptide: (K/R)G(F/Y)(G/A)FVX(F/Y). We used a variety of homology searches to classify all of the RRM proteins of the three kinetoplastids Trypanosoma brucei, Trypanosoma cruzi, and Leishmania major. All three organisms have similar sets of RRM-containing protein orthologues, suggesting common posttranscriptional processing and regulatory pathways. Of the 75 RRM proteins identified in T. brucei, only 13 had clear homologues in other eukaryotes, although 8 more could be given putative functional assignments. A comparison with the 18 RRM proteins of the obligate intracellular parasite Encephalitozoon cuniculi revealed just 3 RRM proteins which appear to be conserved at the primary sequence level throughout eukaryotic evolution: poly(A) binding protein, the rRNA-processing protein MRD1, and the nuclear cap binding protein.
Organisms of the order Kinetoplastida are flagellated protists with a microscopically visible complex of mitochondrial DNA. They include a number of pathogens of animals and plants. Among these organisms are Leishmania major, which causes a range of diseases in the tropics and Europe; Trypanosoma brucei, which causes sleeping sickness in Africa; and Trypanosoma cruzi, which is the etiological agent of Chagas' disease in Latin America. All three parasites undergo a “digenetic” life cycle involving transmission from one mammal to the next by an arthropod vector.
Some aspects of RNA metabolism in trypanosomes and leishmanias are similar to those of other eukaryotes, whereas others are quite deviant. As in other organisms, multiple stable and structural RNAs must be processed, modified, and assembled into ribonucleoprotein complexes. Some of these complexes are themselves involved in further RNA-processing reactions. Perhaps the most remarkable aspect of trypanosomatid RNA metabolism is the editing of kinetoplast mRNA transcripts through the addition and deletion of uridine residues (46). Almost as surprising, however, is the dependence of the parasites on posttranscriptional mechanisms to control the levels and translation efficiencies of mRNAs. Trypanosomatid protein-coding genes are arrayed in long polycistronic transcription units, for which specific promoters have proved elusive (34, 35). Monocistronic mature RNAs are generated by trans splicing (at the 5′ end) and polyadenylation (at the 3′ end) (30). The cotranscription of many open reading frames, irrespective of the cell's requirement for differing levels of the final gene products, results in an almost exclusive dependence upon posttranscriptional mechanisms for the regulation of gene expression (8). Although control could theoretically act during mRNA processing or export from the nucleus, most mRNAs tested so far have been found to be regulated primarily at the levels of mRNA degradation and translation; the signals determining the regulation are usually located in the 3′ untranslated regions (UTRs) (8).
Posttranscriptional processing and degradation of RNAs are effected by enzymes and ribonucleoprotein complexes and regulated by trans-acting factors that bind the RNAs (14). In mammalian cells, several hundred unstable mRNAs contain AU-rich elements (AREs) in their 3′ UTRs. Proteins implicated in the regulation of ARE-RNA stability fall into several classes, depending on the RNA-binding domains: these include KH domains, zinc fingers, and RNA recognition motif (RRM) domains (45). Similar AREs are present in various regulated mRNAs in kinetoplastids, and there is evidence that the mechanisms of regulation, including the involvement of RRM-containing regulatory proteins, may resemble those in mammals (12, 44).
The RRM comprises about 90 amino acid residues. It often contains the signature RNP1 sequence motif, (K/R)G(F/Y)(G/A)FVX(F/Y). Proteins with RRMs are involved in a large number of processes through specific interactions with RNA (14), but a subset of RRMs can also interact with other proteins (27). The signature sequence RNP1, together with a second one known as RNP2, form the central part of the RRM beta-sheet which is involved in RNA binding. In this article, we describe the proteins containing RRM domains from the three sequenced trypanosomatid genomes and compare the gene set with those from other eukaryotes, particularly the minimal eukaryote Encephalitozoon cuniculi.
MATERIALS AND METHODS
Databases.
The databases utilized in this work were obtained from sequencing projects of the three trypanosomatids. The main databases used were the T. cruzi database (TcBr version 2.0), the T. brucei database (Tb927 versions 2.1 and 3), and the L. major database (LmjF version 3), but the final results were updated using the July 2005 versions, including homologues derived from aligning the genomes of all three species (5, 15, 16, 24). The European Molecular Biology Open Software Suite (EMBOSS) (http://www.ebi.ac.uk/emboss/) and various other programs available from GeneDB were used to extract sequences from the databases.
Sequence searches.
A selection of RRM-containing sequences from trypanosomes, Saccharomyces cerevisiae, and Homo sapiens were used in BLAST searches against trypanosomatid databases. BLAST hits having log E values of <9 were used in further analysis. The motif search utility within GeneDB was also used to detect RNA-binding proteins. The searches were continued until no more potential proteins with RRMs could be located.
All RRM-containing protein sequences were run against the Pfam (ftp://ftp.sanger.ac.uk/pub/databases/Pfam/) set of domains, hidden Markov model profiles, and Prosite databases (ftp://bo.expasy.org/databases/prosite/tools/ps_scan/). Potential RNA-binding proteins were identified using the following RNA recognition motifs: Pfam PF00076, Smart SM00360, Prosite PS50102, and Prosite PS00030. All of the T. brucei protein sequences were then compared against the entire EMBL database using BLASTP and against the Saccharomyces cerevisiae genome. A functional assignment was made for a predicted protein only when a BLAST search using the T. brucei sequence gave the corresponding functionally characterized protein from another eukaryote with a low (usually less than 10−13) P value and a reciprocal BLAST search with the characterized protein sequence also gave the T. brucei protein as the best match. Matches within the RRMs alone usually gave probabilities in the range of 10−11 to 10−12 and were not used for functional assignments; matches outside the RRM, and the presence of appropriate domains, were also required. We did not infer function because of matches with proteins for which function had been predicted on the basis of homology or was designated “putative.”
Proteins with clear functional assignments were named according to the homologue in yeast or mammals or using published names. Other proteins were named according to the following scheme, where x is a number: One RRM, RBPx; two RRMs, DRBDx; three RRMs, TRRMx; RRM(s) and an SR domain(s), RBSRx. Since the number of RNA-binding domains recognized depended on the cutoff used, and sometimes the apparent numbers of domains were not the same in all three organisms, these names are only an approximate guide.
Alignments and orthologues.
Each of the sequences obtained for one kinetoplastid species was used as a query in reciprocal multiple BLAST searches against the two other genomes in order to identify orthologues. The assignments were confirmed manually by examining the protein lengths and domain structures; then, orthologous proteins were assigned similar names. Multiple sequence alignments were performed using CLUSTALX v1.81 (47) with the default alignment parameters, and this information was also used in orthologous identification. The orthologue assignment was then checked against orthologues assigned based on aligning all three genomes (clusters of orthologous groups [COGs]) (15). Nearly all assignments were consistent. The exceptions were found exclusively among families of genes encoding proteins of unknown function with a single RRM, such as the RBP7 family; often, the more divergent orthologue had acquired additional sequence.
NJ trees.
Protein distances were calculated by Clustal algorithms using the DNAStar package (species-specific trees) or the CLUSTALX v1.81 program (comparative tree). For the calculation, the neighbor-joining (NJ) method was used. Percent divergences between all pairs of sequence from the multiple alignments were calculated, and the NJ method was applied to the distance matrix. The unrooted tree obtained was plotted using Treeview software (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html) (39).
RESULTS
Overview of RRM-containing proteins in trypanosomes.
Predicted proteins containing RRM domains were identified from the complete predicted proteomes of T. brucei, T. cruzi, and L. major (tri-tryp). A total of 139 RRM-type proteins were found in T. cruzi, 75 in T. brucei, and 80 in L. major. When these were classified as orthologues by sequence similarity and by comparing the positions of the genes on all three genomes (see Materials and Methods), 77 different RRM proteins were found. In most cases, the T. cruzi genome contains two or more paralogues, presumably because the CL Brener clone used for sequencing has a hybrid genotype (31). In addition, some genes were present in two or more copies in either T. brucei or L. major as a consequence of tandem duplications. A complete list is provided in the supplemental material. Two genes were unique to T. cruzi, three were unique to L. major, and seven were found in T. cruzi and T. brucei but not L. major.
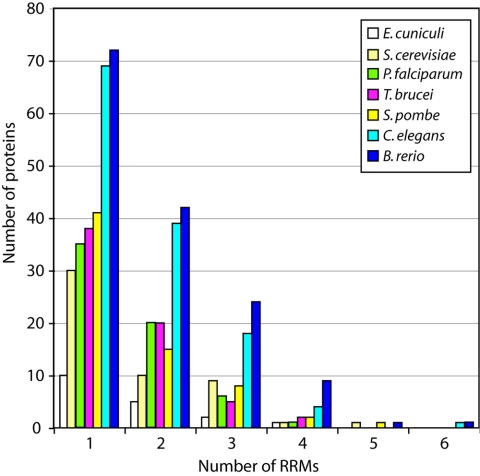
We compared the protein set from T. brucei with those of several other organisms, relying mainly on annotation available from the Pfam database (Table 1 and Fig. 1). Overall, the number of proteins with RRM domains—and the number of domains in the proteins—increased with genome complexity. We have previously noted that in comparison with S. cerevisiae, trypanosomes have more proteins that are less than 200 residues long and have a single RRM (24). The broader comparison, however, suggests that trypanosomes are not remarkable in their RRM protein content. The only irregularity visible in Fig. 1 is that the two parasites Plasmodium falciparum and T. brucei had a slight excess of proteins with two RRMs, with a corresponding deficit in three-RRM proteins, compared with the two yeasts.
TABLE 1.
RRM-containing proteins in selected genomes
| Genome | No. of RRM proteins | No. of RRMs | Total no. of ORFs | % proteins with RRM | RRM no./ORF no. |
|---|---|---|---|---|---|
| Encephalitozoon cuniculi | 18 | 30 | 1,909 | 0.94 | 0.016 |
| Schizosaccharomyces pombe | 67 | 107 | 4,956 | 1.35 | 0.022 |
| Plasmodium falciparum | 61 | 95 | 5,251 | 1.16 | 0.018 |
| Saccharomyces cerevisiae | 50 | 86 | 6,218 | 0.80 | 0.014 |
| Leishmania major | 78 | 120 | 8,277 | 0.94 | 0.014 |
| Trypanosoma brucei | 67 | 100 | 8,599 | 0.78 | 0.012 |
| Brachydanio rerio | 150 | 273 | 9,249 | 1.62 | 0.030 |
| Drosophila melanogaster | 177 | 304 | 16,175 | 1.09 | 0.019 |
| Caenorhabditis elegans | 130 | 223 | 21,926 | 0.59 | 0.010 |
| Homo sapiens | 308 | 542 | 35,844 | 0.86 | 0.015 |
FIG. 1.
Comparison of RRM proteins in Encephalitozoon cuniculi, Trypanosoma brucei, Saccharomyces cerevisiae, Plasmodium falciparum, Schizosaccharomyces pombe, Caenorhabditis elegans, and Brachydanio rerio. The proteins were classified into groups according to the number of RRMs.
Phylogenetic analysis of the kinetoplastid RRM proteins.
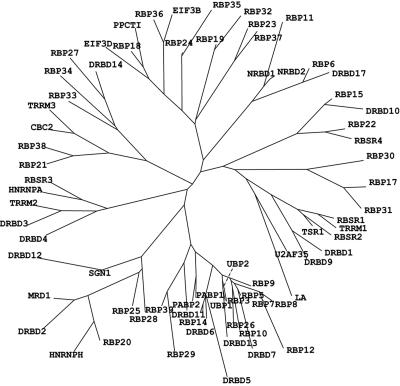
We next performed multiple sequence alignments and plotted a cladogram of the RRM proteins in the hope of discerning functional or evolutionary relationships. This analysis was most useful for the identification of homologues, both within and between species; virtually all of the results were consistent with the recently published comparison of all three genomes, which included the conservation of gene order between species and the clustering of homologues as tandem repeats as additional criteria (COG analysis) (15) (see Table S1 in the supplemental material). The phylogenetic analysis for T. brucei is shown in Fig. 2, and trees of the proteins in the three parasites are supplied in the supplemental material. The phylogenetic analysis does not, strictly speaking, show evolutionary relationships, since the proteins contain a variety of conserved domains of various origins. Nevertheless, some proteins with similar likely functions were grouped together. Diagrams of the T. brucei RRM proteins are shown in Fig. 3 and 4. In the figures, the proteins are grouped partly according to the presence of additional motifs and possible functions and partly according to the single-organism and tri-tryp cladograms. In the discussion below, we have grouped the proteins according to their possible functions; it is, however, important to note that many RNA-binding proteins are in fact involved at multiple stages of RNA maturation, transport, function, and decay.
FIG. 2.
Phylogenetic tree of the T. brucei RNA-binding-domain proteins. Alignments of the 75 sequences from T. brucei were performed with the ClustalW algorithm using the DNAStar package, and the radial tree was plotted with TreeView software.
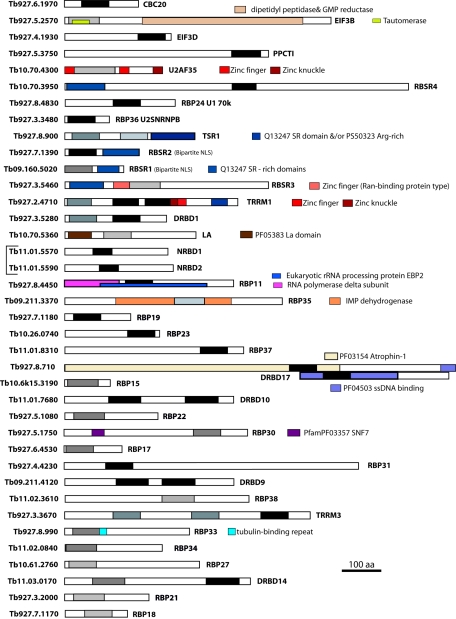
FIG. 3.
Domain structures of T. brucei proteins found mainly in the lower half of Fig. 2. Locus numbers are in the left column, and protein names are on the right. Different domains are specific colors as shown on the adjacent blocks; the strongest RNA-binding-domain matches are black, and the weakest are light gray. Proteins encoded by adjacent genes are bracketed.
FIG. 4.
Domain structures of T. brucei proteins found mainly in the upper half of Fig. 2. The key is as for Fig. 3. DRBD17 did not fit and so is shown broken into two portions.
Proteins predicted to be involved in splicing.
Since all trypanosome mRNAs are likely to be trans spliced, the composition of the splicing complex and the mechanism of splice acceptor site identification are of considerable interest. Several of the proteins involved are shown in Fig. 4. The U2 snRNP auxiliary factor (U2AF) is a heterodimer consisting of 65- and 35-kDa subunits that binds to the polypyrimidine tract located on the 5′ side of the 3′ acceptor splice site from mRNA precursors (pre-mRNAs), and it forms part of the spliceosome complex (27). U2AF35 has already been characterized in T. cruzi; the assignment is based on sequence alignments and localization in nuclear speckles (48). The predicted sequence lacks several residues that in other organisms were shown to be necessary for the interaction with U2AF65 (48). The trypanosome protein has a C-terminal zinc finger that replaces an SR domain found in other eukaryotic U2AF35 proteins, but neither motif is present at the corresponding position in the Leishmania homologue.
If, indeed, trypanosomes have a U2AF65, with different requirements for the U2AF35 interaction, it might be RBSR4. RBSR4 has only one RRM (by Pfam analysis using the default cutoffs), whereas a U2AF65 should have three (27), but the localization of the RBSR4 protein, and protein-protein interaction patterns, are consistent with a role in splicing (M. Levin, personal communication). In other organisms, the third RRM domain of U2AF65 is involved in protein-protein interactions (27); a search of the T. brucei database using the consensus for this variant domain yielded no results.
T. brucei RBP24 (TbRBP24) has been shown to be the functional equivalent of the 70-kDa component of the U1 snRNP (40, 41). We could not have annotated this on the grounds of sequence identity alone, since most E values were around 10−10, which was higher than many matches based only on RRM domain similarities, and also, the predicted protein is a third of the expected size. This illustrates the fact that we could easily have missed further functional assignments of other proteins because of the difficulties in deciding which matches were significant. Another example is the very small RBP36, which is annotated as U2 snRNPb in the database (24): although the P values when searching individual genomes are low (e.g., about 10−7 for S. cerevisiae), the match to U2 snRNP B" is consistent across species.
Trypanosome trans splice acceptor sites, like cis acceptor sites of other eukaryotes, are preceded by polypyrimidine tracts (4). Both DRBD3 and DRBD4 give BLASTP outputs listing polypyrimidine tract binding proteins (PTBs), which are principally known as splicing regulators but which have also been implicated in the control of polyadenylation and of the use of internal ribosomal entry sites during translation initiation (summarized in reference 38). PTBs have four RRMs, and each of them can bind RNA independently without any apparent influence of the linker sequences (38), but the solution structure reveals extensive interactions between the third and fourth RRMs (38). Intriguingly, DRBD3 aligns better with N-terminal RRMs 1 and 2 of PTB, while DRBD4 matches RRMs 3 and 4. It is tempting to speculate that the two trypanosome proteins may jointly perform the function of PTB.
At the top of Fig. 4 is CBC20, the 20-kDa nuclear cap binding protein (CBC2 in yeast; CBC20 in other organisms), which is important in RNA processing and nuclear transport. In the nucleus, CBC20 is usually found associated with CBC80 (a rather poorly conserved protein that does not have an RRM). The trypanosome CBC20 homologue has been demonstrated to be present in the T. brucei cap-binding complex (29), and to be associated with importin alpha and three novel subunits, none of which is obviously homologous to CBP80; the trypanosome cap-binding complex was also shown to be essential in early steps of trans splicing (29).
At the top of Fig. 3 are two possible hnRNP-like proteins, similar to those found in mammals but not in yeast. The protein which is annotated as hnRNPA (24) has an N-terminal glycine-rich region with a few RGG motifs and a single RRM; mammalian hnRNPA has, in contrast, two RRMs and a more extensive glycine-rich domain that contains proportionately more RGG motifs and is at the C terminus. The hnRNPH homologue is considerably more similar to the mammalian counterparts. Both hnRNPA and hnRNPH family proteins affect several aspects of RNA processing. hnRNPH is required for splicing enhancer activity in some viral messages (7), and both proteins have been implicated as regulators of alternative splicing of endogenous mRNAs (19, 20). Regulated shuttling of hnRNPA into the cytoplasm (1) means that this protein could also be involved in the control of cytoplasmic processes, such as translation (6).
In addition to these proteins, there are several with serine-arginine-rich domains, which are suggestive of an involvement in splicing (22). RBSR3 also has a Ran-binding-type zinc finger. TRRM1, a protein with three RRMs, a zinc finger and a zinc knuckle, and an SR domain, was shown to be located in nuclear speckles (32). RBSR1 and RBSR2 have bipartite nuclear localization signals; RBSR3 has a zinc finger, suggestive of binding to the nuclear import factor Ran. TSR1 was previously shown to be in the nucleus of T. brucei and to be capable of interacting with two different parts of the spliced leader RNA in the yeast three-hybrid system (23); its role is as yet unclear, but it is clearly not, as originally thought, a direct homologue of U1 70k.
Processing of stable and nuclear RNAs.
The two related nuclear-RNA-binding proteins NRBD1 and NRBD2 (previously p34 and p37) (51) were shown to be involved in the import and/or assembly pathway of 5S rRNA during ribosome biogenesis in trypanosomes (43). The two genes are neighbors on chromosome 11 and are likely to be products of gene duplication. The multiple RNA-binding domain 1 protein (MRD1) is involved in rRNA processing in yeast (25). The La protein is implicated in the maturation of various RNAs, including the SL RNA and tRNA (17, 33). RBP11 has domains suggestive of involvement in rRNA processing or transcription. Each of three related and chromosomally linked proteins with two RRMs, DRBD6A, 6B, and 11, has a bipartite nuclear localization signal, suggesting a nuclear function.
Translation.
There are two poly(A) binding proteins (PABPs). PABPs are involved throughout the lifetime of polyadenylated mRNAs, with roles including stabilization and the interaction of the poly(A) tail with the translation initiation complex. A similar function in transcript stabilization has also been demonstrated in trypanosomatid extracts (37); poly(A) binding was demonstrated for L. major and T. cruzi PABP1 (2, 3) and T. brucei PABP2 (21). One of the two L. major PABP2 genes is classified as a singleton in the published COG analysis (15), having been isolated on a nonsyntenic chromosome.
We found two possible components of eukaryotic initiation translation factor 3 (EIF3). The EIF3β homologue EIF3B is clear, whereas the putative EIF3D homologue is less convincing. In humans, EIF3 is a factor composed of 11 subunits which interacts with a 40S ribosomal subunit and is involved in the formation of the 43S preinitiation complex (28, 42). Yeast SGN1 is thought to influence translation (50), so the trypanosome homologue may also have this function. DRBD1 matches TIA-like RNA-binding proteins, but this may be only a consequence of the arrangement of the RRMs. TIA proteins have been implicated in both translational silencing and control of alternative splicing (10, 18). Finally, peptidyl-prolyl cis-trans isomerase (PPCTI) is involved in protein refolding and is ubiquitous in living organisms, although more than one class of proteins with this function exists (36).
Possible regulators of mRNA turnover.
Several of the proteins in Fig. 3 are notable for the presence of low-complexity sequences, such as glutamine-rich or glycine-rich regions. The T. cruzi RBP family proteins were shown to have binding specificity for G- or U-rich RNA (9), and UBP1 and UBP2 have been implicated in posttranscriptional control of gene expression in both T. cruzi and T. brucei. AREs and G-rich elements are present in the 3′ UTR of the small mucin gene of T. cruzi and confer stage-specific mRNA degradation/stability (11). T. cruzi UBP1 (TcUBP1) and TcUBP2 bind to the 3′ UTRs of small mucin gene RNAs and promote RNA stabilization, forming a ribonucleoprotein complex that includes poly(A) binding protein (13). The UBP1 and UBP2 genes were probably produced by a gene duplication event that occurred before the divergence of trypanosomes and leishmanias, as they are present as neighbors on orthologous chromosomes in all three species. We have recently found that TbUBP1 and TbUBP2 are also implicated in posttranscriptional gene regulation in T. brucei (C. Hartmann, unpublished data). In TcUBP1, an extra beta hairpin enlarges the surface of RNA binding in the RRM domain (49). The functions of the other single-RRM proteins in this group (from UBP1 to DRBD13 in Fig. 3) remain to be elucidated. In BLASTP alignments against the EMBL databases, the proteins of known function with the best matches are often from the ELAV family, but this is only because the single RRMs of the trypanosome proteins align with the three RRMs of the ELAV proteins.
The minimal set of RRM proteins.
Our results with the three kinetoplastids had revealed 12 RRM proteins which had clearly been conserved from kinetoplastids to yeast or mammals (Table 2). These were two poly(A) binding proteins; splicing factors U1 70k, U2AF35, and U2AF65; rRNA-processing protein MRD1; peptidyl-prolyl cis-trans isomerase; hnRNPH; translation factor SGN1; nuclear cap binding protein; and translation factors EIF3B and EIF3D. This prompted us to ask if these proteins comprised a minimal set required for eukaryotic gene expression. To answer this, we compared the trypanosome gene set with that of Encephalitozoon cuniculi, a microsporidian parasite with an obligately intracellular lifestyle and a minimal genome (26). E. cuniculi has a quarter of the number of proteins in the other unicellular eukaryotes considered, including T. brucei, and only 18 RRM domain proteins. We compared the sequences with those of several other unicellular eukaryotes—S. cerevisiae, Schizosaccharomyces pombe, Entamoeba histolytica, Plasmodium falciparum, and T. brucei—using GeneDB to screen the individual genomes and applying an E-value cutoff of 10−15 in order to exclude matches based only on the RRMs (which generally gave values of about 10−10). This revealed only three RRM proteins that were present in all organisms tested: the nuclear cap binding protein (Q8SW46_ENCCU), poly(A) binding protein (Q8SR30_ENCCU), and Mrd1p (Q8SRD9_ENCCU). All of the organismal genomes tested, except that of T. brucei, also encoded a homologue of S. cerevisiae Mot2p (Q8SQS6_ENCCU for E. cuniculi), and Q8SSA1_ENCCU has a serine-rich region and aligns with an rRNA-processing protein present in the two yeasts and Entamoeba. E. cuniculi is probably a reduced derivative of a fungal ancestor, and P. falciparum contains a plastid of algal origin; consistent with this, a few of the other E. cuniculi RRM proteins showed their best matches to S. pombe, S. cerevisiae, and P. falciparum, with E values between 10−10 and 10−15. The annotated database does not contain E. cuniculi RRM-containing translation elongation factors EIF3B and -D: it may be that the genes have diverged beyond recognition. Although the genome of E. cuniculi is annotated to include 13 mRNAs with short introns (26), the putative U2AF35 (Q8SQL5_ENCCU) lacks classical RRMs and the large subunit (Q8SRX8_ENCCU) has a C-terminal RNA recognition motif that is too diverged to be recognized by Pfam. The E. cuniculi peptidyl-prolyl cis-trans isomerase is of a prokaryotic type, with no RRM. None of the E. cuniculi RRM proteins had zinc fingers or glutamine-rich domains, and no clear SR domains were present.
TABLE 2.
Accession numbers of conserved proteinsa
GenBank numbers except for T. brucei, which are GeneDB locus numbers.
From these results, it would seem that the minimal set of conserved RRM-containing proteins at the primary sequence level consists of poly(A) binding protein, MRD1, and the nuclear cap binding protein. Even in E. cuniculi, over half of the RRM proteins appeared to be organism specific.
DISCUSSION
Of the 75 T. brucei RRM proteins, only 18 can be assigned a function with any confidence. There are two poly(A) binding proteins, U2AF35, MRD1, PPCTI, hnRNPH, SGN1, CBC20, U1 70k (RBP24), and EIF3B, and also possible homologues of EIF3D, U2AF65, and hnRNPA. Experimental functional evidence is available concerning six proteins that are specific to trypanosomatids: NRBD1 and NRBD2, TRRM1, TSR1, and UBP1 and UBP2. This leaves over 50 proteins with no known function. The additional domains, such as zinc fingers, glutamine-rich regions, and SR-rich regions, have diverse functions but can be of only limited utility in predictions; however, it is tempting to speculate that the SR domain proteins might be involved in the regulation of cis or trans splicing or the selection of alternative splice sites. The comparison with E. cuniculi suggested that the presence of organism-specific RRM proteins may be ubiquitous, which implies that the RRM proteins have coevolved with their RNA targets. RNA target evolution will occur most readily in molecules that are relatively free of structural or sequence constraints. Since the kinetoplastids have only two or three cis-spliced genes, and regulation of trans splicing has not been documented, we hypothesize that many of the kinetoplastid-specific RRM proteins will bind untranslated regions of mRNAs and will be involved in regulating mRNA degradation and translation.
Supplementary Material
Acknowledgments
We thank Christiane Hertz-Fowler for extensive assistance with GeneDB and Mariano Levin (INGEBI, Buenos Aires) for communicating unpublished results. Noreen Williams (SUNY, Buffalo) proposed the new names for p34 and p37.
This work was supported by grants from the National Institutes of Health, grant AI060645-01, and the Agencia Nacional de Promoción Científica y Tecnológica (Argentina) to A.C.F. The research of A.C.F. was also supported in part by an International Research Scholars Grant from the Howard Hughes Medical Institute. J.D.G. is a research fellow from the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina, and A.C.F. is a researcher from CONICET, Argentina.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Allemand, E., S. Guil, M. Myers, J. Moscat, J. F. Caceres, and A. R. Krainer. 2005. Regulation of heterogenous nuclear ribonucleoprotein A1 transport by phosphorylation in cells stressed by osmotic shock. Proc. Natl. Acad. Sci. USA 102:3605-3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates, E. J., E. Knuepfer, and D. F. Smith. 2000. Poly(A)-binding protein I of Leishmania: functional analysis and localisation in trypanosomatid parasites. Nucleic Acids Res. 28:1211-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batista, J. A. N., S. M. R. Teixeira, J. E. Donelson, L. V. Kirchhoff, and C. Martins de Sá. 1994. Characterization of Trypanosoma cruzi poly(A)-binding protein and its genes. Mol. Biochem. Parasitol. 67:301-312. [DOI] [PubMed] [Google Scholar]
- 4.Benz, C., D. Nilsson, B. Andersson, C. Clayton, and D. L. Guilbride. 2005. Messenger RNA processing sites in Trypanosoma brucei. Mol. Biochem. Parasitol. 143:125-134. [DOI] [PubMed] [Google Scholar]
- 5.Berriman, M., et. al. 2005. The genome of the African trypanosome, Trypanosoma brucei. Science 309:416-422. [DOI] [PubMed] [Google Scholar]
- 6.Bonnal, S., F. Pileur, C. Orsini, F. Parker, F. Pujol, A. C. Prats, and S. Vagner. 2005. Heterogeneous nuclear ribonucleoprotein A1 is a novel internal ribosome entry site trans-acting factor that modulates alternative initiation of translation of the fibroblast growth factor 2 mRNA. J. Biol. Chem. 280:4144-4153. [DOI] [PubMed] [Google Scholar]
- 7.Caputi, M., and A. M. Zahler. 2002. SR proteins and hnRNP H regulate the splicing of the HIV-1 tev-specific exon 6D. EMBO J. 21:845-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clayton, C. E. 2002. Developmental regulation without transcriptional control? From fly to man and back again. EMBO J. 21:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Gaudenzi, J. G., I. D'Orso, and A. C. C. Frasch. 2003. RNA recognition motif-type RNA-binding proteins in Trypanosoma cruzi form a family involved in the interaction with specific transcripts in vivo. J. Biol. Chem. 21:18884-18894. [DOI] [PubMed] [Google Scholar]
- 10.Dixon, D. A., G. C. Balch, N. Kedersha, P. Anderson, G. A. Zimmerman, R. D. Beauchamp, and S. M. Prescott. 2003. Regulation of cyclooxygenase-2 expression by the translational silencer TIA-1. J. Exp. Med. 198:475-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Orso, I., and A. C. C. Frasch. 2001. Functionally different AU- and G-rich cis elements confer developmentally-regulated mRNA stability by interaction with specific RNA-binding proteins. J. Biol. Chem. 276:15783-15793. [DOI] [PubMed] [Google Scholar]
- 12.D'Orso, I., and A. C. C. Frasch. 2001. TcUBP-1, a developmentally regulated U-rich RNA-binding protein involved in selective mRNA destabilization in trypanosomes. J. Biol. Chem. 276:34801-34809. [DOI] [PubMed] [Google Scholar]
- 13.D'Orso, I., and A. C. C. Frasch. 2002. TcUBP-1, an mRNA destabilizing factor from trypanosomes, homodimerizes and interacts with novel AU-rich element- and poly(A)-binding proteins forming a ribonucleoprotein complex. J. Biol. Chem. 277:50520-50528. [DOI] [PubMed] [Google Scholar]
- 14.Dreyfuss, G., V. N. Kim, and N. Kataoka. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3:195-205. [DOI] [PubMed] [Google Scholar]
- 15.El-Sayed, N. M., et al. 2005. Comparative genomics of trypanosomatid parasitic protozoa. Science 309:404-409. [DOI] [PubMed] [Google Scholar]
- 16.El-Sayed, N. M., et al. 2005. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science 309:409-415. [DOI] [PubMed] [Google Scholar]
- 17.Foldynova-Trantirkova, S., Z. Paris, N. R. Sturm, D. A. Campbell, and J. Lukes. 2005. The Trypanosoma brucei La protein is a candidate poly(U) shield that impacts spliced leader RNA maturation and tRNA intron removal. Int. J. Parasitol. 35:359-366. [DOI] [PubMed] [Google Scholar]
- 18.Forch, P., O. Puig, C. Martinez, B. Seraphin, and J. Valcarcel. 2002. The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J. 21:6882-6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garneau, D., T. Revil, J. F. Fisette, and B. Chabot. 2005. Heterogeneous nuclear ribonucleoprotein F/H proteins modulate the alternative splicing of the apoptotic mediator Bcl-x. J. Biol. Chem. 280:22641-22650. [DOI] [PubMed] [Google Scholar]
- 20.Han, K., G. Yeo, P. An, C. B. Burge, and P. J. Grabowski. 19 April 2005, posting date. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol. 3:e158. [Online.] doi: 10.1371/journal.pbio.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hotchkiss, T. L., G. E. Nerantzakis, S. C. Dills, L. Shang, and L. K. Read. 1999. Trypanosoma brucei poly(A) binding protein I cDNA cloning, expression, and binding to 5′ untranslated region sequence elements. Mol. Biochem. Parasitol. 98:117-129. [DOI] [PubMed] [Google Scholar]
- 22.Huang, Y., and J. A. Steitz. 2005. SRprises along a messenger's journey. Mol. Cell 17:613-615. [DOI] [PubMed] [Google Scholar]
- 23.Ismaili, N., D. Pérez-Morga, P. Walsh, A. Mayeda, A. Pays, P. Tebabi, A. R. Krainer, and E. Pays. 1999. Characterization of a SR protein from Trypanosoma brucei with homology to RNA-binding cis-splicing proteins. Mol. Biochem. Parasitol. 102:103-115. [DOI] [PubMed] [Google Scholar]
- 24.Ivens, A. C., et. al. 2005. The genome of the kinetoplastid parasite, Leishmania major. Science 309:436-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin, S. B., J. Zhao, P. Bjork, K. Schmekel, P. O. Ljungdahl, and L. Wieslander. 2002. Mrd1p is required for processing of pre-rRNA and for maintenance of steady-state levels of 40 S ribosomal subunits in yeast. J. Biol. Chem. 277:18431-18439. [DOI] [PubMed] [Google Scholar]
- 26.Katinka, M. D., et al. 2001. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414:451-455. [DOI] [PubMed] [Google Scholar]
- 27.Kielkopf, C. L., S. Lucke, and M. R. Green. 2004. U2AF homology motifs: protein recognition in the RRM world. Genes Dev. 18:1513-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolupaeva, V. G., A. Unbehaun, I. B. Lomakin, C. U. Hellen, and T. V. Pestova. 2005. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA 11:470-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, H., and C. Tschudi. 2005. Novel and essential subunits in the 300-kilodalton nuclear cap binding complex of Trypanosoma brucei. Mol. Cell. Biol. 25:2216-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang, X., A. Haritan, S. Uliel, and S. Michaeli. 2003. trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot. Cell 2:830-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Machado, C. A., and F. J. Ayala. 2001. Nucleotide sequences provide evidence of genetic exchange among distantly related lineages of Trypanosoma cruzi. Proc. Natl. Acad. Sci. USA 98:7396-7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manger, I. D., and J. C. Boothroyd. 1998. Identification of a nuclear protein in Trypanosoma brucei with homology to RNA-binding proteins from cis-splicing systems. Mol. Biochem. Parasitol. 97:1-11. [DOI] [PubMed] [Google Scholar]
- 33.Marchetti, M. A., C. Tschudi, H. Kwon, S. L. Wolin, and E. Ullu. 2000. Import of proteins into the trypanosome nucleus and their distribution at karyokinesis. J. Cell Sci. 113:899-906. [DOI] [PubMed] [Google Scholar]
- 34.Martinez-Calvillo, S., D. Nguyen, K. Stuart, and P. J. Myler. 2004. Transcription initiation and termination on Leishmania major chromosome 3. Eukaryot. Cell 3:506-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinez-Calvillo, S., S. Yan, D. Nguyen, M. Fox, K. Stuart, and P. J. Myler. 2003. Transcription of Leishmania major Friedlin chromosome 1 initiates in both directions within a single region. Mol. Cell 11:1291-1299. [DOI] [PubMed] [Google Scholar]
- 36.Maruyama, T., R. Suzuki, and M. Furutani. 2004. Archaeal peptidyl prolyl cis-trans isomerases (PPIases) update 2004. Front. Biosci. 9:1680-1720. [DOI] [PubMed] [Google Scholar]
- 37.Milone, J., J. Wilusz, and V. Bellofatto. 2004. Characterization of deadenylation in trypanosome extracts and its inhibition by poly(A)-binding protein Pab1p. RNA 10:448-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oberstrass, F. C., S. D. Auweter, M. Erat, Y. Hargous, A. Henning, P. Wenter, L. Reymond, B. Amir-Ahmady, S. Pitsch, D. L. Black, and F. H. Allain. 2005. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science 309:2054-2057. [DOI] [PubMed] [Google Scholar]
- 39.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 40.Palfi, Z., W. S. Lane, and A. Bindereif. 2002. Biochemical and functional characterization of the cis-spliceosomal U1 small nuclear RNP from Trypanosoma brucei. Mol. Biochem. Parasitol. 121:233-243. [DOI] [PubMed] [Google Scholar]
- 41.Palfi, Z., B. Schimanski, A. Gunzl, S. Lucke, and A. Bindereif. 2005. U1 small nuclear RNP from Trypanosoma brucei: a minimal U1 snRNA with unusual protein components. Nucleic Acids Res. 33:2493-2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pestova, T. V., and V. G. Kolupaeva. 2002. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 16:2906-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pitula, J., W. T. Ruyechan, and N. Williams. 2002. Two novel RNA binding proteins from Trypanosoma brucei are associated with 5S rRNA. Biochem. Biophys. Res. Commun. 290:569-576. [DOI] [PubMed] [Google Scholar]
- 44.Quijada, L., C. Hartmann, C. Guerra-Giraldez, M. Drozdz, H. Irmer, and C. E. Clayton. 2002. Expression of the human RNA-binding protein HuR in Trypanosoma brucei induces differentiation-related changes in the abundance of developmentally-regulated mRNAs. Nucleic Acids Res. 30:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raineri, I., D. Wegmueller, B. Gross, U. Certa, and C. Moroni. 2004. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 32:1279-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simpson, L., S. Sbicego, and R. Aphasizhev. 2003. Uridine insertion/deletion RNA editing in trypanosome mitochondria: a complex business. RNA 9:265-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vazquez, M., C. Atorrasagasti, N. Bercovich, R. Volcovich, and M. J. Levin. 2003. Unique features of the Trypanosoma cruzi U2AF35 splicing factor. Mol. Biochem. Parasitol. 128:77-81. [DOI] [PubMed] [Google Scholar]
- 49.Volpon, L., I. D'Orso, C. R. Young, A. C. Frasch, and K. Gehring. 2005. NMR structural study of TcUBP1, a single RRM domain protein from Trypanosoma cruzi: contribution of a beta hairpin to RNA binding. Biochemistry 44:3708-3717. [DOI] [PubMed] [Google Scholar]
- 50.Winstall, E., M. Sadowski, U. Kühn, E. Wahle, and A. B. Sachs. 2000. The Saccharomyces cerevisiae RNA-binding protein Rbp29 functions in cytoplasmic mRNA metabolism. J. Biol. Chem. 275:21817-21826. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, J., W. Ruyechan, and N. Williams. 1998. Developmental regulation of two nuclear RNA binding proteins, p34 and p37, from Trypanosoma brucei. Mol. Biochem. Parasitol. 92:79-88. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.