Abstract
Eukaryotic cells require mitochondrial compartments for viability. However, the budding yeast Saccharomyces cerevisiae is able to survive when mitochondrial DNA suffers substantial deletions or is completely absent, so long as a sufficient mitochondrial inner membrane potential is generated. In the absence of functional mitochondrial DNA, and consequently a functional electron transport chain and F1Fo-ATPase, the essential electrical potential is maintained by the electrogenic exchange of ATP4− for ADP3− through the adenine nucleotide translocator. An essential aspect of this electrogenic process is the conversion of ATP4− to ADP3− in the mitochondrial matrix, and the nuclear-encoded subunits of F1-ATPase are hypothesized to be required for this process in vivo. Deletion of ATP3, the structural gene for the γ subunit of the F1-ATPase, causes yeast to quantitatively lose mitochondrial DNA and grow extremely slowly, presumably by interfering with the generation of an energized inner membrane. A spontaneous suppressor of this slow-growth phenotype was found to convert a conserved glycine to serine in the β subunit of F1-ATPase (atp2-227). This mutation allowed substantial ATP hydrolysis by the F1-ATPase even in the absence of the γ subunit, enabling yeast to generate a twofold greater inner membrane potential in response to ATP compared to mitochondria isolated from yeast lacking the γ subunit and containing wild-type β subunits. Analysis of the suppressing mutation by blue native polyacrylamide gel electrophoresis also revealed that the α3β3 heterohexamer can form in the absence of the γ subunit.
The mitochondrial compartment is the site of oxidative phosphorylation and numerous other metabolic processes and is required for cell viability, even when the compartment is compromised by the loss of the mitochondrial genome (mtDNA) (1). An essential feature of mitochondria is an energized inner mitochondrial membrane, which allows for insertion of key proteins and, depending on environmental and genetic conditions, the synthesis of ATP4− from ADP3− and Pi (5, 10, 13, 29). Cells that contain an intact mitochondrial genome, referred to as ρ+, can generate an energized inner membrane by creating a membrane potential (ΔΨM) in three ways: (i) by pumping protons into the intermembrane space as electrons are passed to O2 via the electron transport chain, (ii) by coupling the pumping of protons into the intermembrane space to ATP hydrolysis by the F1Fo-ATPase, and (iii) by exchanging ATP4− for ADP3− through the adenine nucleotide translocator (ANT). Because four subunits of the electron transport chain and three subunits of the Fo portion of the F1Fo-ATPase are encoded by mtDNA, viable cells lacking a mitochondrial genome (ρ° cells) must maintain ΔΨM by the exchange of ATP4− for ADP3− through ANT (14). ADP3− is provided by the hydrolysis of ATP4−, catalyzed by the remaining F1 portion of the ATPase (F1-ATPase).
Numerous observations illustrate the dependence of ρ° cells upon ATP hydrolysis and ANT activity in different organisms. For example, Saccharomyces cerevisiae (budding yeast) tolerates loss of mtDNA (petite-positive phenotype), but loss-of-function mutations in the major isoform of ANT (e.g., the point mutation encoded by op1) render yeast petite-negative (19). Yeast cells bearing mutations that inactivate F1-ATPase or the mitochondrial AAA protease (yme1Δ) also exhibit a petite-negative phenotype (7, 14, 18, 35, 37). The yme1Δ petite-negative phenotype can be suppressed by point mutations that create changes of certain residues in the α and γ subunits of F1-ATPase, leading to increased ATP hydrolysis activity and an elevated ΔΨM (17, 37). Some of the changes occur in the same residues that have also been shown to allow the petite-negative yeast Kluyveromyces lactis to grow without mtDNA (8). Finally, as in yeast, ANT and F1-ATPase activity are essential in ρ° human cells (3, 6).
F1-ATPase activity in S. cerevisiae is severely compromised by deletion of ATP3, which encodes the γ subunit of F1-ATPase.atp3Δ yeast cells also quantitatively lose their mtDNA and grow extremely slowly (20). Presumably, there is sufficient ATP hydrolysis in the mitochondrial matrix to support at least the minimal ΔΨM required for viability. The loss of mtDNA in atp3Δ yeast is likely the result of the dissipation of the mitochondrial inner membrane electrical potential that is created by an assembled proton-leaking, γ-deficient F1Fo-ATPase (23). The slow-growth phenotype of atp3Δ yeast is suppressed by loss of the gene encoding either the α subunit (atp1Δ) or the β subunit (atp2Δ) (20). In atp1Δ atp3Δ and atp2Δ atp3Δ yeast cells, the F1Fo-ATPase never assembles, so a proton leak is not formed. Therefore, mtDNA is stable, and the membrane potential is maintained by the electron transport chain (23).
Based upon the model for generation of ΔΨM in ρ° yeast, we hypothesized that the slow-growth phenotype of atp3Δ yeast can also be overcome by improved ATP hydrolysis activity in the mitochondrial matrix as opposed to mutations that stabilize mtDNA. Utilizing an unbiased genetic approach, we designed a suppressor selection strategy to isolate a mutation(s) that allows vigorous growth of atp3Δ ρ° yeast. The selection strategy yielded one spontaneous suppressing mutation that changed a conserved residue near the active site of F1-ATPase, leading to a more active, γ-deficient F1-ATPase. The observed increase in ATP hydrolysis activity and consequent increase in ΔΨM are consistent with the model that a threshold of ATP hydrolysis is required in the mitochondrial matrix for formation of the essential membrane potential in ρ° yeast. Analysis of the suppressing mutation by blue native polyacrylamide gel electrophoresis (BN-PAGE) also revealed that the α3β3 heterohexamer of the F1-ATPase forms in the absence of the γ subunit, providing additional insight into the assembly of the F1Fo-ATPase.
MATERIALS AND METHODS
Strains and media.
All S. cerevisiae strains were derived from D273-10B, and the genotypes of the strains used in this study are summarized in Table 1 (34). Standard genetic techniques were used to construct and analyze various yeast strains (30). Yeast strains were grown in complete glucose (YPD) medium containing 2% glucose, 2% Bacto peptone, 1% yeast extract, 40 mg/liter adenine, 40 mg/liter tryptophan; complete ethanol-glycerol (YPEG) medium containing 3% glycerol, 3% ethanol, 2% Bacto peptone, 1% yeast extract, 40 mg/liter adenine, 40mg/liter tryptophan; complete raffinose (YPRaf) medium containing 2% raffinose, 2% Bacto peptone, 1% yeast extract, 40 mg/liter adenine, 40 mg/liter tryptophan; or synthetic dextrose (SD) medium containing 2% glucose and 6.7 g/liter yeast nitrogen base without amino acids (Difco), supplemented with the appropriate nutrients (34). Where indicated, ethidium bromide (EtBr) was added at a concentration of 25 μg/ml (37), or Geneticin was added at a concentration of 300 μg/ml.
TABLE 1.
Yeast strains
| Strain | Genotypea | Source or reference |
|---|---|---|
| PTY44 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 [ρ+, TRP1] | 34 |
| JTY3 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 atp3-Δ1::kanMX4 [ρ°] | This study |
| JTY6 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 atp2-227 atp3-Δ1::kanMX4 [ρ°] | This study |
| TCY55 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 atp2-227 [ρ+, TRP1] | This study |
| TCY60 | MATα ura3-52 ade2 leu2-3,112 trp1-Δ1 atp2-Δ1::kanMX4 atp3-Δ1::kanMX4 [ρ+, TRP1] | This study |
| TCY46 | MATα ura3-52 ade2 leu2-3,112 trp1-Δ1 atp1-Δ1::kanMX4 [ρ+, TRP1] | This study |
| TCY64 | MATα ura3-52 lys2 leu2-3,112 trp1-Δ1 atp1-Δ1::kanMX4 atp3-Δ1::kanMX4 [ρ+, TRP1] | This study |
| TCY102 | MATα ura3-52 ade2 leu2-3,112 trp1-Δ1 atp1-Δ1::kanMX4 atp2-227 atp3-Δ1::kanMX4 [ρ+, TRP1] | This study |
| TCY103 | MATα ura3-52 ade2 leu2-3,112 trp1-Δ1 atp1-Δ1::kanMX4 atp2-227 [ρ+, TRP1] | This study |
| TCY38 | MATα ura3-52 ade2 leu2-3,112 trp1-Δ1 his3-Δ1::hisG opl [ρ+, TRP1] | This study |
The mitochondrial genotype is bracketed.
Nucleic acid techniques.
All manipulations of DNA were performed using standard techniques (26). Restriction enzymes and DNA modification enzymes were purchased from New England Biolabs (Beverly, MA). Plasmid DNA was prepared from Escherichia coli by boiling lysis (26).
Null mutations of ATP1, ATP2, and ATP3 were created by homologous gene replacement using DNA fragments generated by PCR. Genomic DNA prepared from a yeast deletion library (Research Genetics) containing the appropriate null allele (replaced with the KanMx4 cassette) was used as a template. Oligonucleotides used in the PCR to generate DNA for the disruptions were the following: 5′-CCATCTTTCCCATTGACGTT-3′(forward primer) and 5′-CTTGCAGGC GATATTTCCTT-3′ (reverse primer) for ATP1, 5′-AAGTGCTCCTCCTCAAGTCAA-3′ (forward primer) and 5′-AACAAGAAGGAAGCAGGGAA-3′ (reverse primer) for ATP2, and 5′-ACGTGATGGAGAACCAATGA-3′ (forward primer) and 5′-GCGAGAAGGACTTTCAAAAAA-3′ (reverse primer) for ATP3. PCR-generated DNAs were used to transform the yeast strain PTY44. Transformants resistant to Geneticin (Sigma Chemical Corp.) were putative null alleles of ATP1, ATP2, or ATP3 and were verified by PCR.
ATP2 (pATP2) and atp2-227 (pATP2-227) were amplified by PCR and cloned into the BamHI and SacI sites of pRS316 (CEN6, ARS, and URA3). Genomic DNA from appropriate yeast strains was used for PCRs using the following oligonucleotides: 5′-CGGATTGGTACCTTTGGATCCGGACAGGCCAATTGATAAA-3′ (forward primer) and 5′-GTGGGATCCTTTGAGCTCTTATCGCAGGTTGTGTGCTT-3′ (reverse primer). The primers were located 192 bp upstream and 319 bp downstream of the ATP2 open reading frame.
Isolation of mitochondria.
Mitochondrial isolation was performed essentially as described (9, 38). Yeast were grown in YPRaf medium, treated with zymolyase (Seikagaku America) to create spheroplasts and broken with a Dounce homogenizer, and mitochondria were collected by differential centrifugation. Mitochondrial yield was determined by spectrophotometric analysis (A280).
BN gel electrophoresis.
BN-PAGE was performed as previously described (25, 27, 28). Isolated mitochondrial pellets (200 to 300 μg of protein) were lysed in 35 μl of ice-cold digitonin buffer (1% [wt/vol] digitonin, 20 mM Tris-HCl, pH 7.4, 50 mM NaCl, 10% [vol/vol] glycerol). After a clarifying spin, 5 μl of sample buffer (5% [wt/vol] Serva Blue [Invitrogen], 100 mM Bis-Tris, pH 7.0, 500 mM ɛ-aminocaproic acid, 25% glycerol) was added to the supernatants. The samples (100 μg of protein for ρ+ yeast and 150 μg of protein for ρ° yeast) were separated on a 4 to 10% polyacrylamide gradient gel at 4°C. Atp2p and Atp1p were detected using antisera that were a gift from Trevor Lithgow. Signals were detected using the ECL detection method (Amersham).
Determination of mitochondrial F1Fo-ATPase activity and mitochondrial membrane potential.
ATPase activities were determined using isolated mitochondria essentially as previously described (36). Studies were performed in parallel, with and without 10 μg of oligomycin (Sigma). Each reaction was performed in triplicate from the same isolated mitochondria preparation. The reaction mixtures contained 120 μg of mitochondria and were incubated at 37°C for 12 min.
The effect of the addition of ATP upon the inner mitochondrial membrane potential was assayed by monitoring the potential-dependent quenching of the fluorescent dye rhodamine-123 (Molecular Probes, Eugene, OR) (11, 14). Fluorescence was monitored using a FluoroMax-2 spectrofluorometer operating in steady-state mode. Samples were excited at 502 nm, and emission was measured at 525 nm. Each reaction was performed using 250 μg of mitochondria in a final volume of 2.5 ml. At the indicated time, ATP was added to a final concentration of 1 mM. A total of 50 ng of valinomycin in ethanol (50 μl) was subsequently added at the indicated times. All experiments were performed in triplicate.
RESULTS
atp2-227 suppresses the slow-growth phenotype of atp3Δ yeast.
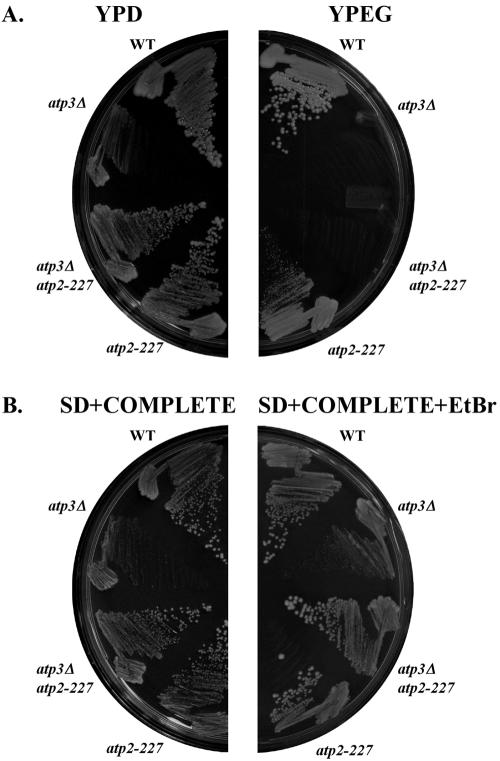
atp3Δ yeast cells grow slowly, even on rich glucose medium. Suppressing mutations of this phenotype were selected in complete SD medium containing EtBr, which induces the quantitative loss of mtDNA (31). Fifty independent cultures were grown to saturation, and approximately 1 × 107 cells/culture were plated to separate solid growth medium plates containing glucose as a carbon source. A single faster-growing colony was isolated (atp3Δ atp2-227), indicating that such suppressing mutations are rare. Yeast containing atp2-227 grew at a rate comparable to wild-type ρ+ cells on fermentable carbon sources (YPD medium and SD plus complete medium), regardless of the presence or absence of the γ subunit (Fig. 1, ATP3 versus atp3Δ). Similarly, atp2-227 atp3Δ ρ° and atp2-227 ATP3 ρ° yeast cells grow at a rate similar to wild-type ρ° yeast (Fig. 1B, SD+COMPLETE+EtBr).
FIG. 1.
Suppression of the atp3Δ slow-growth phenotype by atp2-227. The indicated strains were streaked on YPD and YPEG media (A) and on synthetic dextrose SD medium lacking or containing 25μg/ml EtBr (B) and incubated for 5 days at 30°C. Growth of yeast in the presence of EtBr induces the quantitative loss of mtDNA (12, 31). Strains: wild type (WT), PTY44; atp3Δ, JTY3; atp2-227 atp3Δ, JTY6; atp2-227, TCY55.
atp3Δ and atp3Δ atp2-227 yeast cells failed to grow on the nonfermentable carbon source (YPEG medium) because cells lacking the γ subunit of F1Fo-ATPase are incapable of respiratory growth (24, 37) (Fig. 1A). atp3Δ yeast cells containing the suppressing atp2-227 mutation were quantitatively ρ°, like unsuppressed atp3Δ yeast, after reintroduction of mtDNA upon backcrossing (data not shown). atp2-227 ATP3 yeast showed a dramatic reduction in growth on YPEG medium compared to wild-type yeast (Fig. 1A). This intrinsic slow-growth phenotype of atp2-227 ρ+ yeast on YPEG medium was not the result of increased ρ° formation, since cultures of both wild-type ρ+ and atp2-227 yeast contained approximately 98% ρ+ cells.
atp2-227 encodes a mutant form of the β subunit of F1Fo-ATPase.
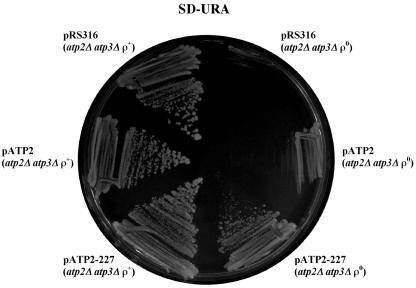
The respiratory-deficient phenotype of atp2-227 ρ+ yeast was used to clone the wild-type gene corresponding to the suppressing mutation. Plasmids that complemented the atp2-227 respiratory defect were identified and sequenced. The complementing clones encoded ATP2, the structural gene for the β subunit of the F1Fo-ATPase. The wild-type and suppressing ATP2 alleles were cloned by PCR into a replicating plasmid and expressed in yeast. atp2Δ atp3Δ yeast cells bearing a plasmid expressing the atp2-227 suppressing allele grew more vigorously in the absence of mtDNA than did the same yeast strain containing a plasmid expressing the wild-type ATP2 allele or a plasmid control (Fig. 2).
FIG. 2.
Rescue of atp3Δ slow-growth phenotype by atp2-227. atp2Δ atp3Δ yeast cells were transformed with pRS316 (vector control), pATP2 (wild-type ATP2), and pATP2-227 (atp2-227). Transformed strains were grown on plates of SD medium lacking uracil (URA) and lacking (ρ+) or containing (ρ°) 25 μg/ml EtBr and then streaked to SD medium lacking uracil and incubated for 5 days at 30°C to compare growth.
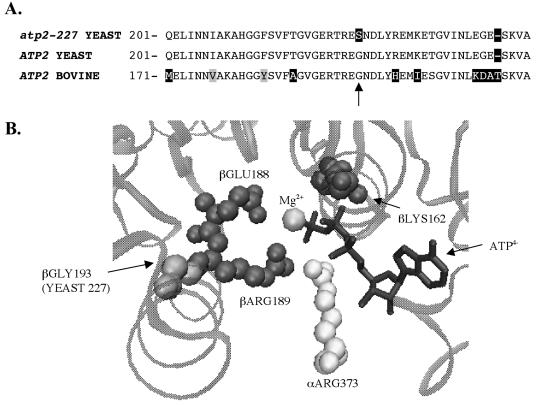
Analysis of the sequence for atp2-227 reveals a point mutation in codon 227 (GGT → AGT) that causes a change from glycine to serine. This codon is located in a highly conserved region of the β subunit (33) (Fig. 3A). Probing databases with the BLASTP algorithm identified only two orthologs out of 500 hits that do not contain a glycine at this position (2). The ortholog found in the purple sea urchin Strongylocentrotus purpuratus contains a serine in this position, like the atp2-227 allele, while one of several reported sequences for Anopheles gambiae contains a valine at the equivalent residue. Utilizing the high-resolution X-ray crystal structure data for the bovine ATP synthase, the equivalent glycine (193) residue was found to be near the ATP hydrolysis/synthesis active site (1, 15) (Fig. 3B). The location of this residue led us to hypothesize that the mutation suppressed the slow-growth phenotype of atp3Δ yeast by increasing ATP hydrolysis activity.
FIG. 3.
atp2-227 encodes a change from glycine to serine at residue 227 of the β-subunit of the F1-ATPase. (A) Predicted amino acid sequence of residues 201 to 249 of the yeast wild-type ATP2 and atp2-227 alleles aligned with the comparable region of bovine ATP2 using CLUSTALW (33). Black boxes indicate nonconserved amino acid substitutions, and gray boxes indicate conservative substitutions. The arrow marks position 227. (B) A three-dimensional rendering of the bovine F1-ATPase active site with the equivalent glycine residue (yeast residue 227 is equivalent to bovine βGLY193) was created using the program VMD, version 1.8.2 (15).
atp2-227 increases ATP hydrolysis activity in atp3Δ yeast.
The ATP hydrolysis activity measured in isolated mitochondria is summarized in Table 2. The ATP hydrolysis assays were performed in the presence and absence of the F1Fo-ATPase proton-translocation inhibitor, oligomycin (36). Wild-type ρ+ cells showed an approximate fourfold reduction in ATP hydrolysis in the presence of oligomycin, indicating that much of the ATP hydrolysis was a result of an assembled F1Fo-ATPase. atp2-227 ρ+ yeast cells also showed a fourfold reduction in ATP hydrolysis upon the addition of oligomycin, indicating that the mutation does not impact assembly of an intact F1Fo-ATPase. However, atp2-227 ρ+ yeast showed an overall reduction of F1Fo-ATPase activity relative to wild-type ρ+ yeast, indicating that the assembled F1Fo-ATPase has impaired catalytic ability, which is consistent with the reduced growth of atp2-227 ρ+ yeast on nonfermentable carbon sources (Fig. 1A). Mitochondria prepared from yeast that lack mtDNA show an overall reduction in ATP hydrolysis compared to mitochondria prepared from ρ+ yeast, and the ATPase activity is insensitive to oligomycin. Mitochondria isolated from atp3Δ ρ° yeast have very low ATPase activity, reduced to approximately one-third the level found in mitochondria of wild-type ρ° yeast. In contrast, mitochondria from atp2-227 atp3Δ ρ° yeast exhibit a comparable rate of ATP hydrolysis with that found in mitochondria prepared from wild-type ρ° yeast and show a statistically significant threefold increase of activity relative to that measured in atp3Δ ρ° yeast. Finally, atp2Δ atp3Δ ρ+ yeast showed virtually no ATP hydrolysis activity, consistent with the absence of a functional F1-ATPase. We conclude that the improved hydrolysis of ATP by F1-ATPase containing β subunits encoded by atp2-227 in yeast lacking the γ subunit is the likely mode of suppression of the slow-growth phenotype of atp3Δ ρ° yeast.
TABLE 2.
ATPase activity of isolated mitochondria
| Strain | Relevant genotype | ATP hydrolysis activity (μmol of Pi/min/mg)
|
Total ATP hydrolysis activity (% of control level)a | |
|---|---|---|---|---|
| With oligomycin | Without oligomycin | |||
| PTY44 | ATP3 ATP2 ρ+ | 1.33 ± 0.09 | 5.89 ± 0.03 | 100 |
| TCY55 | ATP3 atp2-227 ρ+ | 1.02 ± 0.09 | 4.09 ± 0.38 | 69.4 |
| JTY6 | atp3Δ atp2-227 ρ° | 1.06 ± 0.12 | 1.12 ± 0.07b | 19.0 |
| PTY44 (ρ°) | ATP3 ATP2 ρ° | 0.84 ± 0.05 | 1.00 ± 0.13 | 16.9 |
| JTY3 | atp3Δ ATP2 ρ° | 0.40 ± 0.11 | 0.33 ± 0.01b | 5.6 |
| TCY60 | atp3Δ atp2Δ ρ+ | 0.0130 ± 0.0030 | 0.0122 ± 0.0028 | 0.2 |
ATPase activity was normalized to wild-type (PTY44) in the absence of oligomycin.
The ATPase activity levels of the two strains are statistically different as determined by a t test of independent samples, with P < 0.0001.
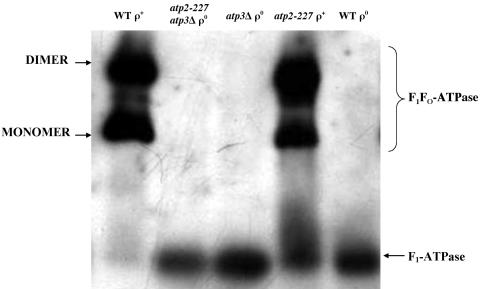
F1Fo-ATPase assembly in atp2-227,atp3Δ, and atp3Δ atp2-227 yeast.
Previous efforts found that a mutation in yeast that changed glycine-227 to aspartate impaired F1Fo-ATPase assembly, leading to a dramatic reduction in ATP hydrolysis activity in the presence of the γ subunit (22). In contrast, we found that the substitution of serine for glycine at position 227 resulted in only 25% less activity than wild-type F1Fo-ATPase and similarl sensitivity to inhibition by oligomycin. To directly address the physical state of F1Fo-ATPase, BN-PAGE was used to assess assembly of the multisubunit complex (Fig. 4). Isolated mitochondria were subjected to BN-PAGE, blotted to nitrocellulose membranes, and probed with antibody for the β subunit of F1Fo-ATPase (anti-Atp2p) (Fig. 4). All samples showed bands at the gel front, presumably indicating that free β subunits were present in each of the yeast strains (data not shown). The wild-type ρ+ and atp-227 ρ+ yeast showed bands that correspond to the monomer and dimer forms of the F1Fo-ATPase (4, 32). Therefore, the change of glycine to serine at residue 227 in the yeast F1Fo-ATPase does not significantly impact enzyme-complex assembly.
FIG. 4.
F1Fo-ATPase assembly in atp2-227, atp3Δ, and atp3Δ atp2-227 yeast. A total of 100 μg (ρ+ yeast) or 150 μg (ρ° yeast) of mitochondria isolated from the indicated strains in 1% digitonin buffer was resolved by BN-PAGE on a 4 to 10% polyacrylamide gel. Proteins were transferred to nitrocellulose and probed with antibodies against Atp2p. The band corresponding to the monomer of the F1Fo-ATPase migrated with a molecular weight slightly smaller than thyroglobulin (669 kDa; data not shown). Strains: wild type (WT) ρ+, PTY44 ρ+; atp2-227 atp3Δ ρ°, JTY6; atp3Δ ρ°, JTY3; atp2-227 ρ+, TCY55; WT ρ°, PTY44 ρ°.
None of the mitochondria isolated from ρ° yeast cells contained assembled F1Fo-ATPase, consistent with the absence of the Fo-portion, which is largely encoded by mtDNA (Fig. 4). However, all three ρ° mitochondrial preparations exhibited a band that has previously been attributed to an F1-ATPase (3, 4, 32). The band is of similar size, regardless of the presence (wild-type ρ°) or absence (atp3Δ ρ° and atp3Δ atp2-227 ρ°) of the γ subunit. When probed with antisera directed against the α subunit of F1Fo-ATPase (anti-Atp1p), all of the ρ° yeast cells showed a similar band corresponding to a free F1-ATPase (data not shown). Based upon BN-PAGE, the α3β3 heterohexamer of the F1-ATPase can assemble without the γ subunit when the β subunit is encoded by either ATP2 or atp2-227.
atp2-227 requires the α subunit (ATP1) of F1Fo-ATPase and a fully functional ANT for suppression of the slow-growth phenotype of atp3Δ yeast.
Based upon the proximity of the altered residue to the α subunit and the ability of the α3β3 ring to assemble in atp3Δ yeast, we hypothesized that the substitution of serine for glycine at position 227 in the β subunit alters the conformation of the F1-ATPase active site by interacting with residues of both the α and β subunits. Consequently, suppression of the atp3Δ slow-growth phenotype by atp2-227 would require the α subunit. Yeast cells lacking both the α (atp1Δ) and γ subunits (atp3Δ) of the F1Fo-ATPase remain ρ+ and grow faster than atp3Δ ρ° yeast (21) (Fig. 5A). However, atp1Δ atp3Δ yeast cells grow extremely slowly without mtDNA (Fig. 5B). The suppressing mutation atp2-227 was introduced into yeast lacking both the α and γ subunits of F1-ATPase to assess its ability to suppress the extreme-slow-growth phenotype of these yeast cells when they are ρ°. As previously observed, atp2-227 atp3Δ yeast grew better than atp3Δ yeast on both media (both are quantitatively ρ°) (Fig. 5A). However, the suppressing mutation did not affect the growth of atp1Δ ρ+ or atp1Δ atp3Δ ρ+ yeast (Fig. 5A). In addition, the presence of the altered β subunit encoded by atp2-227 did not improve growth of atp1Δ ρ° or atp1Δ atp3Δ ρ° yeast (Fig. 5B). Consequently, atp2-227 does not disrupt the interaction between α and β subunits but in fact requires this interaction to suppress the atp3Δ growth defect.
FIG. 5.
atp2-227 atp3Δ ρ° and atp2-227 ρ° yeast require the α subunit (ATP1) of the F1-ATPase for vigorous growth. The indicated strains were streaked to SD medium lacking (A) and containing (B) 25 μg/ml EtBr and incubated for 5 days at 30°C. Strains: wild type (WT), PTY44; atp3Δ, JTY3; atp2-227, TCY55; atp2-227 atp3Δ, JTY6; atp1Δ, TCY46; atp1Δ atp2-227, TCY103; atp1Δ atp3Δ, TCY64; atp1Δ atp2-227atp3Δ, TCY102.
Tetrad analysis of genetic crosses of haploid yeast containing atp3Δ, op1 (a loss-of-function point mutation of ANT), and atp2Δ or atp2-227 was performed to determine if ANT activity was required for suppression of the atp3Δ slow-growth phenotype by atp2-227. First, 100% of yeast spores bearing both atp3Δ and op1 were nonviable (n = 19). Similarly, 100% of atp3Δ op1 atp2-227 spores were nonviable (n = 5). These results are consistent with our hypothesis that atp3Δ ρ° and atp3Δ atp2-227 ρ° yeast require ANT to generate a sufficient ΔΨM for viability.
atp2-227 increases ΔΨM in yeast lacking the γ subunit (atp3Δ) of F1Fo-ATPase.
Mitochondria isolated from atp2-227 atp3Δ ρ° yeast have increased ATPase activity compared to mitochondria prepared from atp3Δ ρ° yeast. Theoretically, this improved ATP hydrolysis provides an increased amount of ADP3− to exchange for ATP4− through ANT, increasing theinner membrane potential and consequently improving growth. To test this hypothesis, changes of ΔΨM in isolated mitochondria were monitored in response to added ATP by measuring the uptake of the fluorescent dye, rhodamine 123 (11). These changes were recorded after the addition of mitochondria, ATP, and the ionophore valinomycin, which dissipated the ΔΨM generated by hydrolysis of ATP. Rhodamine 123 fluorescence was quenched in response to ATP4− in the absence of mitochondria but showed little response to valinomycin after an initial increase in intensity (<1%) (data not shown). Therefore, the response to valinomycin can be used to compare the ΔΨM of mitochondria prepared from different strains of yeast.
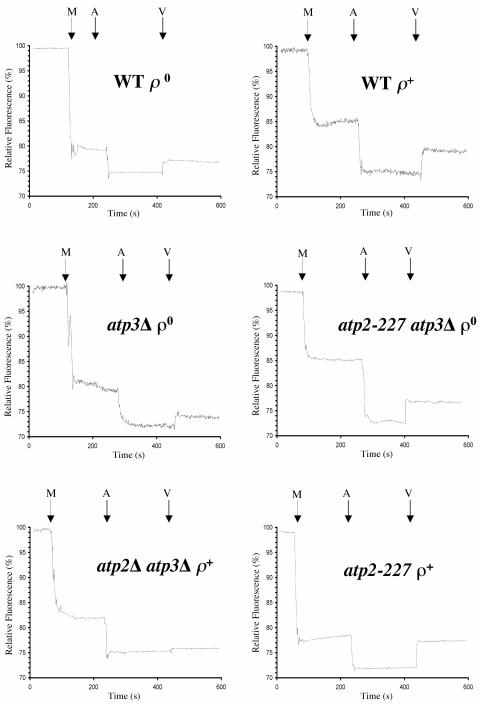
Mitochondria prepared from wild-type ρ+ yeast had a 5% increase in relative fluorescence upon the addition of valinomycin, compared to a 2% increase in fluorescence under similar conditions with mitochondria prepared from wild-type ρ° cells (Fig. 6A). Mitochondria isolated from cells that lack the γ subunit (atp3Δ) exhibited a 2% increase in relative fluorescence upon valinomycin addition, similar to mitochondria prepared from wild-type ρ° yeast (Fig. 6B). Mitochondria isolated from atp2-227 atp3Δ ρ° yeast had a 5% increase in fluorescence upon valinomycin treatment, equivalent to that found in mitochondria prepared from wild-type ρ+ yeast (Fig. 6B). Mitochondria prepared from yeast lacking β and γ subunits (atp2Δ atp3Δ) showed essentially no response to valinomycin (Fig. 6C), indicating that very little ATP hydrolysis occurred, consistent with the inability to grow without mtDNA (Fig. 2). Finally, mitochondria isolated from atp2-227 ρ+ yeast also showed a 5% increase in fluorescence in response to valinomycin (Fig. 6C). The greater response of mitochondria prepared from atp3Δ atp2-227 ρ° yeast to valinomycin indicates that atp3Δ atp2-227 ρ° yeast cells have a greater ΔΨM than atp3Δ ρ° yeast.
FIG. 6.
atp2-227 atp3Δ ρ° yeast generates a greater inner mitochondrial membrane potential in response to ATP compared to atp3Δ ρ° yeast. At the indicated times, 250 μg of mitochondria isolated from the indicated strains (M), ATP to a final concentration of 1 mM (A), and 50 ng of valinomycin in 95% ethanol (V) were added to a final volume of 2.5 ml of reaction buffer containing 1 μM rhodamine 123 (Molecular Probes) (11, 14). The relative fluorescence was monitored using a FluoroMax-2 spectrofluorometer operating in the steady-state mode. The samples were excited at 502 nm, and emission was measured at 525 nm. Strains: wild type (WT) ρ+, PTY44 ρ+; WT ρ°, PTY44 ρ°; atp3Δ ρ°, JTY3; atp2-227 atp3Δ ρ°, JTY6; atp2Δ atp3Δ ρ+, TCY60; atp2-227 ρ+, TCY55.
DISCUSSION
For yeast that lack mtDNA, and consequently a functional electron transport chain and ATP synthase, the major role of the F1-ATPase is to generate ADP3− in support of the electrogenic exchange of ADP3− for ATP4− through ANT in ρ° yeast. Yeast cells lacking the γ subunit of F1-ATPase quantitatively lose mtDNA and grow extremely slowly, likely because ATP hydrolysis activity in the mitochondrial matrix is severely compromised. By employing an unbiased genetic approach, we identified a suppressing mutation in the β subunit (atp2-227) that improves growth of atp3Δ ρ° yeast (Fig. 1) by improving ATP hydrolysis activity (Table 2) and consequently increasing ΔΨM (Fig. 6). In addition, we have demonstrated that atp3Δ ρ°and atp2-227 atp3Δ ρ° yeast cells assemble a functional F1-ATPase (Fig. 4) and require both an interaction with the α subunit and the presence of a fully function ANT for growth (Fig. 5) (see Results).
The arginine residue (αArg373) of the bovine α subunit, which is part of the conserved arginine-finger domain of the active site, is proposed to sense the presence or absence of theγ-phosphate of the bound ATP (16). Because of the relative proximity of the β subunit residue changed by atp2-227 to the active site of the enzyme (corresponding to G193S in the bovine enzyme) (Fig. 3B), we hypothesize that the introduction of a serine at position 227 changes the active site either through alteration of the hydrogen bonding network or steric effects, potentially affecting either the position of αArg373 and/or the βArg189. Such an alteration of the active site may lead to improved ATP hydrolysis in the absence of the γ subunit. This postulated repositioning of key active-site residues in the atp2-227 allele would be the structural basis for suppression of the slow-growth phenotype of atp3Δ yeast. The corresponding alteration of the interaction between the active site formed by the α and β subunits and the rotating γ subunit may also explain the reduced ability of strains bearing the atp2-227 allele to grow on carbon sources that require oxidative phosphorylation.
Finally, the isolation of the spontaneous suppressing mutation and its subsequent characterization also provide further insight into the assembly of the F1Fo-ATPase. Previously, deletions in the α and β subunits were shown to suppress the slow-growth phenotype of atp3Δ yeast (23). The current model of suppression suggests that the Fo-portion of the ATPase “senses” the α3β3 heterohexamer during assembly, and the important proton-channel subunit encoded by ATP6 is added to the complex. Once formed, the uncoupled, γ-less enzyme freely passes protons down an electrochemical gradient, dissipating ΔΨM and ultimately destabilizing mtDNA. In the work presented here, we provide BN-PAGE evidence suggesting that the α3β3 heterohexamer of the F1-ATPase is assembled in the presence or absence of the γ subunit, supporting this model. This suggests that an interaction between the α and β subunits and the Fo-portion of the ATPase, independent of the γ subunit, is a key step during assembly of the ATP synthase enzyme-complex.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (GM068066).
We thank Sharon Ackerman for assistance with the protocol for the ATP hydrolysis assay, Trevor Lithgow for generously providing antisera, and Michael T. Ryan for the BN-PAGE protocol. We are also grateful to Mary Brownson and Jeremy Tiller for technical assistance and to Brian Francis, Mary Thorsness, and Karen White for critical review of the manuscript.
REFERENCES
- 1.Abrahams, J. P., A. G. W. Leslie, R. Lutter, and J. Walker. 1994. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature (London) 370:621-628. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Appleby, R. D., W. K. Porteous, G. Hughes, A. M. James, D. Shannon, Y. H. Wei, and M. P. Murphy. 1999. Quantitation and origin of the mitochondrial membrane potential in human cells lacking mitochondrial DNA. Eur. J. Biochem. 262:108-116. [DOI] [PubMed] [Google Scholar]
- 4.Arnold, I., K. Pfeiffer, W. Neupert, R. A. Stuart, and H. Schagger. 1998. Yeast mitochondrial F1F0-ATP synthase exists as a dimer: identification of three dimer-specific subunits. EMBO J. 17:7170-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker, K. P., and G. Schatz. 1991. Mitochondrial proteins essential for viability mediate protein import into yeast mitochondria. Nature (London) 349:205-208. [DOI] [PubMed] [Google Scholar]
- 6.Buchet, K., and C. Godinot. 1998. Functional F1-ATPase essential in maintaining growth and membrane potential of human mitochondrial DNA-depleted rho° cells. J. Biol. Chem. 273:22983-22989. [DOI] [PubMed] [Google Scholar]
- 7.Chen, X. J., and G. D. Clark-Walker. 1999. Alpha and beta subunits of F1-ATPase are required for survival of petite mutants in Saccharomyces cerevisiae. Mol. Gen. Genet. 262:898-908. [DOI] [PubMed] [Google Scholar]
- 8.Clark-Walker, G. D., P. M. Hansbro, F. Gibson, and X. J. Chen. 2000. Mutant residues suppressing rho°-lethality in Kluyveromyces lactis occur at contact sites between subunits of F(1)-ATPase. Biochim. Biophys. Acta 1478:125-137. [DOI] [PubMed] [Google Scholar]
- 9.Daum, G., P. C. Böhni, and G. Schatz. 1982. Import of proteins into mitochondria. Energy-dependent uptake of precursors by isolated mitochondria. J. Biol. Chem. 257:13028-13035. [PubMed] [Google Scholar]
- 10.Dunn, C. D., and R. E. Jensen. 2003. Suppression of a defect in mitochondrial protein import identifies cytosolic proteins required for viability of yeast cells lacking mitochondrial DNA. Genetics 165:35-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emaus, R. K., R. Grunwald, and J. J. Lemasters. 1986. Rhodamine 123 as a probe of transmembrane potential in isolated rat-liver mitochondria: spectral and metabolic properties. Biochim. Biophys. Acta 850:436-448. [DOI] [PubMed] [Google Scholar]
- 12.Fox, T. D., L. S. Folley, J. J. Mulero, T. W. McMullin, P. E. Thorsness, L. O. Hedin, and M. C. Costanzo. 1991. Analysis and manipulation of yeast mitochondrial genes. Methods Enzymol. 194:149-165. [DOI] [PubMed] [Google Scholar]
- 13.Gasser, S. M., G. Daum, and G. Schatz. 1982. Import of proteins into mitochondria. Energy-dependent uptake of precursors by isolated mitochondria. J. Biol. Chem. 257:13034-13041. [PubMed] [Google Scholar]
- 14.Giraud, M. F., and J. Velours. 1997. The absence of the mitochondrial ATP synthase delta subunit promotes a slow growth phenotype of rho− yeast cells by a lack of assembly of the catalytic sector F1. Eur. J. Biochem. 245:813-818. [DOI] [PubMed] [Google Scholar]
- 15.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33-38, 27-28. [DOI] [PubMed] [Google Scholar]
- 16.Kagawa, R., M. G. Montgomery, K. Braig, A. G. Leslie, and J. E. Walker. 2004. The structure of bovine F(1)-ATPase inhibited by ADP and beryllium fluoride. EMBO J. 23:2734-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kominsky, D. J., M. P. Brownson, D. Updike, and P. E. Thorsness. 2002. Genetic and biochemical basis for viability of yeast lacking mitochondrial genomes. Genetics Society of America, Bethesda, M.D. [DOI] [PMC free article] [PubMed]
- 18.Kominsky, D. J., and P. E. Thorsness. 2000. Expression of the Saccharomyces cerevisiae gene YME1 in the petite-negative yeast Schizosaccharomyces pombe converts it to a petite-positive yeast. Genetics 154:147-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovacova, V., J. Irmlerova, and L. Kovac. 1968. Oxidative phosphorylation in yeast, IV: combination of a nuclear mutation affecting oxidative phosphorylation with cytoplasmic mutation to respiratory deficiency. Biochim. Biophys. Acta 162:157-163. [DOI] [PubMed] [Google Scholar]
- 20.Lai-Zhang, J., and D. M. Mueller. 2000. Complementation of deletion mutants in the genes encoding the F1-ATPase by expression of the corresponding bovine subunits in yeast S. cerevisiae. Eur. J. Biochem. 267:2409-2418. [DOI] [PubMed] [Google Scholar]
- 21.Lai-Zhang, J., Y. Xiao, and D. M. Mueller. 1999. Epistatic interactions of deletion mutants in the genes encoding the F1-ATPase in yeast Saccharomyces cerevisiae. EMBO J. 18:58-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang, Y., and S. H. Ackerman. 1996. Characterization of mutations in the beta subunit of the mitochondrial F1-ATPase that produce defects in enzyme catalysis and assembly. J. Biol. Chem. 271:26522-26528. [DOI] [PubMed] [Google Scholar]
- 23.Mueller, D. M. 2000. Partial assembly of the yeast mitochondrial ATP synthase. J. Bioenerg. Biomembr. 32:391-400. [DOI] [PubMed] [Google Scholar]
- 24.Paul, M.-F., S. Ackerman, J. Yue, G. Arselin, J. Velours, and A. Tzagoloff. 1994. Cloning of the yeast ATP3 gene coding for the γ-subunit of F1 and characterization of atp3 mutants. J. Biol. Chem. 269:26158-26164. [PubMed] [Google Scholar]
- 25.Ryan, M. T., H. Muller, and N. Pfanner. 1999. Functional staging of ADP/ATP carrier translocation across the outer mitochondrial membrane. J. Biol. Chem. 274:20619-20627. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Schagger, H., W. A. Cramer, and G. von Jagow. 1994. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217:220-230. [DOI] [PubMed] [Google Scholar]
- 28.Schagger, H., and G. von Jagow. 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199:223-231. [DOI] [PubMed] [Google Scholar]
- 29.Schleyer, M., B. Schmidt, and W. Neupert. 1982. Requirement of a membrane potential for the posttranslational transfer of proteins into mitochondria. Eur. J. Biochem. 125:109-116. [DOI] [PubMed] [Google Scholar]
- 30.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Slonimski, P. P., G. Perrodin, and J. H. Croft. 1968. Ethidium bromide induced mutation of yeast mitochondria: complete transformation of cells into respiratory deficient non-chromosomal “petites.” Biochem. Biophys. Res. Commun. 30:232-239. [DOI] [PubMed] [Google Scholar]
- 32.Soubannier, V., J. Vaillier, P. Paumard, B. Coulary, J. Schaeffer, and J. Velours. 2002. In the absence of the first membrane-spanning segment of subunit 4(b), the yeast ATP synthase is functional but does not dimerize or oligomerize. J. Biol. Chem. 277:10739-10745. [DOI] [PubMed] [Google Scholar]
- 33.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thorsness, P. E., and T. D. Fox. 1993. Nuclear mutations in Saccharomyces cerevisiae that affect the escape of DNA from mitochondria to the nucleus. Genetics 134:21-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thorsness, P. E., K. H. White, and T. D. Fox. 1993. Inactivation of YME1, a member of the ftsH-SEC18-PAS1-CDC48 family of putative ATPase-encoding genes, causes increased escape of DNA from mitochondria in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:5418-5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tzagoloff, A. 1979. Oligomycin-sensitive ATPase of Saccharomyces cerevisiae. Methods Enzymol. 55:351-358. [DOI] [PubMed] [Google Scholar]
- 37.Weber, E. R., R. S. Rooks, K. S. Shafer, J. W. Chase, and P. E. Thorsness. 1995. Mutations in the mitochondrial ATP synthase gamma subunit suppress a slow-growth phenotype of yme1 yeast lacking mitochondrial DNA. Genetics 140:435-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yaffe, M. P. 1991. Analysis of mitochondrial function and assembly. Methods Enzymol. 194:627-643. [DOI] [PubMed] [Google Scholar]