Abstract
In most retroviruses, the first nucleotide added to the tRNA primer becomes the right end of the U5 region in the right long terminal repeat (LTR); the removal of this tRNA primer by RNase H defines the right end of the linear double-stranded DNA. Most retroviruses have two nucleotides between the 5′ end of the primer binding site (PBS) and the CA dinucleotide that will become the end of the integrated provirus. However, human immunodeficiency virus type 1 (HIV-1) has only one nucleotide at this position, and HIV-2 has three nucleotides. We changed the two nucleotides (TT) between the PBS and the CA dinucleotide of the Rous sarcoma virus (RSV)-derived vector RSVP(A)Z to match the HIV-1 sequence (G) and the HIV-2 sequence (GGT), and we changed the CA dinucleotide to TC. In all three mutants, RNase H removes the entire tRNA primer. Sequence analysis of RSVP(HIV2) proviruses suggests that RSV integrase can remove three nucleotides from the U5 LTR terminus of the linear viral DNA during integration, although this mutation significantly reduced virus titer, suggesting that removing three nucleotides is inefficient. However, the results obtained with RSVP(HIV1) and RSVP(CATC) show that RSV integrase can process and integrate the normal U3 LTR terminus of a linear DNA independently of an aberrant U5 LTR terminus. The aberrant end can then be joined to the host DNA by unusual processes that do not involve the conserved CA dinucleotide. These unusual events generate either large duplications or, less frequently, deletions in the host genomic DNA instead of the normal 5- to 6-base duplications.
After the viral core enters a susceptible host cell, the single-strand viral RNA genome is reverse transcribed into a linear double-stranded DNA (dsDNA). Reverse transcription (RT) is absolutely dependent on the two distinct enzymatic activities of RT: a DNA polymerase that can copy both DNA and RNA templates and an RNase H that cleaves the RNA strand of RNA:DNA duplexes. The synthesis of minus-strand DNA is initiated from a cellular tRNA primer that is base paired at the primer binding site (PBS) located immediately adjacent to the U5 sequence in the viral genome. The tRNA used to prime reverse transcription in human immunodeficiency virus type 1 (HIV-1) is tRNALys (25, 30), and in Rous sarcoma virus (RSV) it is tRNATrp (2, 22, 26, 28, 31). Plus-strand DNA synthesis is initiated from a polypurine tract (PPT) primer that is generated by specific RNase H cleavages of the RNA genome adjacent to U3.
The ends of the unintegrated viral DNA are defined by the sites where RNase H removes the plus-strand and minus-strand primers. More specifically, the left (U3) long terminal repeat (LTR) terminus of the linear dsDNA molecule is defined by the removal of PPT primer. In the case of HIV-1, the PPT sequence is an important determinant for the proper generation and removal of the PPT primer by HIV-1 RNase H (7, 17, 18, 23, 26). The right (U5) LTR terminus of the linear dsDNA molecule is defined by the removal of the tRNA used to initiate minus-strand DNA synthesis. For most retroviruses, cleavage occurs at the junction between the RNA primer and the first DNA nucleotide added by RT. However, the tRNALys primer is cleaved by HIV-1 RNase H between the terminal rA and the adjacent rC of the tRNA primer, one nucleotide from the RNA-DNA junction (10, 24, 32).
Following reverse transcription, the linear dsDNA molecule is transported to the nucleus, where it is the precursor to the integrated provirus. A virally encoded integrase (IN) recognizes sequences at the ends of the linear molecule and catalyzes integration in a two-step reaction. In the first step, integrase removes a specific number of nucleotides, usually two, from each of the 3′ ends of the linear viral DNA (6, 9, 16, 27). The processed ends are then joined to host chromosomal DNA by integrase, creating a duplication of a short sequence from the target site, which flanks the integrated provirus as a direct repeat of 4 to 6 bp (8, 14, 15, 27). However, the sequence of HIV-2 proviruses suggest that retroviral integrases do not always remove two nucleotides from the end of linear viral DNA; there are three nucleotides between the PBS and the conserved CA found at the U5 boundary of the HIV-2 provirus. Integration of HIV-2 DNA is accompanied by the asymmetric loss of two and three nucleotides, respectively, from the U3 and U5 ends of the linear dsDNA prior to integration (33).
We were interested in asking how sequence differences between the PBS and the CA dinucleotide would affect the ability of the RNase H of RSV to create the U5 LTR terminus of a linear DNA and integrase to insert the resulting linear DNA into the host genome. We changed the sequence (TT) that is normally present between the PBS and the CA dinucleotide of the RSV-derived vector, RSVP(A)Z (21), to match the HIV-1 sequence (G) and the HIV-2 sequence (GGT). We also changed the CA dinucleotide of RSVP(A)Z to TC to ask how this mutation affects integration. In all three of the mutants, RNase H removes the entire tRNA. We recovered the full length of proviruses flanked by cellular genomic DNA and analyzed the integration of viral DNAs with aberrant U5 LTR termini. Sequence analysis of RSVP(HIV2) suggests that RSV integrase can remove three nucleotides from the U5 LTR terminus of the linear viral DNA during integration, like the IN of HIV-2, although this mutation significantly reduced virus titer, which suggests that RSV IN removes three nucleotides inefficiently. Sequence analysis of RSVP(HIV1) and RSVP(CATC) proviruses shows that RSV integrase can process and integrate the normal U3 LTR terminus of a linear DNA even if the other end of the linear DNA is aberrant. The aberrant U5 LTR terminus can be joined to the host DNA by unusual processes that do not involve the conserved CA dinucleotide. Furthermore, these unusual events generate either large duplications or, less frequently, deletions, instead of the normal 5- to 6-base duplications in the host genomic DNA.
MATERIALS AND METHODS
Plasmid construction.
KS(MluI) was derived from the pBluescript II KS phagemid by replacing a SacI-to-SpeI segment with two complementary oligonucleotides that included the following cloning sites: AatII, NcoI, and MluI. To introduce mutations at the end of the U5 sequence, a 0.9-kb MluI-to-SacI fragment containing the PPT, both LTRs, and the PBS region of RSVP(A)Z was inserted into the MluI/SacI site of the KS(MluI) plasmid, which generated KS/RSVP(A)Z. In vitro site-directed mutagenesis was carried out with KS/RSVP(A)Z as a template and the appropriate set of primers using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) following the manufacturer's recommendations. Sequence analysis of the resulting plasmids (KS/CATC, KS/HIV1, and KS/HIV2) was performed to ensure that only desired mutations were introduced. The MluI-SacI fragments of KS/CATC, KS/HIV1, and KS/HIV2 were used to replace the corresponding MluI-SacI fragment in RSVP(A)Z, which generated RSVP(CATC), RSVP(HIV1), and RSVP(HIV2), respectively.
To make it easy to measure virus titer, the gfp gene was placed under the control of the cytomegalovirus (CMV) promoter from the pLEGFP-N1 plasmid (Clontech, Palo Alto, CA) and was introduced immediately upstream of the plasmid-recovery cassette, giving rise to RSVP/CMV-GFP. However, the genome of RSVP/CMV-GFP was too large to be efficiently packaged into viral particles. To make RSVP/CMV-GFP smaller, part of the env gene was removed by cleaving with PciI (which removed ∼1.26 kb), generating RSVP/GFP(Δenv). Finally, the PciI-MluI fragment of RSVP/GFP(Δenv) was used to replace the corresponding PciI-MluI fragments in RSVP(CATC), RSVP(HIV1), and RSVP(HIV2), generating RSVP/GFP(CATC), RSVP/GFP(HIV1), and RSVP/GFP(HIV2), respectively.
The env gene expression plasmid, CMV-env(A), was constructed as follows. The 2-kb KpnI-to-ClaI fragment containing the env region of RSVP(A)Z was introduced into the KpnI/NotI site of pDsRed2-N1 plasmid (Clontech) by three-piece ligation with duplex oligonucleotides containing the ClaI and NotI restriction sites. The CMV promoter was introduced into the plasmid immediately upstream of the cloned env gene.
The D64K mutant was generated by subcloning the 862-bp PmlI-to-KpnI DNA fragment from RSVP(A)Z into a pBluescript II SK+ derivative that had a PmlI polylinker inserted into the multicloning site. The D64K mutation was introduced into the subcloned fragment by a QuickChange site-directed mutagenesis kit (Stratagene) using the primers D64For (5′-ACAGATATGGCAGACAAAGTTTACGCTTGAGCCTA) and D64Rev (5′-TAGGCTCAAGCGTAAACTTTGTCTGCCATATCTGT). The mutation was confirmed by sequencing and introduced into RSVP(A)Z using the PmlI and KpnI restriction enzyme sites.
Cells, transfection, and infection.
DF-1, a continuous line of chicken fibroblasts, was derived from EV-O embryos (12, 29). 293 cells expressing the tva receptor (293-tva) were kindly provided by John A. Young. The cells were maintained in Dulbecco's modified Eagle medium (GIBCO, Carlsbad, Calif.) supplemented with 5% fetal bovine serum, 5% newborn calf serum, 100 U of penicillin per ml, and 100 μg of streptomycin (Quality Biological, Inc., Gaithersburg, Md.) per ml. DF-1 cells were incubated at 39°C with 5% CO2, and 293-tva cells were incubated at 37°C with 5% CO2. Cells were passaged 1:5 at confluence with trypsin DeLarco (pH 6.8). Plasmid DNA encoding RSVP(HIV1), RSVP(HIV2), and RSVP(CATC) was introduced into DF-1 cells by using the calcium phosphate transfection kit (Invitrogen, Carlsbad, Calif.) following the manufacturer's recommendations. DF-1 cells were incubated with medium containing 15% glycerol for 5 min at 39°C 16 h after transfection. The cells were washed twice with phosphate-buffered saline and incubated in fresh medium for 48 h. The 48-h supernatants were harvested and subjected to low-speed centrifugation to remove cellular debris. A portion of the infectious virions was used to infect fresh DF-1 cells or 293-tva cells. Selection for zeocin resistance was initiated 48 h postinfection with 300 μg/ml of zeocin (Invitrogen). The zeocin titers were similar to the green fluorescent protein (GFP) titers (see the next section).
Measurement of virus titer.
Viral stocks generated by cotransfection with the various RSVP/GFP(Δenv) vectors and CMV-env(A) were titered on 293-tva cells, and the percentage of GFP-positive cells was quantitated by flow cytometry 48 h after infection. The values were normalized to the amount of p27 antigen present in the viral stocks, as measured by p27 antigen capture enzyme-linked immunosorbent assay. The relative titer was determined by normalizing the resulting values to wild-type RSVP/GFP(Δenv).
Recovery of 2-LTR circle junctions.
Genomic DNA was isolated from the infected cells ca. 48 h after infection by using a DNeasy tissue kit (QIAGEN). A portion of the recovered DNA was used to transform ElectroMax DH10B or DH5α (Invitrogen) by electroporation. Electroporation was performed as described previously (21).
Lac repressor-mediated recovery of integrated retroviral DNA.
Genomic DNA was isolated from infected cells that survived zeocin selection using a QIAamp DNA blood maxi kit (QIAGEN). To recover full-length proviruses, 100 to 200 μg of genomic DNA was digested with DraI. Lac repressor-mediated viral DNA recovery was carried out essentially as described previously (21). Briefly, the digested DNA was incubated with purified Lac repressor protein, and the DNA-Lac repressor protein mixture was filtered through a nitrocellulose membrane. The enriched DNA was eluted with 10 mM isopropyl-β-d-thiogalactopyranoside and precipitated with ethanol. The precipitated DNA was ligated with T4 DNA ligase (30 U/200 μl; Roche, Indianapolis, Ind.) for 18 h at 16°C. The ligated DNA was then used to transform Escherichia coli as described above. After a 3- to 4-h recovery period in SOC medium at 37°C, the transformed bacteria were plated onto low-salt Luria-Bertani plates containing 50 μg of zeocin per ml.
Sequencing of 2-LTR circle junctions and integration sites.
Recovered plasmids were directly sequenced using the PBS primer for the 2-LTR circle junction and the U3 integration sites (5′-ACTATCACGTCGGGGTCACCA) and the PPT primer for the U5 integration sites (5′-AGGAGTCCCCTTAGGATATAG). A gag-up primer was used to confirm that the mutations we introduced were present in the recovered plasmids (5′-CCGACGGTACTCAGCTTCTGC). In those cases in which the provirus was flanked by substantial duplications, we sequenced through the ends of the duplications to demonstrate that the duplicated regions were present on both ends of the provirus. We also did additional sequencing to determine the structure of the complex proviruses. Human and chicken genomic sequences were analyzed by BLAT searches (http://genome.ucsc.edu/cgi-bin/hgBlat).
RESULTS
The U5-end mutations affect virus titer.
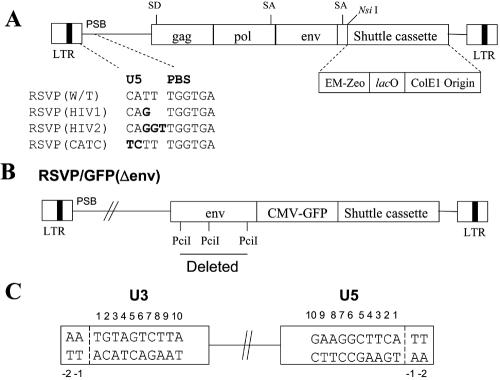
We changed the sequence of RSVP(A)Z (TT) between the PBS and the CA dinucleotide normally found at the U5 end of the provirus to match the sequence in the HIV-1 (G) and HIV-2 (GGT) genome. This should help us understand how RT and IN collaborate to create a linear DNA and insert it into the host genome. We also changed the CA dinucleotide of RSVP(A)Z to TC to ask how this affects integration (Fig. 1A).
FIG. 1.
Schematic drawing showing the structure of the RSVP vectors. (A) Schematic representation of the recovery cassette and the mutants. Mutations are indicated in boldface. EM-Zeo, EM-7 promoter-zeocin resistance gene; lacO, lac operator. (B) RSVP/GFP(Δenv). The gfp gene, under the control of the CMV promoter, was introduced immediately upstream of the recovery cassette. The portion that was deleted from env is indicated. (C) Schematic numbering of the nucleotide positions in the ends of the provirus. The last nucleotide on each end of the provirus is number 1.
To measure virus titer, we inserted the gfp gene under the control of the CMV promoter immediately upstream of the plasmid cassette. However, the introduction of the CMV promoter and the gfp gene made the viral genome too large to be packaged efficiently (∼10.25 kb). To make the viral genome smaller, a part of the env gene (∼1.26 kb) was removed (Fig. 1B). To provide an envelop glycoprotein in trans, the plasmid CMV-env was constructed in which the envA gene is expressed under the control of the CMV promoter. Viruses with genomes containing the U5-end mutation were generated by cotransfecting DF-1 cells with plasmids that contain viral genomes with the U5-end mutations and the CMV-env plasmid. The virions were used to infect 293-tva cells, and GFP expression was measured using fluorescence-activated cell sorting; the titers were normalized relative to the amount of p27 in the infecting viral stock. RSVP(HIV1) and RSVP(CATC) decreased virus titer to about 46% and 36% of the wild-type level, respectively. RSVP(HIV2) decreased virus titer to about 3.5% of the wild-type level. Similar results were obtained with zeocin selection, which suggests that the measured titer depends on the integration of the viral DNA (data not shown).
Recovery and analysis of 2-LTR circle junctions.
To recover 2-LTR circle junctions, the RSVP vectors containing the U5-end mutations were transfected into DF-1 cells. The virions were harvested and used to infect fresh DF-1 cells. DNA was prepared from the infected cells and used to transform E. coli; zeocin-resistant cells were selected.
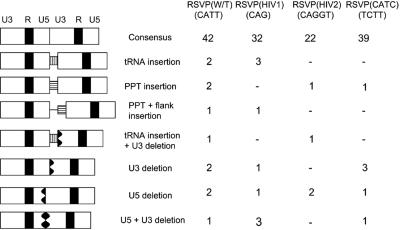
Analysis of the 2-LTR circle junctions from the mutants is shown in Fig. 2. Although the 2-LTR circle junctions obtained from the mutants included both insertions and deletions, the frequency of the aberrant 2-LTR circle junctions was not significantly different from that seen in infections with viruses with a wild-type sequence adjacent to the PBS. Moreover, the proportion of consensus sequence junctions that contained the entire sequence from both ends of the linear viral DNA was the same for the mutants and wild type. This result suggests that, for these mutants, the RNase H of RSV RT usually removes the entire tRNA during plus-strand synthesis.
FIG. 2.
2-LTR circle junctions isolated from infected cells. A consensus circle junction is shown at the top. Different types of aberrant 2-LTR circle junctions are shown. The PBS is indicated by a white box with black vertical bars; the PPT is a white box with black horizontal bars; the deletions are indicated by black jagged boxes. W/T, wild type.
Recovery of the U5 LTR terminus of integrated viral DNA.
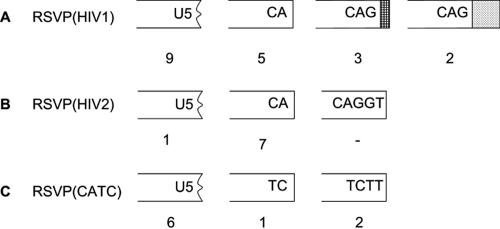
To analyze the ability of integrase to process and insert viral DNA with aberrant ends, genomic DNA was isolated from infected 293-tva cells and digested with NsiI. This allowed us to recover the right (U5) junction between the U5 end of the provirus and host genomic DNA. One NsiI site comes from the RSVP genome; the second NsiI site comes from the adjacent cellular DNA. The NsiI-digested DNAs were enriched for viral sequences by binding to the Lac repressor protein; the enriched DNA was self ligated and used to transform E. coli. Rescued plasmids were directly sequenced with a primer from the U5 sequence in the viral vector. Sequence analyses of RSVP(HIV2) showed that a CA dinucleotide was present at the ends of the viral DNA in seven of eight cases (Fig. 3B). This result implies that the RSV integrase successfully removed three bases from the U5 end of the linear viral DNA during integration. However, sequence analyses of RSVP(HIV1) showed three types of integration events: (i) there were junctions involving CA, (ii) there were events in which viral DNA was inserted at other positions in U5, causing a deletion of a portion of the U5 sequence, and (iii) there were junctions in which there was an integration of an unprocessed U5 end (Fig. 3A). We also found tRNA insertions in three proviral clones. In the case of RSVP(CATC), most of the integrations involved deletions of the U5 sequence; however, we obtained two clones in which the last two nucleotides at the U5 end of the linear viral DNA were not removed (Fig. 3C). We also obtained complex insertions, some of which may have involved autointegration events (Fig. 3A). To obtain a more complete picture of the aberrant insertions, we recovered complete proviruses with cellular DNA attached to both ends.
FIG. 3.
Recovery of the 3′ end of integrated viral DNA. Deletions in U5 are indicated by a black jagged end. Insertions of tRNA are shown by a box with cross bars; viral DNA insertions are shown by a box with dots; deletions are shown by a jagged end.
Recovery of full-length integrated viral DNA.
To recover the full-length proviruses, genomic DNAs were isolated from infected 293-tva cells and DF-1 cells and was digested with DraI. Because there is no DraI site in RSVP, both DraI sites must come from the adjacent cellular DNA. Full-length integrated viral DNA was recovered as described in Materials and Methods.
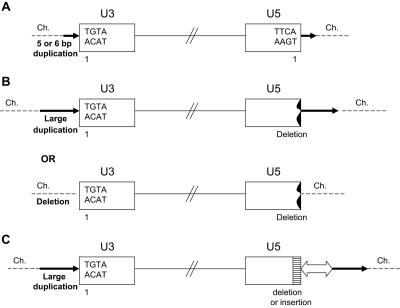
Based on sequence analysis, the proviruses from the three mutant viruses can be sorted into three types of integration events, as shown in Fig. 4. The first is a normal integration event in which one, two, or three nucleotides are removed from each end of the linear viral DNA (described below) and the provirus is inserted into the host genome, creating a 5- or 6-base duplication at the target site (Fig. 4A). However, we also found two types of aberrant integration events. In the first type, the U3 LTR terminus appears to be the result of a normal integration event; the U3 LTR terminus of the viral DNA was always joined to the host genome at the normal CA dinucleotide. However, on the U5 LTR terminus of the integrated viral DNA, the junction did not involve the canonical CA. When the integration of the U5 LTR terminus did not involve the canonical CA, instead of a 5- or 6-nucleotide duplication of host sequences there were usually large duplications, and more rarely deletions, of the host sequences (Fig. 4B). The second type of aberrant integration event was complex. In some cases there were additional viral sequences; in one case the insertion involved sequences from more than one host chromosome (Fig. 4C).
FIG. 4.
Three types of proviruses. (A) Normal provirus; (B) provirus with an aberrant integration at the U5 LTR terminus. Deletions in U5 are indicated by a black jagged end. Target duplications (a long black arrow) or deletions (no arrow) are indicated (see the legend to Fig. 5 for details). (C) Proviruses with complex structures. Deletions or insertions in U5 are indicated by a white box with black horizontal bars. Additional insertions of other viral sequences or host genome are shown with a white arrow. The direction of the arrowhead indicates the orientation of the extra viral sequences relative to the viral genome. Large duplications of host DNA are indicated by a long black arrow. The direction of the arrow corresponds to the numbering of the host genomic DNA. Ch., chromosome.
In the case of RSVP(HIV2), the integration-specific CA sequence was present in both the U3 and U5 ends of all of the proviruses and there were, in all cases, 5- or 6-bp duplications at the target site (Table 1). The same result was obtained both with 293-tva cells and DF-1 cells, suggesting that RSV integrase can process three bases from the U5 LTR terminus of the linear viral DNA during integration, albeit with reduced efficiency.
TABLE 1.
Recovery of full-length integrated viral DNA
| Provirus | U5a | Size of duplication (bp) | No. of cases |
|---|---|---|---|
| 293-tva, RSVP(HIV1) | 1 | 6 | 2 |
| 1 | 5 | 1 | |
| Variable | Variable | 4 | |
| DF-1, RSVP(HIV1) | 1 | 6 | 7 |
| 1 | 5 | 2 | |
| Variable | Variable | 4 | |
| 293-tva, RSVP(HIV2) | 1 | 6 | 3 |
| 1 | 5 | 1 | |
| DF-1, RSVP(HIV2) | 1 | 6 | 10 |
| 1 | 5 | 1 | |
| 293-tva, RSVP(CATC) | Variable | Variable | 7 |
| DF-1, RSVP(CATC) | Variable | Variable | 2 |
| DF-1, RSVP(HIV1) | Variable | Variableb | 2 |
| DF-1, RSVP(CATC) | 16 | 16b | 1 |
The last viral nucleotide at each end of the proviruses is indicated by a number, using the numbering system in Fig. 1C. For U3, the last nucleotide was a 1 in all cases.
Size of deletion.
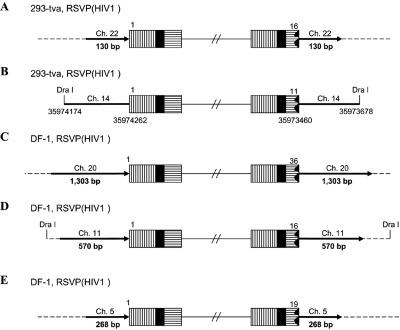
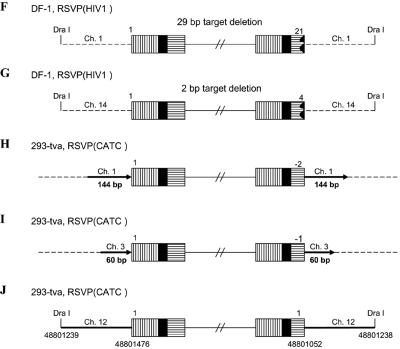
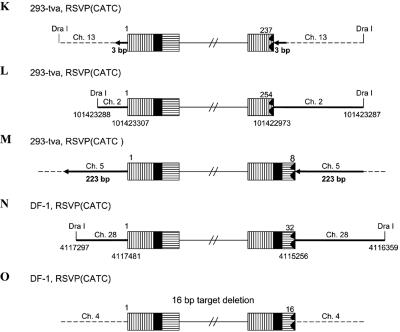
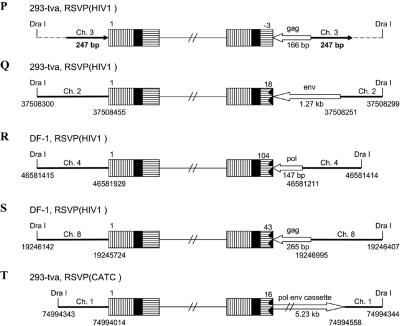
In the case of RSVP(HIV1), the canonical CA sequence was present at the U5 host-virus junction in about 70% of the proviruses we isolated (Table 1). In these proviruses, there was a 5- or 6-bp duplication at the target site, indicating that RSV integrase can remove a single nucleotide from the U5 LTR terminus of the linear viral DNA during integration. In about 30% of the proviruses, there was a CA sequence at the U3 LTR terminus of the provirus; however, the terminus of the U5 LTR was deleted, and there were duplications of the host sequences at the target site, ranging in size from hundreds to thousands of nucleotides, instead of the normal 5- or 6-bp duplication. For most of the proviruses (Fig. 5A and C to E), we sequenced through the duplicated regions at both ends of the proviruses into the nonduplicated host sequences unambiguously demonstrating the presence of the duplicated regions. However, in one case (Fig. 5B) the placement of the DraI sites means that, if there were a duplication of the host sequence flanking the viral DNA, the duplicated sequence would be lost in the DraI digest. Because there is no clear demonstration that the flanking host sequence is duplicated, it is possible, for viral DNAs having the structure shown in Fig. 5B, that the viral DNA results from an abortive integration event in which the normal U3 end integrated properly and an aberrant event involving the U5 end created a circular DNA which had a piece of host DNA between the two LTRs (see Discussion). In two proviruses, we found small deletions at the end of U5, and there were small deletions rather than a duplication of the host sequences, one of which was only 2 bp (Fig. 5F and G). These results suggest that RSV integrase is able to process a linear viral DNA with one nucleotide beyond the CA and appropriately integrate the processed end into host DNA. However, this type of linear viral DNA substrate appears to be difficult for integrase to process, and we also found proviruses in which the aberrant end did not appear to be processed or inserted by IN. Some of target duplications showed a complicated joining of viral and host DNA (Fig. 5P to S). One provirus isolated from 293-tva cells showed a normal structure at the U3 host-virus DNA junction. The CA sequence at the U3 LTR terminus was joined to human chromosome 3. However, at the U5 LTR terminus, there was a tRNA insertion immediately followed by part of the gag sequence (166 bp) that was then joined to human chromosome 3, creating a 247-bp duplication (Fig. 5P). Another provirus (Fig. 5Q) isolated from 293-tva cells showed a similar structure. The CA sequence at the U3 LTR terminus was joined to human chromosome 2. However, at the U5 LTR terminus of provirus, there was an 18-bp deletion at the end of U5 immediately followed by part of the env sequence (∼1.27 kb), which was then joined to human chromosome 2. Because there is a DraI site in the host sequence, we cannot unambiguously show that the host sequence was duplicated, nor can we be sure that this DNA was derived from a provirus. One viral DNA that was isolated from DF-1 cells showed that the U3 LTR CA was joined to chicken chromosome 4 (Fig. 5R). However, at the U5 LTR terminus, there was a 104-bp deletion followed by part of the pol sequence (147 bp) that was then joined to chicken chromosome 4. We are not sure that the host sequences are duplicated due to the placement of the DraI site. Another viral DNA isolated from DF-1 cells had the U3 LTR CA sequence appropriately joined to chicken chromosome 8 (Fig. 5S). However, there was a 43-bp deletion at the end of U5 immediately followed by a short segment from part of the gag sequence (265 bp), which was then joined to chicken chromosome 8. The placement of the DraI site means that we cannot be sure the host sequences are duplicated or that the DNA was from an integrated provirus. In all of these cases, the extra piece of viral DNA was inserted in the opposite orientation relative to the proviral DNA (see Discussion).
FIG. 5.
Structure of aberrant proviruses. U3 is shown by a box with horizontal bars. R is indicated by a black box. U5 is shown by a box with vertical bars. Deletions in U5 are indicated by a black jagged end. Additional insertions are shown as a white arrow. Large duplications of host sequences are indicated by long black arrows (not to scale). The direction of the arrow corresponds to the numbering of the host genomic DNA. When there are additional viral sequences, they are indicated by an open arrow; the direction of the arrow indicates the orientation of the viral DNA. In some cases in which the nature of the viral DNA is ambiguous (see the text), the positions of DraI sites and the virus-host DNA junctions are indicated by nucleotide position numbers from a BLAT search. Ch., chromosome.
Similar aberrant proviruses were obtained with RSVP(CATC). In all cases, the normal CA sequence was present at the U3 junction with host DNA. However, there were large duplications or small deletions in the host sequence at the target site, and the junctions occurred at various positions in U5 (Table 1, Fig. 5H to O). In one provirus, the mutated TC sequence that replaced the normal CA sequence was present at the U5 LTR junction with host DNA (Fig. 5J). However, we cannot define the exact junction, because the last six nucleotides at the U5 LTR terminus were identical with the first six nucleotides of the host DNA (discussed below; see also Table 3). In two cases, there was a complicated junction at the U5 terminus. One viral DNA isolated from 293-tva cells had the U3-terminal CA sequence joined appropriately to human chromosome 1. However, there was a 16-bp deletion at the end of U5 immediately followed by a long segment of viral DNA (∼5.23 kb), which was then joined to human chromosome 1. The placement of the DraI site means that we cannot be sure the host sequences are duplicated or that the viral DNA was from a provirus (Fig. 5T). Another viral DNA isolated from DF-1 cells showed that two nucleotides of U5 were retained beyond the mutated TC sequence. The viral sequence, which corresponds to a complete copy of the end of the linear viral DNA, was joined to chicken chromosome 5 (154 bp), which was then joined to chromosome 2. Because there is a DraI site in the host DNA, we cannot be sure this viral DNA is from a proviral insertion that generated a duplication of the host genome (Fig. 5U). These results confirm previous data showing that the CA sequence at the end of viral DNA is critical for proper integration. In addition, these results suggest that when RSV integrase is given a linear DNA with one normal end and one aberrant end, it can process and integrate the normal end independent of the aberrant end.
TABLE 3.
Microhomology at the host and viral DNA junctions in aberrant provirusesa
| Provirusb | U3 | U5 | U5 (first junction) | U5 (second junction) |
|---|---|---|---|---|
| A | taca | tgcaTG | ||
| B | tacA | tgaGCA | ||
| C | taca | ccctgA | ||
| D | taca | tgcatG | ||
| E | taca | acctgc | ||
| F | tacA | gcacCT | ||
| G | taca | aAGGCT | ||
| H | taca | tttctt | ||
| I | taca | ctttcT | ||
| J | taca | GCTTTC | ||
| K | taCA | gtgcAT | ||
| L | tacA | aggaGA | ||
| M | taca | gcAGAA | ||
| N | taCA | gacgAC | ||
| O | taca | tgcatG | ||
| P | taCA | tcagtG | cggtct | |
| Q | taca | cttgCA | gcaACA | |
| R | taca | tacaaT | ccctcG | |
| S | tacA | gtTGAT | cggctG | |
| T | taca | tgcaTG | ctggTA | |
| U | taca | tttctt | ctTTGT |
Four nucleotides from the U3 end junction and six nucleotides from the U5 end junction are shown. A boldface capital letter indicates sequences showing microhomology with the host DNA.
Letters A to U correspond to the same letters in Fig. 5.
The aberrant U5 host DNA junctions often involve microhomology.
We compared the normal virus-host DNA junctions with the aberrant U5-host junctions to see whether homology might play a role in the formation of the aberrant junctions. If homology is not involved in generating the virus-host junction, the presence of a microhomology should conform to the chance of the presence of matching nucleotides. This is exactly what was found at the normal junctions (Table 2). However, when the aberrant U5 host junctions were examined, there was, in most cases, a microhomology from one to six nucleotides between the viral and host sequences at the junction (Table 3). Of 27 aberrant ends, only 4 showed no homology between the viral sequence and the host at the junction. This result differs significantly from the results seen at the normal junctions, increasing the likelihood that the aberrant junctions were not generated by viral integrase.
TABLE 2.
Frequency of microhomology at the normal integration junctions
| Microhomology | Expected | Observed | ||
|---|---|---|---|---|
| 1 Nucleotide (A) | 1/4 | 25% | 14/46 | 30% |
| 2 Nucleotides (CA) | 1/16 | 6.2% | 3/46 | 6.5% |
| 3 Nucleotides (5′ end, ACA; 3′ end, TCA) | 1/64 | 1.6% | 1/46 | 2.2% |
Recovery of integrated viral DNA generated in the presence of enzymatically inactive integrase.
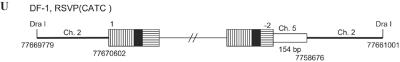
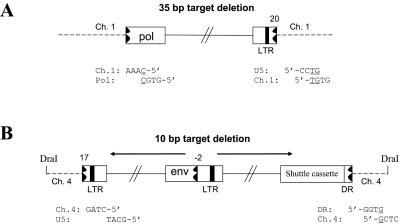
Based on the idea that the aberrant U5-host DNA junctions were not generated by viral integrase, we recovered proviruses from infections with a virus mutated at the integrase active site (D64K) and compared the virus-host DNA junctions generated by this integrase active site mutant with the aberrant U5-host DNA junctions found in proviruses from the other viral mutants. The virions containing a D64K mutation were harvested and used to infect fresh DF-1 cells. Very few colonies were obtained after zeocin selection. Two integrated viral DNAs were recovered; their structures are shown in Fig. 6. In one case, only a portion of the viral DNA was inserted into chicken chromosome 1. The U3 end of the provirus was in the pol gene, and the U5 end had a deletion in U5. Neither end of the provirus involved a CA dinucleotide. In addition, there was a 35-bp deletion of host sequence at the integration site (Fig. 6A). In the second case, the U3 end of the provirus was joined to vector sequences. This segment, from env to the right LTR, was in the reverse orientation compared to the rest of the viral DNA. There was a deletion in U5 of the inverted segment; this partially deleted U5 sequence was joined to chicken chromosome 4. On the U5 end, the entire LTR was absent. Instead, part of the direct repeat (DR) sequence was joined to chicken chromosome 4. There were no CA dinucleotides on either end of the provirus, and there was a 10-bp deletion of host sequence at the integration site (Fig. 6B). Although we have only four junctions, three of four involve microhomology. Presumably, the two aberrant proviruses generated in the absence of active IN are similar to the majority of aberrant proviruses reported by Hagino-Yamagishi et al.; however, we did not recover any oligomeric insertions (11).
FIG. 6.
Integrated viral DNA from a virus mutated at the active site of integrase (D64K). Deletions are indicated by a black jagged end. Positions of the deletions are indicated by number, as in Fig. 1C. Four nucleotides at the virus-host DNA junctions are shown, and a microhomology with the host DNA is underlined. The orientation of the viral sequences is indicated by black arrows. DR, direct repeat.
DISCUSSION
Our finding that the RNase H of RSV removes the entire tRNA primer regardless of mutations at the U5 terminus shows that the RNase H of RSV RT and the RNase H of HIV-1 RT treat similar nucleic acid substrates differently. Because these mutants give rise to viral DNAs with aberrant U5 ends, we asked how these aberrant viral DNAs were handled by RSV IN. We tested three different mutants: RSVP(HIV1), which has only a single nucleotide after the CA; RSVP(HIV2), which has three nucleotides; and RSCP(CATC), in which the CA dinucleotide has been mutated to TC. Proviruses recovered from infections with the RSVP(HIV1) mutant showed that RSV IN can remove a single nucleotide beyond the CA from the linear viral DNA and properly insert the resulting linear DNA into the host genome, creating the expected 5- to 6-bp duplication at the target site. However, in about 30% of proviruses we isolated, the U5 LTR termini of the proviruses usually were deleted and there usually were large duplications and, more rarely, deletions at the target site instead of the normal 5- to 6-bp duplication. We also found more complex insertions that had extra viral sequences (discussed below). We obtained similar aberrant proviruses when we mutated the CA dinucleotide, which is usually involved in joining the viral DNA to host sequences.
Colicelli and Goff (4, 5) showed, by making mutations in the 3′ end of U5 of a murine leukemia virus (MLV), that MLV integrase could, with reduced efficiency, remove a single nucleotide beyond the conserved CA and could process viral DNAs with two extra nucleotides (removing a total of four). However, they found that removing both nucleotides between the PBS and the conserved CA blocked viral replication. Based primarily on analysis of autointegrations, they reported, for the viruses that were able to replicate with too few or too many nucleotides between the PBS and the CA, that most of the autointegration events were normal in the sense that there was a small duplication at the target sequence and that most (but not all) of the insertions occurred at the canonical CA. They did not report that these mutants gave rise to insertions equivalent to the aberrant integrations we found with the RSVP mutants; however, it is possible, if such events occurred among the MLV autointegrants, that it was difficult to recognize them as integration events.
It should be pointed out that the proviral DNAs we recovered and analyzed came from integration events in which the proviral DNA was inserted into the host genome in a way that allowed the cell to survive and the provirus to be expressed. Because the cultured cells are pseudodiploid, it is likely that we can recover proviruses in which there is significant damage to the target chromosome. However, it is also possible that there are aberrant insertions that give rise to inserted viral DNAs that cannot be expressed and/or rescued using the techniques we employ.
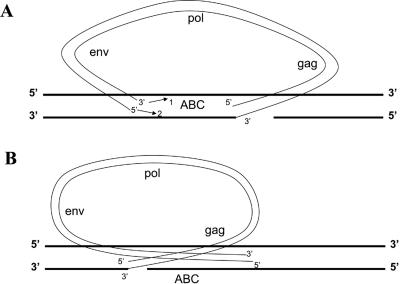
How is the linear viral DNA integrated when one end of the linear DNA cannot be properly processed or inserted by integrase? It would appear for RSVP(HIV1) and RSVP(CATC) that the normal end of the linear viral DNA is appropriately processed and inserted by integrase and the aberrant end is inserted separately, probably by host factors. However, in a number of cases, the insertion of the aberrant end does not involve the canonical CA that integrase requires, which argues against the involvement of integrase for those insertion events. The aberrant insertions often involve microhomology which normal integrations catalyzed by IN do not. Although we recovered only a few viral insertions made in the absence of enzymatically active integrase, the virus-host DNA junctions made in the absence of active integrase resemble aberrant U5-host DNA junctions in that there are deletions of the ends of the viral DNA and the junctions that appear to involve microhomologies. Moreover, for the aberrant ends not inserted at the canonical CA, the insertion event does not generate the 5- to 6-bp duplication in the host genome that is characteristic of the normal concerted insertions that involve integrase. Taken together, the data support, but do not prove, a model in which the normal end is inserted by integrase in a reaction similar to the single-end insertions seen in in vitro integrase assays (13, 19) and the aberrant end is inserted into the host genome by host enzymes. However, in an RSV-based in vitro integration assay that led to efficient concerted integration events with a substrate with a wild-type sequence at the ends, mutating the concerted CA reduced the concerted integrations and did not appear to produce single-end insertions (1). This in vitro result is similar to the result reported by Chen and Engleman, who used an HIV-1-based in vitro assay (3). They reported that, with linear DNA substrates in which there was one normal end and one end that had the canonical CA mutated, IN appropriately removed two nucleotides from the normal end but not the mutant end and was then unable to carry out a single-end insertion reaction. Our results show that, in vivo, RSV IN can, in some cases, process and insert a wild-type end normally in a reaction that does not appear to be concerted. In a simple version of a one-end IN insertion model, one might expect that the secondary event creating the second (aberrant) junction would be equally likely to occur on either side of the initial integrase-mediated single-end insertion. This would mean that these aberrant insertions cause approximately an equal number of duplications and deletions (Fig. 7). However, we found duplications much more frequently than we found deletions. This suggests that there is directionality in the insertion of the aberrant end. One way for this to happen would be if the initial complex between virus and host DNA involves integrase bound to both ends of the virus DNA in a complex that resembles the complex that carries out the normal concerted integration reaction. If this complex then inserts the normal end but cannot insert the aberrant end, it is possible that the integrase complex can dissociate into two pieces without releasing either the virus or the host DNA. This could allow the integrase bound to the aberrant end to diffuse along the host chromosome in the direction that would generate duplications (Fig. 7). In this model, the actual joining of virus DNA and host DNA at the aberrant end would still involve host enzymes. Because we also find deletions, it is likely that the aberrant end can occasionally be released from the chromosome into which the normal end was inserted. If the aberrant end of the linear DNA is released from the chromosome into which the normal end is inserted, it would then presumably be free and could insert either right or left of the original insertion or, more rarely, into another chromosome.
FIG. 7.
Models for the generation of duplications and deletions in host DNA flanking the aberrant proviruses. Viral DNA is indicated by thin black lines, and host DNA is shown by thick black lines. (A) Models to explain the duplication and capture of host sequence. Arrow 1 shows that only the 3′ end is joined. This will cause the provirus to be inserted and host sequence “ABC” to be duplicated flanking provirus. Arrow 2 shows that only the 5′ end is joined. Host sequence “ABC” is captured between the LTRs of a circular viral DNA. Both 5′ and 3′ ends (arrow 1 and arrow 2) are joined. Host sequence “ABC” is captured between the LTRs of a circular viral DNA, and the host chromosome is broken. (B) Models to explain the deletion of host sequences. In all cases (3′ only, 5′ only, and both 3′ and 5′ joined), a provirus is inserted with the loss of the host sequence “ABC.”
The viral DNAs in which there are duplicated host sequences flanking the viral sequences derive from inserted proviruses. However, it is possible that the viral DNAs shown in Fig. 5B, J, L, N, and Q to U derive from an abortive integration event in which the U3 end was inserted normally, joining the 3′ end of the viral DNA to the host DNA. If the 3′ strand of the U5 end were joined to the opposite host DNA strand (as happens in a normal integration event), the viral DNA would be inserted into the chromosome (Fig. 7A). However, if the 5′ strand of the U5 end was then joined to the host DNA, the viral DNA could be released as a circular DNA that contained host sequences between the LTRs. This type of circular viral DNA, with host DNA between the LTRs, would also arise if both strands (5′ and 3′) of the U5 were joined to host DNA; however, joining both viral strands to both host strands would break the host chromosome (Fig. 7A). Circular forms arising from abortive integration events of this type would not necessarily involve host DNAs that contained a DraI site. However, all of the ambiguous viral DNAs we isolated (Fig. 5B, J, L, N, and Q to U) were linked to host sequences that contained at least one DraI site, suggesting that at least some of these events probably derive from integrated proviruses in which there was a host DNA duplication that contained at least one DraI site, and the duplicated host sequences were lost in the DraI digest.
As shown in Fig. 7B, for the events that give rise to deletions of the host sequence it does not matter whether the U5 joining event involves the 5′ or the 3′ strand (or both strands). The fact that all of the three types of the secondary joining reactions (3′ strand only, 5′ strand only, or both strands) that occur to the right side of the initial IN-mediated reaction (as shown in Fig. 7B) lead to the insertion of a provirus with a deletion of host sequences, where only one of the reactions (3′ strand only) that happens to the left (shown in Fig. 7A) leads to the generation of a provirus flanked by a duplication of host DNA sequences, reinforces the idea that there is directionality in the reaction that generated the proviruses with aberrant ends.
Proviruses recovered from the RSVP(HIV2) mutant showed that RSV IN can remove three nucleotides beyond the CA and properly integrate the processed ends. However, this mutation significantly reduced virus titer to only 3.5% of the wild-type level, which is much lower than the titer of the other mutants. Why does this mutant show such a low titer, even though RSV IN can correctly process and integrate the aberrant end? We propose that ends of the RSVP(HIV2) viral DNA are processed inefficiently by RSV IN and that only a small portion of linear DNA is properly processed and used for integration. It has been reported for MLV that some mutations in one end of the linear viral DNA interfere with the processing of both ends by IN (20). With the RSVP(HIV2) mutant, we did not observe any proviruses that had deletions of the U5 LTR terminus with a large duplication of host sequences at the target site, even though such aberrant integrations were found with other mutant viruses that had a higher titer, which lends support to the idea that the processing of both ends of the RSVP(HIV2) linear DNA might be aberrant in this mutant despite the fact that the other mutants appear to be able to process and insert a normal DNA end independent of an aberrant end.
We suspect but cannot prove that the aberrant integrations involve host enzymes. The fact that aberrant integrations were similar in a mammalian cell (293-tva) and a chicken cell (DF-1) suggests that this phenomenon is not limited to a particular cell type, and if, as we suggest, cellular enzymes are involved, these cellular components are probably part of the general host cell DNA repair machinery. The fact that many of the insertion events we recovered appear to specifically join the 3′ end of the viral DNA in a reaction that involves microhomology suggests that the reaction may involve host DNA polymerases. This model is also supported by the observation that most of the viral DNA sequences appended to the aberrant U5 ends are of opposite polarity to the proviral DNA. The junctions between the (inverted) viral DNAs and the normal proviruses also involve microhomology, making it likely that the extra (inverted) viral sequences were copied by a polymerase from a second copy of minus-strand viral DNA.
Acknowledgments
We are grateful to Stan Kaczmarczyk for providing the env construct and to Hilda Marusiodis for help in preparing the manuscript.
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and the National Institute for General Medical Sciences.
REFERENCES
- 1.Aiyar, A., P. Hindmarsh, A. M. Skalka, and J. Leis. 1996. Concerted integration of linear retroviral DNA by the avian sarcoma virus integrase in vitro: dependence on both long terminal repeat termini. J. Virol. 70:3571-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cen, S., H. Javanbakht, S. Kim, K. Shiba, R. Craven, A. Rein, K. Ewalt, P. Schimmel, K. Musier-Forsyth, and L. Kleiman. 2002. Retrovirus-specific packaging of aminoacyl-tRNA synthetase with cognate primer tRNAs. J. Virol. 76:13111-13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, H., and A. Engleman. 2001. Asymmetric processing of human immunodeficiency virus type 1 cDNA in vivo: implications for functional end coupling during the chemical steps of DNA transportation. Mol. Cell. Biol. 21:6758-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colicelli, J., and S. P. Goff. 1985. Mutants and pseudorevertants of Moloney murine leukemia virus with alterations at the integration site. Cell 42:537-580. [DOI] [PubMed] [Google Scholar]
- 5.Colicelli, J., and S. P. Goff. 1988. Sequence and spacing requirements of a retrovirus integration site. J. Mol. Biol. 199:47-59. [DOI] [PubMed] [Google Scholar]
- 6.Craigie, R., T. Fujiwara, and F. Bushman. 1990. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell 62:829-837. [DOI] [PubMed] [Google Scholar]
- 7.Dash, C. J., W. Rausch, and S. F. Le Grice. 2004. Using pyrrolo-deoxycytosine to probe RNA/DNA hybrids containing the human immunodeficiency virus type-1 3′ polypurine tract. Nucleic Acids Res. 32:1539-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhar, R., W. L. McClements, L. W. Enquist, and G. F. Vande Woude. 1980. Nucleotide sequences of integrated Moloney sarcoma provirus long terminal repeats and their host and viral junctions. Proc. Natl. Acad. Sci. USA 77:3937-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujiwara, T., and R. Craigie. 1989. Integration of mini-retroviral DNA: a cell-free reaction for biochemical analysis of retroviral integration. Proc. Natl. Acad. Sci. USA 86:3065-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furfine, E. S., and J. E. Reardon. 1991. Human immunodeficiency virus reverse transcriptase ribonuclease H: specificity of tRNALys3-primer excision. Biochemistry 30:7041-7046. [DOI] [PubMed] [Google Scholar]
- 11.Hagino-Yamgish, K., L. A. Donehower, and H. E. Varmus. 1987. Retroviral DNA integrated during infection by an integration-deficient mutant of murine leukemia virus is oligomeric. J. Virol. 61:1964-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Himly, M., D. N. Foster, I. Botoli, J. S. Iacovoni, and P. K. Vogt. 1998. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leucosis viruses. Virology 248:295-304. [DOI] [PubMed] [Google Scholar]
- 13.Hindmarsh, P., M. Johnson, R. Reeves, and J. Leis. 2001. Base-pair substitutions in avian sarcoma virus U5 and U3 long terminal repeat sequences alter the process of DNA integration in vitro. J. Virol. 75:1131-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes, S. H., A. Mutschler, J. M. Bishop, and H. E. Varmus. 1981. A Rous sarcoma virus provirus is flanked by short direct repeats of a cellular DNA sequence present in only one copy prior to integration. Proc. Natl. Acad. Sci. USA 78:4299-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ju, G., and A. M. Skalka. 1980. Nucleotide sequence analysis of the long terminal repeat (LTR) of avian retroviruses: structural similarities with transposable elements. Cell 22:379-386. [DOI] [PubMed] [Google Scholar]
- 16.Julias, J. G., M. J. McWilliams, S. G. Sarafianos, W. G. Alvord, E. Arnold, and S. H. Hughes. 2004. Effects of mutations in the G tract of the human immunodeficiency virus type 1 polypurine tract on virus replication and RNase H cleavage. J. Virol. 78:13315-13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katzman, M., R. A. Katz, A. M. Skalka, and J. Leis. 1989. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J. Virol. 63:5319-5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McWilliams, M. J., J. G. Julias, S. G. Sarafianos, W. G. Alvord, E. Arnold, and S. H. Hughes. 2003. Mutations in the 5′ end of the human immunodeficiency virus type 1 polypurine tract affect RNase H cleavage specificity and virus titer. J. Virol. 77:11150-11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moreau, K., C. Faure, S. Violot, P. Gouet, G. Verdier, and C. Ronfort. 2004. Mutational analyses of the core domain of avian leukemia and sarcoma viruses integrase: critical residues for concerted integration and multimerization. Virology 318:566-581. [DOI] [PubMed] [Google Scholar]
- 20.Murphy, J. E., and S. P. Goff. 1992. A mutation at one end of Moloney murine leukemia virus DNA blocks cleavage of both ends by the viral integrase in vivo. J. Virol. 66:5092-5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh, J., J. G. Julias, A. L. Ferris, and S. H. Hughes. 2002. Construction and characterization of a replication-competent retroviral shuttle vector plasmid. J. Virol. 76:1762-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peters, G. G., and J. Hu. 1980. Reverse transcriptase as the major determinant for selective packaging of tRNA's into avian sarcoma virus particles. J. Virol. 36:692-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Powell, M. D., and J. G. Levin. 1996. Sequence and structural determinants required for priming of plus-strand DNA synthesis by the human immunodeficiency virus type 1 polypurine tract. J. Virol. 70:5288-5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pullen, K. A., and L. K. Ishimoto, and J. J. Champoux. 1992. Incomplete removal of the RNA primer for minus-strand DNA synthesis by human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 66:367-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ratner, L., and W. Haseltine, R. Patarca, K. J. Livak, B. Starcich, S. F. Josephs, E. R. Doran, J. A. Rafalski, E. A. Whitehorn, K. Baumeister, L. Ivanoff, S. R. Petteway, Jr., M. L. Pearson, J. A. Lautenberger, T. S. Papas, J. Ghrayeb, N. T. Chang, R. C. Gallo, and F. Wong-Staal. 1985. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature 313:277-284. [DOI] [PubMed] [Google Scholar]
- 26.Rattray, A. J., and J. J. Champoux. 1989. Plus-strand priming by Moloney murine leukemia virus. The sequence features important for cleavage by RNase H. J. Mol. Biol. 208:445-456. [DOI] [PubMed] [Google Scholar]
- 27.Roth, M. C., P. L. Schwartzberg, and S. P. Goff. 1989. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell 58:47-54. [DOI] [PubMed] [Google Scholar]
- 28.Sawyer, R. C., and J. E. Dahlberg. 1973. Small RNAs of Rous sarcoma virus: characterization by two-dimensional polyacrylamide gel electrophoresis and fingerprint analysis. J. Virol. 12:1226-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaefer-Klein, J., I. Givol, E. V. Barsov, J. M. Whitcomb, M. VanBrocklin, D. N. Foster, M. J. Federspiel, and S. H. Hughes. 1998. The EV-O derived cell line DF-1 supports the efficient replication of avian leucosis-sarcoma viruses and vectors. Virology 248:305-311. [DOI] [PubMed] [Google Scholar]
- 30.Wain-Hobson, S., P. Sonigo, O. Danos, S. Cole, and M. Alizon. 1985. Nucleotide sequence of the AIDS virus, LAV. Cell 40:9-17. [DOI] [PubMed] [Google Scholar]
- 31.Waters, L. C., and B. C. Mullin. 1977. Transfer RNA in RNA tumor viruses. Prog. Nucleic Acid Res. Mol. Biol. 20:131-160. [DOI] [PubMed] [Google Scholar]
- 32.Whitcomb, J. M., R. Kumar, and S. H. Hughes. 1990. Sequence of the circle junction of human immunodeficiency virus type 1: implications for reverse transcription and integration. J. Virol. 64:4903-4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitcomb, J. M., and S. H. Hughes. 1991. The sequence of human immunodeficiency virus type 2 circle junction suggests that integration protein cleaves the ends of linear DNA asymmetrically. J. Virol. 65:3906-3910. [DOI] [PMC free article] [PubMed] [Google Scholar]