Abstract
The release of Bluetongue virus (BTV) and other members of the Orbivirus genus from infected host cells occurs predominantly by cell lysis, and in some cases, by budding from the plasma membrane. Two nonstructural proteins, NS3 and NS3A, have been implicated in this process. Here we show that both proteins bind to human Tsg101 and its ortholog from Drosophila melanogaster with similar strengths in vitro. This interaction is mediated by a conserved PSAP motif in NS3 and appears to play a role in virus release. The depletion of Tsg101 with small interfering RNA inhibits the release of BTV and African horse sickness virus, a related orbivirus, from HeLa cells up to fivefold and threefold, respectively. Like most other viral proteins which recruit Tsg101, NS3 also harbors a PPXY late-domain motif that allows NS3 to bind NEDD4-like ubiquitin ligases in vitro. However, the late-domain motifs in NS3 do not function as effectively in facilitating the release of mini Gag virus-like particles from 293T cells as the late domains from human immunodeficiency virus type 1, human T-cell leukemia virus, and Ebola virus. A mutagenesis study showed that the arginine residue in the PPRY motif is responsible for the low activity of the NS3 late-domain motifs. Our data suggest that the BTV late-domain motifs either recruit an antagonist that interferes with budding or fail to recruit an agonist which is different from NEDD4.
The genus Orbivirus of the Reoviridae comprises several important animal pathogens that are transmitted to mammalian hosts by insect vectors. In tissue culture, orbiviruses such as bluetongue virus (BTV) and African horse sickness virus (AHSV) establish persistent infections in susceptible insect cells without apparent cytopathic effects (75). In contrast, both viruses cause dramatic cytopathic effects in mammalian cells. While the majority of the progeny particles remain cell associated in most mammalian cells, as is commonly observed with reoviruses, there is clear evidence that orbiviruses can also leave the host cells as enveloped particles by budding at the cell membrane (28, 35, 64).
Recent work revealed that retroviruses and most other viruses that bud at the plasma or endosomal membrane recruit cellular proteins for this purpose (12, 45, 52). The cellular proteins normally function in the biogenesis of multivesicular bodies (MVB), which play an important role in protein sorting and degradation and are formed by the budding of vesicles into the lumen of late endosomal compartments, a process that is topologically similar to the formation of an enveloped virus particle (52, 55). The formation of multivesicular bodies requires a multitude of cellular proteins which function in a sequential manner and include, among others, the components of the ESCRT-I, -II, and -III complexes (3, 4, 18, 33). Retroviruses bind to different components of this pathway by means of so-called late-domain motifs, which are highly conserved protein-protein interaction motifs that can be located at different positions within Gag (13, 16, 48, 77). To date, three classes of late-domain motifs (PT/SAP, YPXL/LXXLF, and PPXY) have been identified, of which the PSAP motif recruits the cellular protein Tsg101, a component of the ESCRT-I complex (15, 43, 46, 72). The YPXL and LXXLF motifs bind to AIP-1/Alix, which acts downstream of Tsg101 and appears to bridge ESCRT-I and ESCRT-III complexes (42, 66, 73). The third motif, PPXY, plays a role in recruiting host ubiquitin ligases (7, 8, 17, 34, 38, 40, 54, 70, 73, 79, 80). Although it is clear that the cellular ubiquitination machinery is important for the budding of many animal viruses, current knowledge about the exact targets of the ubiquitin ligases and their interactions with other components of the budding machinery is still very limited (7, 41, 47, 49, 53). A fourth late-domain motif (FPIV) has been described recently for the matrix protein of paramyxoviruses, but its cellular binding partner is not yet known (61). The PTAP and PPXY motifs have also been identified in VP40 of Ebola virus, the matrix protein of vesicular stomatitis virus, and the Z protein of Lassa virus, where they appear to exhibit a similar function to that of the late-domain motifs of retroviruses (9, 21-23, 38, 50, 69, 80). Both motifs are also present within the nonstructural protein NS3 of BTV, but their function has not been studied so far (65).
NS3 and its shorter form, NS3A, which lacks the N-terminal 13 amino acids of NS3, are the only membrane proteins encoded by orbiviruses (78). Interestingly, NS3 and NS3A appear to be associated with smooth intracellular membranes, although they are also present at the plasma membrane (29, 64). Both NS3 and NS3A comprise a long N-terminal domain and a shorter C-terminal cytoplasmic domain, which are connected by two transmembrane domains and a short extracellular domain (5, 71). A single glycosylation site is present in the extracellular domain of BTV NS3, but not in AHSV NS3, and therefore does not seem to be essential for the function of the protein (5, 71, 78). Using yeast two-hybrid technology, we have recently identified the p11 subunit of the cellular calpactin complex as a cellular binding partner of NS3 (6). Even though the exact physiological role of this interaction is still unknown, it is intriguing that p11 binds to NS3 but not to NS3A. While both proteins seem to be equally expressed in cells infected with AHSV (71), NS3 is the predominant form in BTV-infected cells. Both proteins are overexpressed when BTV is adapted to grow in mosquito cells (19), and a strong correlation was observed in this and earlier studies between the presence of NS3/NS3A and the extent of virus release (29). As observed by ourselves and others, NS3 exhibits cytotoxicity when expressed in mammalian or Spodoptera frugiperda cells (14, 71). This cytotoxicity requires membrane association of the protein and depends on the presence of the first transmembrane domain, as shown recently (20). In the same study, it was reported that NS3 does not exhibit a late-domain function, and the authors concluded that NS3 functions as a viroporin, facilitating virus release by inducing membrane permeabilization. While evidence for the viroporin activity of NS3 is compelling and consistent with earlier studies, which show an association of NS3 with areas of membrane perturbation, the late-domain function was not studied in the context of an infectious BTV and therefore cannot be ruled out. Given the presence of late-domain motifs in NS3 and our previous observation that NS3 can function as a release factor for virus-like particles (VLPs) in a heterologous expression system (30), we sought to establish whether this activity requires an interaction of NS3 with Tsg101 or other components of the ESCRT machinery.
MATERIALS AND METHODS
Plasmids and site-directed mutagenesis.
Fragments encoding full-length Tsg101 or the UEV domain of human Tsg101 and its ortholog from Drosophila melanogaster (GenBank locus NM_079396, codons 1 to 151) were obtained by reverse transcription-PCR, using RNAs isolated from 293T or S2 cells as a template, respectively. The fragments were cloned into pGEX2T (Pharmacia) and sequenced to verify the absence of mutations. The sequence encoding green fluorescent protein was amplified from pEGFP (Clontech), transferred into pTriex1 (Novagen), and fused to the 5′ end of the full-length coding sequence of human Tsg101 to generate pGFP-Tsg101. Fragments encoding the WW domains of human NEDD4.1 (codons 184 to 509), WWP1 (codons 338 to 547), Itch (codons 278 to 478), and Alix (codons 1 to 704) were amplified from HeLa cDNA and cloned into pTriex1/GST for expression as glutathione S-transferase (GST)-tagged proteins. The full-length S10 segment of AHSV serotype 6 (AHSV-6) was amplified following the procedure of Potgieter et al. (53) and sequenced to determine the consensus sequence. The coding region was then cloned into pTriex1 (Novagen) and ligated to oligonucleotide duplexes to add a C-terminal FLAG tag. Mutants of BTV-10 NS3 were obtained by the QuickChange method (Stratagene), using a full-length cDNA copy of BTV-10 S10 in pBR322 as a template (37). The region encoding NS3 or NS3A was then amplified by PCR and cloned into pTriex1. The coding sequence of human immunodeficiency virus type 1 (HIV-1) Gag was amplified using plasmid pSPBN-Gag (44) as a template, which contains the full-length Gag sequence from pNL4-3 (2). Standard PCR protocols were used to delete codons 10 to 278 and 486 to 500 and to introduce mutations at codons 470 to 473, changing PTAP to LAAL. The PCR products were cloned into pTriex1 to generate plasmids pH6, pH6Ako, pH6Tko, and pH6ko. The last plasmid was ligated with PCR products spanning amino acids 1 to 117 of NS3 or mutant forms of NS3 to generate plasmids pNS3, pM1, and pM2. Oligonucleotide duplexes spanning 12 to 18 codons were ligated to pH6ko, pH6Ako, and pH6Tko to generate the other plasmids, which were used for assaying budding activity. Full details of the cloning procedures are available upon request.
Cell culture.
HeLa, BSR, and 293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) and antibiotics. Insect cells were maintained in spinner or shaking flasks using SF900II serum-free medium or TC100 (Invitrogen) containing 10% FBS. Spodoptera frugiperda Sf21 cells were infected at a multiplicity of 2 to 5 PFU per cell with a recombinant baculovirus expressing NS3 of BTV-10 (14). Cells were harvested at 48 h postinfection and lysed in either TENT buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) or NP-40 buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40). Lysates were clarified by centrifugation for 20 min at 9,000 × g, and supernatants were used in pull-down assays as described below.
Antisera and immunoblotting.
NS3 was detected with a mouse polyclonal serum (14). Antibodies against p11 and p36 were obtained from Transduction Laboratories, a Tsg101-specific monoclonal antibody was obtained from Santa Cruz, an anti-tubulin antibody was obtained from Sigma, and an anti-p24 monoclonal antibody was obtained from Chemicon. The M2 anti-FLAG antibody (Sigma) was used for detecting tagged AHSV-6 NS3. The BTV nonstructural proteins NS1 and NS2 were identified with a rabbit polyclonal antibody. Samples were resolved by electrophoresis on either 10% or 15% polyacrylamide gels (36, 60), transferred onto Hybond-C Extra membranes (Amersham), and immunodetected (56).
siRNAs.
The following small interfering RNAs (siRNAs) were chemically synthesized: Tsg101 si1 (sense, GCUGAGGGCACUAAUGCAAAAdTdT; antisense, TTTTGCATTAGTGCCCTCAGCdTdG), Tsg101 si2 (sense, CCUCCAGUCUUCUCUCGAGdTdT; antisense, GACGAGAGAAGACUGGAGGdCdA), NS3 siRNA (sense, CAACUUGGGAGCAACUUAAdTdT; antisense, AAAAGUUGCUCCCAAGUUGdCdA), p11 siRNA (sense, GCUGUGGACAAAAUAAUGAAGdTdT; antisense, CUUCAUUAUUUUGUCCACAGCdCdA), p36 control siRNA (sense, CCUUAUGACAUGUUGGAAAdTdT; antisense, UUUCCAACAUGUCAUAAGGdGdC). The siRNAs were obtained from either Operon or MWG-Biotech and were annealed as recommended by the manufacturer prior to use.
siRNA transfections and plaque assays.
HeLa cells were seeded in 24-well plates and transfected at 40 to 60% confluence with 60 nmol of siRNA and 3 μl Oligofectamine (Invitrogen) in serum-free medium. After 12 h, the transfections were repeated and subsequently infected with BTV-10 for 1 h at a multiplicity of infection of 1 to 5 24 h after the first transfection. The infected cells were washed four times with DMEM and incubated in Optimem or DMEM supplemented with 2% FBS. Supernatants were collected after 12 and 24 h, and virus titers were determined by a plaque assay on BSR cells. Briefly, cells were seeded in 12-well plates and incubated for 1 h with diluted virus in 100 μl serum-free DMEM or Optimem. After removal of the inoculum, cells were overlaid with 1 ml of 0.8 to 1% agarose in DMEM containing 2% fetal bovine serum and antibiotics. Plaques were visualized after an incubation period of 2 to 3 days at 35°C by staining with neutral red for several hours. To quantitate the amount of intracellular virus, the cells were lysed by three freeze-thaw cycles. The virus titer in the lysate was determined by a plaque assay as described above. The ratio of extracellular to total virus (extracellular plus intracellular virus) was then calculated and normalized to the ratio obtained with untransfected cells to determine the inhibition of particle release.
Expression and purification of GST fusion proteins.
GST fusion proteins were expressed in Escherichia coli BL21. Briefly, cells were grown at 28°C overnight in LB medium. The next morning, cells were induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and growth was continued for 8 h. The cells were harvested at 4,000 × g for 15 min, resuspended in 50 mM Tris (pH 7.5), 100 mM NaCl buffer, and incubated at 28°C for 15 min in the presence of 100 μg/ml lysozyme. The cells were then lysed by sonication. After centrifugation for 20 min at 8,000 × g, the soluble fraction was incubated with 0.5 ml GST-agarose beads for 2 h at 4°C. The beads were then washed three times with 10 ml phosphate-buffered saline (PBS)-1% Triton X-100 and two times with 10 ml PBS. The washed beads were kept at 4°C in the presence of sodium azide and a cocktail of protease inhibitors (Sigma).
Pull-down experiments.
For pull-down experiments, 293T cells were seeded in six-well plates and transfected when they were nearly confluent with 4 μg plasmid DNA and 10 μl Lipofectamine 2000 for 5 to 8 h. After 24 to 36 h, the cells were harvested, resuspended in 250 μl 50 mM Tris (pH 8), 150 mM NaCl, 10% glycerol, 1% Nonidet P-40 in the presence of a protease inhibitor cocktail (Sigma), and incubated on ice for 30 min. Soluble protein was collected after 15 min of centrifugation at 20,000 × g. Aliquots of the soluble extract were then incubated for 2 to 4 h with GST fusion proteins on a shaking platform. The beads were washed four times with 1 ml lysis buffer containing 0.1% NP-40 and once with 1 ml PBS. For pull-down experiments with NS3 expressed in insect cells, TENT or NP-40 lysis buffer was used during incubation with GST and GST fusion proteins. The respective lysis buffer was also used for washing the agarose-bound protein complexes. After the final wash, an equal volume of sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer was added, and the beads were heated for 15 min at 70°C. The samples were then centrifuged briefly, and equal aliquots of supernatant were run in denaturing 10% or 15% polyacrylamide gels (36, 60).
Confocal microscopy.
For colocalization studies, HeLa cells were seeded onto coverslips in six-well plates and cotransfected with equal amounts of pTriex1-NS3 and pTriex-GFP-Tsg101, using Fugene6 as recommended by the manufacturer (Roche, United Kingdom). At 24 h posttransfection, the cells were washed with serum-free medium, fixed with 4% paraformaldehyde in PBS for 15 min at 4°C, and washed once with 25 mM glycine in PBS and once with PBS. For intracellular staining, cells were permeabilized with 1% Tween 20 in PBS for 5 min at room temperature and washed once with PBS. Cells were then incubated with polyclonal antiserum against NS3 and stained with tetramethyl rhodamine isocyanate-labeled secondary antibody (Sigma) for 1 h at room temperature. The antibodies were applied sequentially at a dilution of 1:100, and the cells were washed three times between each step with PBS. After a final wash with water, cells were mounted with Vectashield (Vector Laboratories, Burlingame, CA). Images were acquired on an LSM510 microscope (Carl Zeiss Ltd., United Kingdom) using a 43× (numerical aperture, 1.4) oil objective. The pinhole was adjusted to allow a field depth of 1 μm. Multichannel acquisition was used to avoid cross talk between the respective channels. Images were processed with the LSM image browser (Carl Zeiss Ltd., United Kingdom).
VLP release assays.
293T cells (0.8 × 106 to 1 × 106) were seeded into six-well plates and transfected with 4 μg plasmid DNA after 8 to 12 h, using Lipofectamine 2000 (Invitrogen) in serum-free medium. After 4 to 8 h, the transfection mixture was replaced with DMEM containing 10% FBS, and the supernatant was harvested at 24 to 48 h posttransfection. Supernatants were clarified by low-speed centrifugation (5 min at 20,000 × g) and passage through 0.45-μm filters (Pall). VLPs released into the medium were then purified and concentrated by centrifugation through 20% sucrose cushions (in 100 mM Tris, pH 8) in a TLS55 rotor (Beckman) for 2 h at 50,000 rpm. Pelleted VLPs were lysed in sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer and analyzed by immunoblotting as described above. To examine the intracellular expression level of Gag fusion proteins, whole-cell lysates were analyzed in parallel.
RESULTS
Late-domain motifs in orbiviruses.
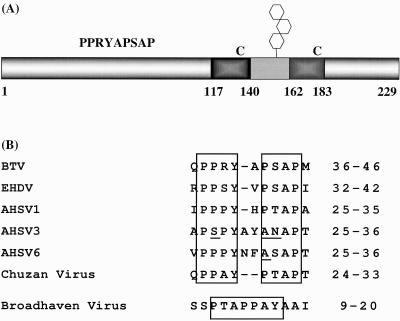
The presence of late-domain motifs in NS3 of BTV was first reported by Strack et al. (65). To determine whether orbiviruses in general encode conventional late-domain motifs, we generated a multiple sequence alignment from published NS3 sequences of 10 different strains of BTV, 3 strains of AHSV, and 1 strain each of epizootic hemorrhagic disease virus (EHDV), Chuzan virus, and Broadhaven virus. All of the NS3 sequences turned out to contain the two most commonly observed late-domain motifs, PPXY and PS/TAP (Fig. 1). In all but one case, the PPXY motif is followed by the PS/TAP motif, with two or fewer amino acids in between. In the case of Broadhaven virus, the PTAP motif precedes and partially overlaps the PPXY motif. Interestingly, we found that only two of the amino acids in each motif are strictly conserved in all NS3 proteins. While both motifs fully match the consensus sequence in all strains of BTV, there seems to be more potential for variation within AHSV NS3 sequences. In two strains of AHSV, the PPXY consensus sequence is replaced by PSXY and the PS/TAP motif is replaced by ANAP. In EHDV and several strains of AHSV, the second motif is replaced by ASAP.
FIG. 1.
Cartoon of BTV NS3 and late-domain motifs in orbivirus NS3 proteins. (A) Cartoon showing the domain structure of NS3 and the location and sequence of the late-domain motifs. Transmembrane domains are indicated by dark gray boxes, and the extracellular domain is shown in light gray. The N- and C-terminal intracellular domains extend from amino acids 1 to 117 and 183 to 229, respectively. Hexagons indicate the single glycosylation site, which is encoded by all BTV strains, and “C” indicates the two conserved cysteine residues. The late-domain motifs extend from amino acids 37 to 45. NS3A is generated by initiation at an in-frame ATG at codon 14 of the NS3 open reading frame. (B) Alignment of late-domain motifs in orbivirus NS3 proteins. Numbers to the right indicate the amino acid positions of the aligned sequences within NS3.
NS3 of BTV binds mammalian and insect cell Tsg101.
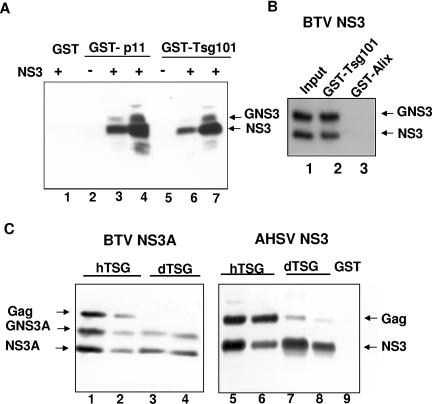
To determine whether BTV NS3 binds to Tsg101, like other viral proteins which harbor the PT/SAP motif, we performed pull-down experiments with recombinant NS3 and the GST-tagged UEV domain of Tsg101, which was shown earlier to mediate the interaction with the PTAP motif. The UEV domain was expressed in E. coli, while NS3 was initially obtained from Sf21 cells which were infected with a recombinant baculovirus that expresses NS3 at a high level but NS3A in a negligible amount (14). For comparison, we also performed pull-down assays with GST-tagged p11, which was previously shown to bind to NS3 (6), and with GST alone as a negative control. As shown in Fig. 2A, NS3 from Sf21 cells bound strongly to Tsg101 and to p11. Since NS3 is expressed as both glycosylated (GNS3) and nonglycosylated (NS3) forms in cell culture (78), both forms of NS3 were detectable in the complexes. No binding of NS3 to the GST control was detectable, demonstrating the specificity of the interaction. To further confirm the interaction between NS3 and Tsg101, we generated a recombinant construct that expressed FLAG-tagged NS3 of another orbivirus, AHSV serotype 6 (AHSV-6), which encodes ASAP instead of PSAP (Fig. 1). When FLAG-tagged AHSV NS3 was used in similar pull-down experiments, the same result was obtained as with BTV NS3 (Fig. 2A and C). The experiment was repeated several times, using NS3 that was expressed transiently in 293T cells and different amounts of GST-tagged proteins, with essentially the same results (data not shown). We also observed that NS3A binds to Tsg101 with a similar strength to that of NS3. The specificity of the interaction of NS3 and Tsg101 was further confirmed in an additional pull-down experiment using GST-tagged Alix, which functions downstream of Tsg101. As shown in Fig. 2B, the GST-tagged Alix completely failed to bind to NS3 expressed in 293T cells, in contrast to Tsg101.
FIG. 2.
Pull-down analysis of the interaction between NS3 and Tsg101. (A) NS3 of BTV-10 was expressed in Sf21 cells, using a recombinant baculovirus, and incubated with GST-agarose beads (lane 1), GST-tagged p11 (lanes 3 and 4), and GST-tagged UEV of human Tsg101 (lanes 6 and 7). Two different buffers (see Materials and Methods) were used for solubilizing NS3 during incubation and for washing of GST-p11 and GST-UEV protein complexes, which was followed by immunoblotting for NS3. (B) NS3 of BTV-10 was expressed in 293T cells and used in pull-down assays with GST-tagged UEV or GST-tagged Alix lacking the C-terminal PRD domain. A whole-cell lysate corresponding to 1% of the amount used for pull-down assays was run in lane 1 for comparison. Note that the transient expression of BTV-10 NS3 in mammalian cells gives rise to two bands, which are formed at approximately equal levels and represent the glycosylated (GNS3) and nonglycosylated (NS3) protein. (C) HIV-1 Gag was expressed in 293T cells, mixed with NS3A of BTV-10 or NS3 of AHSV-6, and used for pull-down assays with GST-tagged UEV of human Tsg101 (hTsg) or its orthologue from Drosophila melanogaster (dTsg). The amount of GST-tagged protein was estimated to be about 10 μg in lanes 1, 3, 5, and 7 and 2 μg in lanes 2, 4, 6, and 8. Western blotting was performed with a mixture of polyclonal antiserum against NS3 and a monoclonal antibody against p24 of HIV Gag.
Since orbiviruses replicate in mammalian as well as insect cells (Culicoides and others) and since Tsg101 is highly conserved from yeast to mammals, we reasoned that NS3 is likely to bind to the insect ortholog of Tsg101. To verify this and to determine whether NS3 binds at similar levels to mammalian Tsg101 and the insect ortholog, we expressed the UEV domain of Drosophila Tsg101 as a GST-tagged protein for use in pull-down experiments. The Drosophila protein was chosen because no genome sequences were available for Culicoides species, which are the natural hosts of BTV. In addition, to further examine the strength of the interaction between NS3 and Tsg101, solubilized NS3A of BTV or NS3 of AHSV was mixed with a lysate of 293T cells expressing full-length Gag of HIV-1 and then used in pull-down experiments with different amounts of GST-tagged UEV of Tsg101 or the tagged insect ortholog. The pull-down experiments showed that HIV Gag binds much more strongly to human Tsg101 than to the Tsg101 ortholog from Drosophila (Fig. 2C). In contrast, NS3 of both BTV and AHSV bound to mammalian Tsg101 and its insect cell ortholog with similar strengths, indicating that in both vertebrate and invertebrate cells, Tsg101 may play a role in facilitating the egress of orbiviruses. The experiment also showed that Tsg101 binds more strongly to HIV Gag than to NS3.
The PSAP motif of NS3 is required for binding of NS3 to Tsg101.
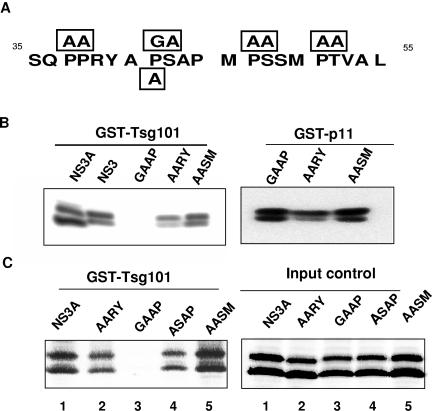
Although studies of other viruses strongly suggested that NS3 binds to Tsg101 by means of the PSAP motif at positions 42 to 45, we noticed that NS3 of BTV-10 contains two additional tetrapeptides, PSSM (amino acids 47 to 50) and PTVA (amino acids 51 to 54), in close proximity. Since both motifs share some sequence similarity with the PS/TAP late-domain motif, we wanted to clarify whether the PSAP sequence is solely responsible for binding Tsg101 or whether the other motifs play a role as well. To this end, we mutated each of the three tetrapeptides individually by introducing double mutations that were expected to diminish the affinity for Tsg101 significantly without changing the overall folding too much (Fig. 3A). Of the three mutants generated, the one that carried a double mutation in the PTVA sequence proved difficult to express and was not analyzed further. To obtain another control that would allow us to distinguish between a localized effect and one that was caused by more far-reaching changes in the tertiary structure, we also replaced two amino acids in the nearby PPXY motif to generate the AARY mutant. In other viruses, the PPXY motif was shown to bind to class I WW domains in NEDD4-like ubiquitin ligases but was not reported to bind to Tsg101. The AARY mutant was therefore not expected to directly influence the binding of NS3 to Tsg101. All mutants were transiently expressed in 293T cells in parallel with wild-type NS3, and detergent-solubilized protein was used in pull-down experiments with GST-tagged UEV from human Tsg101, with GST-tagged p11 as a control. We also verified that the proteins were expressed at approximately equal levels (data not shown). As shown in Fig. 3B, mutation of the PSAP motif abolished the binding of NS3 to Tsg101 but had little effect on binding to p11. In contrast, no significant reduction in binding was observed with the AASM mutant. The AARY control mutant showed some reduction in binding to Tsg101 compared to the wild-type protein. However, this mutant also bound less strongly to p11, which suggests that the observed effect is due to a more pronounced impact of the mutation on the global folding of the protein. In conclusion, the experiment suggests that the PSAP motif is mainly, if not solely, responsible for binding Tsg101.
FIG. 3.
Orbivirus NS3 binds to Tsg101 by means of PSAP or ASAP motifs. (A) Mutations introduced into BTV-10 NS3 for analyzing binding specificity. Five mutants were generated by introducing single or double mutations into NS3 and NS3A. The substitutions are enclosed by rectangles and are aligned with the wild-type sequence of codons 35 to 55 of NS3. (B) Pull-down analysis of the interaction between NS3 and Tsg101. Lysates of 293T cells expressing NS3, NS3A, or one of three different mutants were incubated with GST-UEV (from human Tsg101) or GST-p11, followed by immunoblot analysis for NS3. (C) Pull-down analysis of the interaction between NS3A and Tsg101 (left panel). Lysates of 293T cells expressing NS3A or one of four different mutants were incubated with GST-UEV (from human Tsg101), followed by immunoblot analysis for NS3A. The right panel shows the amounts of input proteins for polyacrylamide gel electrophoresis.
With the exception of those in the AASM control mutant, all of the mutations described above were also transferred into a plasmid which expresses only NS3A. In addition, another mutant was generated in which a single mutation was introduced into the NS3A expression vector, changing PSAP to ASAP. This mutant was designed to test the importance of the proline at position 1 for binding Tsg101, as several orbiviruses, including AHSV-6, do not encode proline at this position. All mutants were expressed in 293T cells at approximately the same level, and lysates were analyzed in pull-down assays as described above, using the UEV domain of human Tsg101 (Fig. 3C) and its ortholog from Drosophila (data not shown). The results were essentially the same as those observed with the NS3 mutants, and no significant difference was apparent between the human and Drosophila UEV domains (data not shown). Notably, both proteins failed to bind the GAAP double mutant but still bound to the ASAP single mutant, though somewhat less strongly than to the PSAP wild-type protein for human UEV.
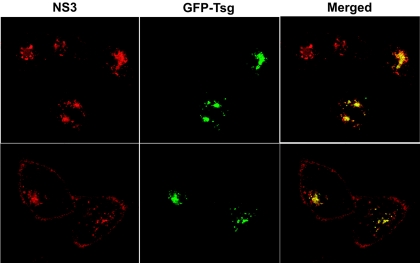
Confocal microscopy shows that NS3 colocalizes with Tsg101.
To provide evidence for a possible interaction between NS3 and Tsg101, in vivo confocal microscopy studies were performed. Expression plasmids for wild-type NS3 and the GAAP mutant of NS3 were transiently transfected into HeLa cells in combination with a plasmid that encodes GFP-tagged Tsg101. The cells were processed for intracellular staining at 24 h posttransfection, which coincides with the peak in the expression level of NS3. As observed earlier, NS3 was expressed at the plasma membrane and at intracellular membranes (Fig. 4). Extensive colocalization with Tsg101 was observed at intracellular compartments, but not on the plasma membrane. In contrast to the wild-type protein, the GAAP mutant did not colocalize with Tsg101 (data not shown). These data suggest that a functional interaction between NS3 and Tsg101 can take place within intracellular vesicles.
FIG. 4.
Confocal images of HeLa cells showing colocalization of NS3 and GFP-tagged Tsg101. Expression plasmids for NS3 and GFP-tagged Tsg101 were cotransfected into HeLa cells. At 24 h posttransfection, the cells were fixed and processed for confocal microscopy. NS3 (red) was detected by indirect immunofluorescence.
The PPXY motif is required for binding NEDD4-like ubiquitin ligases.
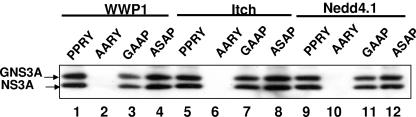
Like most other viral proteins which bind to Tsg101, NS3 contains a PPXY motif in close proximity to the PS/TAP late-domain motif (Fig. 1). The PPXY motif was reported to bind to HECT domain-containing ubiquitin ligases, such as NEDD4 and WWP1 (7, 8, 22, 24, 34, 59, 69, 70, 79, 80). To determine whether NS3 can recruit the same class of ubiquitin ligases, we performed pull-down experiments with the NS3 mutants described above and the GST-tagged WW domains of NEDD4.1, WWP1, and Itch. As shown in Fig. 5, all three proteins bound to wild-type NS3 (lanes 1, 5, and 9) and the GAAP mutant (lanes 3, 7, and 11) but failed to bind the AARY NS3 mutant (lanes 2, 6, and 10), confirming the specificity of the interaction. We did not observe a significant difference in binding to NS3 between the three ubiquitin ligases. We noticed that Itch, and to a lesser extent, WWP1, seemed to bind more strongly to the ASAP mutant (lanes 4 and 8) than to wild-type NS3, which was not the case with NEDD4.1. However, the difference does not seem large enough to be of particular significance.
FIG. 5.
NS3 binds to NEDD4-like ubiquitin ligases in vitro. Lysates of 293T cells expressing NS3A or one of three different mutants were incubated with GST-tagged WW domains of the human ubiquitin ligases WWP1, Itch, and NEDD4.1, followed by immunoblot analysis of bound NS3. The mutants are shown in more detail in Fig. 3.
Depletion of Tsg101 inhibits virus release.
An interaction of NS3 and Tsg101 may be necessary for the release of newly synthesized virus particles from BTV-infected cells, similar to that of enveloped viruses. In order to investigate the biological importance of the interaction between NS3 and Tsg101, we synthesized three siRNAs targeting different sequences in the Tsg101 open reading frame. Two of the siRNAs proved to be highly effective in reducing the expression of Tsg101 when cotransfected into 293T cells together with a plasmid expressing Tsg101 (data not shown). One siRNA was homologous to coding nucleotides 1116 to 1135 (Tsg si1), and the other one was identical to the siRNA described by Garrus et al. (15), except for two mismatches at the 3′ end of the sense strand which were designed to improve the silencing activity (25). We also obtained siRNAs for two different target sequences in BTV-10 NS3, which were effective but less so than the siRNAs designed to suppress Tsg101. For controls, we synthesized siRNAs against the heavy chain (p36) and the light chain (p11) of calpactin I. The latter proved to be very effective in suppressing p11 (data not shown), whereas the siRNA targeting p36 had no effect and was used as a negative control in subsequent experiments. As an additional negative control, we used a 21-nucleotide antisense RNA against a target sequence within Tsg101.
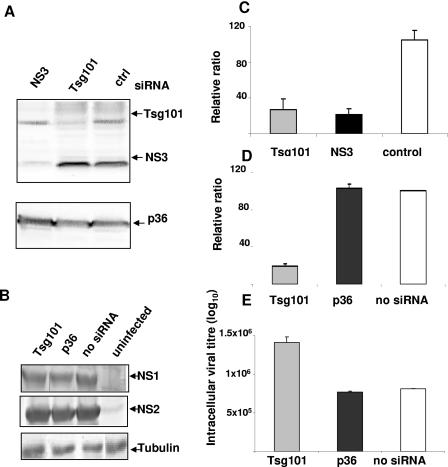
To study the effect of siRNA-induced depletion of Tsg101 on BTV release, HeLa cells were transfected twice with siRNA and subsequently infected with BTV-10. To verify that the siRNAs were effective and specific for their respective targets, cell lysates were analyzed by immunoblotting (Fig. 6A). The transfection of Tsg101 siRNA resulted in a significant reduction of intracellular Tsg101. No effect was observed with the control siRNA. To confirm further, the expression levels of two nonstructural BTV proteins, NS1 and NS2, were investigated in BTV-infected but not siRNA-transfected cells (control) and in cells that had been both siRNA transfected and infected. As shown in Fig. 6B, the levels of expression of the two proteins were similar in all cells, confirming that the siRNAs did not influence viral protein expression. Additionally, the levels of production of a cellular protein (tubulin) were similar in all the cells. This confirms that transfecting cells with siRNA did not make any change in the production of cellular proteins and gives an indication of the amount of viral protein synthesis relative to cellular/total protein.
FIG. 6.
Depletion of Tsg101 inhibits release of BTV-10. HeLa cells were transfected twice with the indicated siRNAs and infected 24 h later with BTV-10. Cell lysates were harvested at 24 h postinfection and analyzed by immunoblotting with antisera specific for Tsg101, NS3, and the heavy chain of annexin II (p36) (A) or with polyclonal antisera against BTV-10 and tubulin (B). (C) Virus titers in the supernatant were determined at 12 h postinfection and plotted as percentages of the titer in untransfected cells. Bars show the means of three independent experiments. (D) The ratio of extracellular to intracellular virus was determined after the cells were lysed and was plotted as a percentage of the ratio of untransfected cells. (E) Logarithmic values of intracellular virus titers of transfected and untransfected cells.
To assess the effect of Tsg101 depletion on virus release, virus titers in the medium were determined 12 and 24 h after infection and were plotted as percentages of the titer in the supernatant of untransfected cells (Fig. 6C). A significant reduction in virus titer was observed with Tsg101 siRNA in repeat experiments. Compared to the negative control, the titer was reduced by 65 to 80% at 12 h postinfection (Fig. 6C). A similar reduction in titer was observed after transfection of NS3 siRNA, whereas cells transfected with control RNA did not show any significant change in virus release compared to untransfected cells. To confirm that the reduction in titer was due to an inhibition of virus release and not the result of interference in the virus replication cycle, we determined the ratio of extra- to intracellular infectious particles. The ratio was essentially the same after transfection of the negative control RNA as that for untransfected cells (Fig. 6D). In contrast, a fourfold reduction was observed after transfection of Tsg101 siRNA. In addition, the intracellular viral titer in cells transfected with Tsg101 siRNA was more than that in untransfected cells or cells transfected with the control siRNA (Fig. 6E). Hence, we concluded that the depletion of Tsg101 inhibits virus release and does not interfere with the replication cycle of BTV.
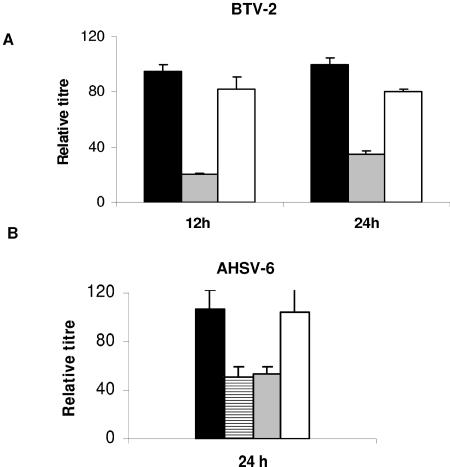
To confirm this conclusion, we also assessed the effect of Tsg101 siRNA on BTV-2 and African horse sickness virus. Very similar results were obtained when the experiment was done with serotype 2 of BTV (Fig. 7A). Importantly, the transfection of Tsg101 siRNA reduced the titer of extracellular virus by about 80% at 12 h postinfection. At 24 h postinfection, the titer was still reduced, but the effect was smaller than that at 12 h, and only a threefold reduction was achieved with Tsg101 siRNA. We then performed the same experiment using AHSV instead of BTV. Although the control RNA did reduce the titer to some extent at 12 h postinfection (data not shown), we still observed an approximately 50 to 60% reduction of virus release from cells transfected with Tsg101 siRNA. The effect was considerably less than that observed with BTV at 12 h postinfection but did not change significantly between 12 and 24 h postinfection (Fig. 7B).
FIG. 7.
Depletion of Tsg101 with siRNA inhibits release of BTV-2 and AHSV-6. HeLa cells were transfected twice with the indicated siRNAs and infected 24 h later with BTV-2 (A) or AHSV-6 (B). Two siRNAs against Tsg101 were used (Tsg si1 [striped bars] and Tsg si2 [gray bars]). Virus titers were determined at 12 and 24 h posttransfection and plotted as percentages of the titer in untransfected cells. For negative controls, p36 siRNA (p36 control) (white bars) and a 21-nucleotide antisense RNA (Tsg control) (black bars) against a target sequence within Tsg101 were used.
VLP release assay.
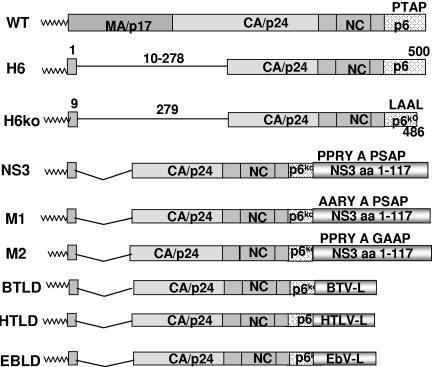
In order to study the late-domain function of NS3 in mammalian cells without having to use infectious virus or a reverse genetics system, which is not available for BTV, we sought to explore whether NS3 has the ability to direct the budding of heterologous virus-like particles. For this purpose, we took advantage of the fact that HIV Gag has the ability to form VLPs that are released if a heterologous late domain is provided in place of the late domain within p6 (43). We first tested several Gag constructs for budding activity, since there are several reports which show a significant release of Gag particles even in the absence of p6 (26, 32, 39, 57). All Gag constructs were placed in a plasmid vector under control of a strong promoter (Fig. 8; see Fig. 10A) and transiently expressed in 293T cells in the absence of other viral proteins. In this context, the p6 domain was not essential for the release of full-length Gag (data not shown). However, as reported by Accola et al. (1), we found that deletion of the N-terminal part of the capsid protein and most of the matrix protein significantly increases the late-domain dependence of particle release (Fig. 9). Of the constructs tested, the one that showed the least residual particle release carried the same deletion of amino acids 10 to 278, with a C-terminal truncation which abolishes binding of Alix (66). In addition, the construct also carries three amino acid substitutions which destroy the Tsg101 binding site. This construct (H6ko) was used in most of the experiments described below for determining the late-domain activity of heterologous sequences, which were fused to the C terminus of the construct via a linker that encodes three alanine residues. To assay their budding activity, the Gag fusion constructs were transfected into 293T cells. VLPs were harvested from the supernatant 24 h after transfection and analyzed by Western blotting. Whole-cell lysates were analyzed in parallel to verify that the Gag chimeras were expressed at approximately equal levels.
FIG. 8.
Schematic illustration of constructs used in VLP release assays. Wild-type HIV-1 Gag is represented at the top by rectangular boxes, and individual Gag proteins are indicated inside the boxes. Vertical lines represent the boundaries between the mature cleavage products. Deletions are indicated by thin lines, and numbers are used to indicate the amino acid position and size of each deletion. Wavy lines represent the myristyl group at the N terminus of Gag. H6 was derived from wild-type Gag by deleting amino acids 10 to 278 and was used as a positive control. H6ko was used as a negative control, as it lacks the C-terminal 14 amino acids of p6 and also carries three substitutions in the PTAP motif, which render the construct defective in the late domain. The construct also carries a C-terminal extension of 18 amino acids derived from vector sequences, which were replaced with oligonucleotides or PCR products to generate the other constructs shown in the cartoons. Codons 33 to 49 of BTV-10 NS3 were appended to p6 to generate BTLD, codons 5 to 16 of Ebola virus VP40 were added to generate EBLD, and codons 117 to 129 of HTLV-1 Gag were added to generate HTLD. PCR products encompassing codons 1 to 117 of wild-type or mutant NS3 were added to generate constructs NS3, M1, and M2. WT, wild type.
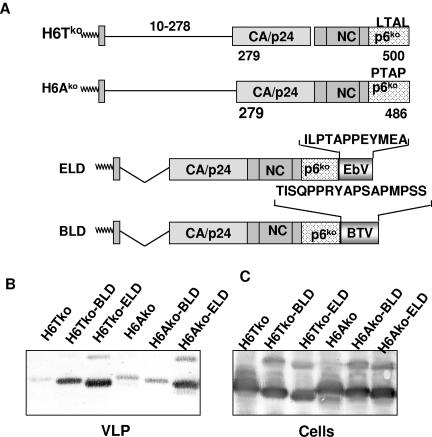
FIG. 10.
Release of virus-like particles from cells expressing HIV Gag constructs. (A) Schematic representation of the HIV Gag construct H6Tko, which carries a knockout mutation in the PTAP motif, and the H6Ako construct, which lacks the binding site for Alix. The late-domain motifs of BTV NS3 and Ebola virus VP40 were appended to the C termini of both constructs, as indicated in the lower panel. Numbers indicate amino acid positions. (B) Gag fusion constructs and parent plasmids were transfected into 293T cells, and VLP release was monitored as described in the legend to Fig. 8. (C) Whole-cell lysates of transfected cells were analyzed with Gag antiserum to verify that the constructs were expressed at equal levels.
FIG. 9.
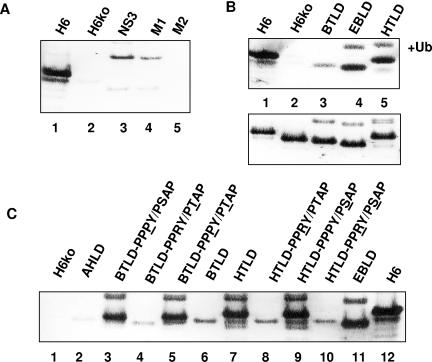
Release of virus-like particles from cells expressing HIV Gag constructs. 293T cells were transfected with the constructs shown in Fig. 8. Virus-like particles were harvested from the supernatant at 24 h posttransfection and analyzed by immunoblotting with an antibody against p24. (A) The NS3 N-terminal domain induces particle release when fused to a late-domain-defective HIV Gag mutant. (B) The NS3 late-domain (BTLD) motifs induce particle release but are less active than the late-domain motifs of Ebola virus (EBLD) and HTLV (HTLD). The upper panel shows particles released into the medium. The lower panel shows whole-cell lysates for comparison to confirm that the proteins were expressed at approximately equal levels. (C) The arginine at position three of the PPRY motif is mainly responsible for the low activity of BTV NS3 in promoting particle release from mammalian cells. Mutated amino acids are underlined in each construct. The first lane shows the negative control (a late-domain-defective Gag mutant). The last lane (lane 12) shows the positive control (a Gag mutant carrying wild-type p6). Note the decrease in electrophoretic mobility when arginine is replaced by proline, which is particularly pronounced in the HTLV background. The construct shown in lane 2 carries amino acids 23 to 38 of AHSV-3 NS3.
In our initial experiments, we fused the full-length N-terminal domain of NS3 to the late-domain-defective Gag construct H6ko. This resulted in a significant increase in budding activity (Fig. 9A, lane 3) which was dependent on the late-domain motifs in NS3. This was demonstrated by control constructs in which the PPRY and PSAP motifs in NS3 were mutated individually to AARY (M1) and GAAP (M2), respectively. Interestingly, only the GAAP mutation (M2) resulted in a complete loss of budding activity (Fig. 9A, lane 5), whereas the AARY mutant (M1) still showed a significant release of particles into the medium (Fig. 9A, lane 4). The budding activity of the H6ko-NS3 fusion construct was much lower than that of the H6 control construct. This was not due to steric constraints or interference with particle assembly, as Gag-NS3 fusion constructs carrying wild-type p6 did not show a significant reduction in budding activity compared to Gag-p6 with no extra sequences fused to it (data not shown). Furthermore, a Gag construct fused to a short peptide containing the NS3 late domain plus a few surrounding amino acids still showed significantly less budding (Fig. 9B, lane 3) than the H6 control construct and two other control constructs carrying the late-domain motifs from Ebola virus VP40 and human T-cell leukemia virus (HTLV) Gag (Fig. 9B, lanes 4 and 5).
The experiment described above indicated that NS3 exhibits late-domain activity in a heterologous context. However, the late-domain motifs in NS3 appeared to be less able to recruit the cellular proteins required for budding than the late-domain motifs in HIV Gag, HTLV Gag, and VP40. This raised the question of the relative importance of each motif. To address this, we changed the PPPY and PTAP late-domain motifs in the H6ko-HTLD construct individually to PPRY and PSAP, respectively, to match the motifs in NS3 and also changed the PPRY and PSAP motifs in the H6ko-BTLD construct to PPPY and PTAP, respectively (Fig. 9C). In addition, a further control (AHLD) was analyzed which is similar to BTLD and HTLD but carries amino acids 23 to 38 of AHSV-3 NS3 appended to the C terminus of H6ko (Fig. 1 and 6). All constructs were transfected into 293T cells in parallel with the control plasmids described above.
As observed in the earlier experiment, the H6ko-BTLD construct showed little budding activity, and no significant change in activity was apparent when the PSAP motif was replaced with PTAP (compare lane 2 with lane 4, lane 3 with lane 5, lane 7 with lane 9, and lane 8 with lane 10). There was also little difference between the Gag fusion proteins carrying PT/SAP motifs and the construct carrying the AHSV-3 late domain (Fig. 9C, lanes 2, 4, 6, 8, and 10). In contrast, a much larger difference was observed between the PPPY and PPRY motifs, regardless of the surrounding amino acids. In fact, introducing the PPPY motif into the BTV NS3 sequence (lanes 3 and 5) seemed to increase the budding activity to nearly the same level as that observed with the Gag-p6 positive control (lane 12), while introduction of the PPRY motif into the HTLV background reduced the budding activity (lanes 8 and 10) to about the same level as that observed with the BTV and AHSV wild-type sequences (lanes 2 and 6). Thus, the presence of arginine at position 3 of the PPXY motif appeared to be mainly responsible for the low budding activity observed with the NS3 wild-type sequence. It is noteworthy in this context that the replacement of arginine with proline at position 3 results in a significant decrease in electrophoretic mobility, which is difficult to explain by a single amino acid substitution alone. At present, we do not know whether the increase in electrophoretic mobility is due to a conformational change, for example, the formation of a left-handed polyproline helix (10, 63, 76), or to a posttranslational modification.
As reported in other studies utilizing mini-Gag constructs, all constructs that carried a PPXY motif gave rise to a higher-molecular-weight band, which presumably represents monoubiquitinated Gag (Fig. 9B and C), and an additional band which represents diubiquitinated Gag (not shown). However, there was no positive correlation between particle release and the extent of Gag ubiquitination, as shown by the BTLD construct, which appeared to be ubiquitinated to about the same extent as the EBLD and HTLD constructs but did not promote particle release (Fig. 9B, lane 3).
In addition to the experiments described above in which we utilized a Gag construct that lacks the binding sites for Alix and Tsg101, we also tested the activity of the NS3 late domains in the context of Gag constructs which retain either the binding site for Alix (H6Tko) or the motif that recruits Tsg101 (H6Ako). In the latter context, the BTV motifs did not provide any additional activity (Fig. 10). This is consistent with the results described above, which show that the PPRY motif is inactive while the PSAP motif in NS3 does not increase the budding activity because the H6Ako construct already contains a functional PTAP motif. In the presence of a functional binding site for Alix, however, the BTV late-domain motifs do increase the budding activity significantly, although to a lesser extent than the late-domain motifs of Ebola virus VP40. In fact, the budding activity of the NS3 motifs appeared to be considerably higher in the single-knockout mutant, which only lacks the PTAP motif, than in the double knockout, which also lacks the binding site for Alix. While this supports the conclusion that the PSAP motif in NS3 is functional and able to recruit Tsg101, it also suggests that the PTAP motif needs to be complemented by binding sites for ubiquitin ligases, Alix, or an uncharacterized protein to fully engage the ESCRT machinery. Our results are also consistent with the notion that the presence of a binding site for Alix might partially obviate the need to recruit ubiquitin ligases.
DISCUSSION
Orbiviruses share a morphological and overall molecular organization of the double capsid structure with the other members of the family Reoviridae. Like the other members of the Reoviridae, orbiviruses seem to lack a lipid envelope, which is unusual among arthropod-borne viruses. Electron microscopic studies of BTV-infected cells revealed both nonenveloped and enveloped particles penetrating the plasma membrane, indicating that the latter become enveloped only transiently (28, 35). The results presented here explain the formation of the enveloped particles. Like other enveloped viruses harboring PT/SAP and PPXY late-domain motifs, orbiviruses seem to usurp the vacuolar protein sorting (VPS) pathway, which normally functions in the formation of multivesicular bodies, to allow virus particles to leave host cells by a budding mechanism. However, this mechanism seems to be inefficient in most mammalian cells since most of the orbivirus progeny particles remain cell associated.
Although there is no direct evidence yet that viruses which replicate in insect cells utilize the VPS pathway, there is good reason to believe they do, as most of the proteins that function in the formation of multivesicular bodies are conserved from yeast to mammals. Our finding that NS3 binds to Tsg101 from Drosophila lends further support to this hypothesis.
One of the intriguing features of BTV is the observation that virions can be extruded through the plasma membranes of mammalian cells as nonenveloped particles. In a recent study, Han and Harty (20) showed that NS3 can act as a viroporin causing the permeabilization of host cell membranes. It is possible that this permeabilization activity facilitates local disruption of the plasma membrane, allowing virus particles to be extruded through a membrane pore without acquiring a lipid envelope. Whether this process requires any of the cellular proteins that participate in the formation of an enveloped viral particle merits further investigation.
While NS3 seems to provide a conventional late-domain function for the formation of an infectious BTV particle, it does exhibit only weak activity compared to other late domains when used to replace the cognate late domain of HIV Gag. This seems to be mainly due to the presence of an arginine at position three of the PPRY motif, as a replacement of arginine by proline, which is typically found at this position in late domains of other viruses, renders the NS3 late domain highly active in human cells.
In other viruses, PPXY motifs have been shown to recruit NEDD4-like ubiquitin ligases (7, 8, 22-24, 34, 59, 79, 80). The function of this interaction has not been elucidated in detail, but the available evidence suggests that NEDD4-like ubiquitin ligases play an important or essential role in the budding process. It is therefore surprising that the PPRY motif binds to NEDD4-like ubiquitin ligases in vitro and also appears to induce the ubiquitination of Gag in vivo but appears to be unable to promote budding in vivo, at least in the context of a mini-Gag particle. In light of available data, we can think of only two explanations for this observation: (i) the PPRY motif recruits an antagonist which interferes with the binding or activity of the cellular budding machinery or (ii) the BTV PPRY late domain is less able than the HTLV PPPY late domain to recruit an essential cellular budding factor. The putative agonist is most likely different from NEDD4, since like the PPPY motif, the PPRY motif appears to promote Gag ubiquitination. Whether the agonist is Tsg101 or an undefined factor remains to be determined.
There is a considerable discrepancy in the literature with regard to the role of Gag ubiquitination and the ability of the PTAP and PPXY motifs to stimulate the release of HIV Gag particles (41, 67), which seems to depend on the context in which the motifs were analyzed. In agreement with other studies that utilized mini-Gag constructs, we observed that the PPXY motif can greatly stimulate the release of mini-Gag particles, which is in marked contrast to its apparent inability to stimulate the release of full-length Gag (41) but consistent with results reported for HTLV, where the PPXY motif has been shown to greatly increase particle release (8, 24, 74). We also observed a positive correlation between the extent of Gag ubiquitination and particle release in the context of a mini-Gag construct carrying the late domains of HTLV and Ebola virus, as reported earlier (41, 67). However, this does not seem to apply to PPXY motifs in general, as the PPRY motif in NS3 seems to induce Gag ubiquitination but does not promote particle release, similar to observations made with full-length Gag (41). This raises the possibility that ubiquitin ligases, which are recruited by the PPXY motif, have another functional target in addition to Gag. While our own results do not allow us to clarify this issue and to resolve the apparent discrepancies between the results obtained with full-length and mini-Gag, we favor the notion that the differences are largely due to the absence or presence of uncharacterized motifs that recruit components of the ESCRT machinery or of other cellular proteins which functionally assist or interfere with ESCRT proteins. The latter might apply to the PPRY motif, as discussed above. However, we cannot exclude other explanations that have been discussed in this context (41). Whatever turns out to be the main reason for the different behaviors of mini- and full-length Gag, there are certainly additional late-domain motifs to be discovered in Gag proteins. In keeping with this notion, we observed that mouse mammary tumor virus Gag harbors an FPVV sequence which is very similar to the FPIV late-domain motif described recently for the matrix protein of Sendai virus (61). Mouse mammary tumor virus seems particularly noteworthy in this context since it contains a PTAP but no PPXY motif, like HIV Gag (11).
Of all the viruses utilizing conventional late domains that we have examined, bluetongue viruses are the only ones that encode arginine at position three of the PPXY motif. This raises the question of why they are different. We believe the answer could lie in the fact that orbiviruses replicate in insects, whereas the other viruses studied do not. It will therefore be interesting to compare the budding activities of the PPPY and PPRY late domains in insect cells.
NS3 is unusual among viral proteins recruiting the MVB pathway because it also functions as a viroporin. While both activities seem to facilitate virus release, the relative contributions of the two activities could differ between different orbiviruses, which could explain why the depletion of Tsg101 inhibits the release of AHSV to a lesser extent than that of BTV. This conclusion is consistent with our observation that cells infected with AHSV displayed a much stronger cytopathic effect at early times after infection than cells infected with BTV, which could reflect a higher intrinsic cytotoxic activity of AHSV NS3 or a higher expression level of NS3 in cells infected with AHSV (71) than in BTV-infected cells (31). In any case, particle release due to cell lysis caused by the viroporin activity of NS3 presumably does not require the function of Tsg101.
Structural studies of a UEV peptide complex as well as mutagenesis of the PTAP motif indicate that P1 contributes least to the binding affinity and specificity of the interaction with Tsg101 (15, 27, 51). This is in agreement with our own studies, which show binding of Tsg101 to NS3 of AHSV even though the conventional PTAP motif was replaced by ASAP. There are, in fact, several other viruses, such as bovine leukemia virus, Rous sarcoma virus, and simian retrovirus 2, which encode AS/TAP instead of PS/TAP in close proximity to a PPXY motif (58, 62, 68) and which might also utilize this motif to help recruit Tsg101.
In conclusion, our study shows that orbivirus NS3 recruits the cellular protein Tsg101, thereby facilitating virus release in mammalian cells and presumably in insect cells as well. In fact, the ability to usurp the MVB pathway is likely to be more important in insect hosts, as orbiviruses establish persistent infections in insect cells without causing significant cytopathic effects. The viroporin activity, on the other hand, might be more important for facilitating virus release from mammalian cells. While full clarification of this issue will have to await the availability of a reverse genetics system, we are currently in the process of identifying insect proteins that interact with NS3. This should allow us to shed more light on the question of whether NS3 is better adapted to engaging insect proteins for facilitating virus release.
Acknowledgments
We thank Craig Williams for critical reading of the manuscript and for help with the figures and Corinne Bolstad for typing.
This work was partly supported by the Wellcome Trust Fund, London, United Kingdom, and by the NIH. C.W. is funded by the Wellcome Trust.
REFERENCES
- 1.Accola, M. A., B. Strack, and H. G. Gottlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babst, M., D. J. Katzmann, E. J. Estepa-Sabal, T. Meerloo, and S. D. Emr. 2002. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell 3:271-282. [DOI] [PubMed] [Google Scholar]
- 4.Babst, M., D. J. Katzmann, W. B. Snyder, B. Wendland, and S. D. Emr. 2002. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell 3:283-289. [DOI] [PubMed] [Google Scholar]
- 5.Bansal, O. B., A. Stokes, A. Bansal, D. H. L. Bishop, and P. Roy. 1998. Membrane organization of bluetongue virus nonstructural glycoprotein NS3. J. Virol. 72:3362-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beaton, A. R., J. Rodriguez, Y. K. Reddy, and P. Roy. 2002. The membrane trafficking protein calpactin forms a complex with bluetongue virus protein NS3 and mediates virus release. Proc. Natl. Acad. Sci. USA 99:13154-13159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blot, V., F. Perugi, B. Gay, M. C. Prevost, L. Briant, F. Tangy, H. Abriel, O. Staub, M. C. Dokhelar, and C. Pique. 2004. Nedd4.1-mediated ubiquitination and subsequent recruitment of Tsg101 ensure HTLV-1 Gag trafficking towards the multivesicular body pathway prior to virus budding. J. Cell Sci. 117:2357-2367. [DOI] [PubMed] [Google Scholar]
- 8.Bouamr, F., J. A. Melillo, M. Q. Wang, K. Nagashima, M. de Los Santos, A. Rein, and S. P. Goff. 2003. PPPYVEPTAP motif is the late domain of human T-cell leukemia virus type 1 Gag and mediates its functional interaction with cellular proteins Nedd4 and Tsg101. J. Virol. 77:11882-11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craven, R. C., R. N. Harty, J. Paragas, P. Palese, and J. W. Wills. 1999. Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J. Virol. 73:3359-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Creamer, T. P. 1998. Left-handed polyproline II helix formation is (very) locally driven. Proteins 33:218-226. [PubMed] [Google Scholar]
- 11.Fasel, N., E. Buetti, J. Firzlaff, K. Pearson, and H. Diggelmann. 1983. Nucleotide sequence of the 5′ noncoding region and part of the gag gene of mouse mammary tumor virus; identification of the 5′ splicing site for subgenomic mRNAs. Nucleic Acids Res. 11:6943-6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freed, E. O. 2004. Mechanisms of enveloped virus release. Virus Res. 106:85-86. [DOI] [PubMed] [Google Scholar]
- 13.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.French, T. J., S. Inumaru, and P. Roy. 1989. Expression of two related nonstructural proteins of bluetongue virus (BTV) type 10 in insect cells by a recombinant baculovirus: production of polyclonal ascitic fluid and characterization of the gene product in BTV-infected BHK cells. J. Virol. 63:3270-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 16.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottwein, E., J. Bodem, B. Muller, A. Schmechel, H. Zentgraf, and H. G. Krausslich. 2003. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J. Virol. 77:9474-9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gruenberg, J., and H. Stenmark. 2004. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 5:317-323. [DOI] [PubMed] [Google Scholar]
- 19.Guirakhoo, F., J. A. Catalan, and T. P. Monath. 1995. Adaptation of bluetongue virus in mosquito cells results in overexpression of NS3 proteins and release of virus particles. Arch. Virol. 140:967-974. [DOI] [PubMed] [Google Scholar]
- 20.Han, Z., and R. N. Harty. 2004. The NS3 protein of bluetongue virus exhibits viroporin-like properties. J. Biol. Chem. 279:43092-43097. [DOI] [PubMed] [Google Scholar]
- 21.Harty, R. N., M. E. Brown, J. P. McGettigan, G. Wang, H. R. Jayakar, J. M. Huibregtse, M. A. Whitt, and M. J. Schnell. 2001. Rhabdoviruses and the cellular ubiquitin-proteasome system: a budding interaction. J. Virol. 75:10623-10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harty, R. N., J. Paragas, M. Sudol, and P. Palese. 1999. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J. Virol. 73:2921-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heidecker, G., P. A. Lloyd, K. Fox, K. Nagashima, and D. Derse. 2004. Late assembly motifs of human T-cell leukemia virus type 1 and their relative roles in particle release. J. Virol. 78:6636-6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hohjoh, H. 2004. Enhancement of RNAi activity by improved siRNA duplexes. FEBS Lett. 557:193-198. [DOI] [PubMed] [Google Scholar]
- 26.Hoshikawa, N., A. Kojima, A. Yasuda, E. Takayashiki, S. Masuko, J. Chiba, T. Sata, and T. Kurata. 1991. Role of the gag and pol genes of human immunodeficiency virus in the morphogenesis and maturation of retrovirus-like particles expressed by recombinant vaccinia virus: an ultrastructural study. J. Gen. Virol. 72:2509-2517. [DOI] [PubMed] [Google Scholar]
- 27.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hyatt, A. D., B. T. Eaton, and S. M. Brookes. 1989. The release of bluetongue virus from infected cells and their superinfection by progeny virus. Virology 173:21-34. [DOI] [PubMed] [Google Scholar]
- 29.Hyatt, A. D., A. R. Gould, B. Coupar, and B. T. Eaton. 1991. Localization of the non-structural protein NS3 in bluetongue virus-infected cells. J. Gen. Virol. 72:2263-2267. [DOI] [PubMed] [Google Scholar]
- 30.Hyatt, A. D., Y. Zhao, and P. Roy. 1993. Release of bluetongue virus-like particles from insect cells is mediated by BTV nonstructural protein NS3/NS3A. Virology 193:592-603. [DOI] [PubMed] [Google Scholar]
- 31.Jensen, M. J., I. W. Cheney, L. H. Thompson, J. O. Mecham, W. C. Wilson, M. Yamakawa, P. Roy, and B. M. Gorman. 1994. The smallest gene of the orbivirus, epizootic hemorrhagic disease, is expressed in virus-infected cells as two proteins and the expression differs from that of the cognate gene of bluetongue virus. Virus Res. 32:353-364. [DOI] [PubMed] [Google Scholar]
- 32.Jowett, J. B., D. J. Hockley, M. V. Nermut, and I. M. Jones. 1992. Distinct signals in human immunodeficiency virus type 1 Pr55 necessary for RNA binding and particle formation. J. Gen. Virol. 73:3079-3086. [DOI] [PubMed] [Google Scholar]
- 33.Katzmann, D. J., M. Babst, and S. D. Emr. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106:145-155. [DOI] [PubMed] [Google Scholar]
- 34.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. USA 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.King, B. M., and M. A. Alders. 1985. Morphology of bluetongue virus-infected Aedes albopictus (C6/36) cell culture. Prog. Clin. Biol. Res. 178:289-294. [PubMed] [Google Scholar]
- 36.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of the bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 37.Lee, J. W., and P. Roy. 1986. Nucleotide sequence of a cDNA clone of RNA segment 10 of bluetongue virus (serotype 10). J. Gen. Virol. 67:2833-2837. [DOI] [PubMed] [Google Scholar]
- 38.Licata, J. M., M. Simpson-Holley, N. T. Wright, Z. Han, J. Paragas, and R. N. Harty. 2003. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J. Virol. 77:1812-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo, L., Y. Li, S. Dales, and C. Y. Kang. 1994. Mapping of functional domains for HIV-2 gag assembly into virus-like particles. Virology 205:496-502. [DOI] [PubMed] [Google Scholar]
- 40.Martin-Serrano, J., S. W. Eastman, W. Chung, and P. D. Bieniasz. 2005. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J. Cell Biol. 168:89-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin-Serrano, J., D. Perez-Caballero, and P. D. Bieniasz. 2004. Context-dependent effects of L domains and ubiquitination on viral budding. J. Virol. 78:5554-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin-Serrano, J., A. Yarovoy, D. Perez-Caballero, and P. D. Bieniasz. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. USA 100:12414-12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 44.McGettigan, J. P., S. Sarma, J. M. Orenstein, R. J. Pomerantz, and M. J. Schnell. 2001. Expression and immunogenicity of human immunodeficiency virus type 1 Gag expressed by a replication-competent rhabdovirus-based vaccine vector. J. Virol. 75:8724-8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morita, E., and W. I. Sundquist. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395-425. [DOI] [PubMed] [Google Scholar]
- 46.Myers, E. L., and J. F. Allen. 2002. Tsg101, an inactive homologue of ubiquitin ligase E2, interacts specifically with human immunodeficiency virus type 2 Gag polyprotein and results in increased levels of ubiquitinated Gag. J. Virol. 76:11226-11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, and U. Schubert. 2000. Ubiquitination of HIV-1 and MuLV Gag. Virology 278:111-121. [DOI] [PubMed] [Google Scholar]
- 48.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perez, M., R. C. Craven, and J. C. de la Torre. 2003. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc. Natl. Acad. Sci. USA 100:12978-12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pornillos, O., S. L. Alam, D. R. Davis, and W. I. Sundquist. 2002. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat. Struct. Biol. 9:812-817. [DOI] [PubMed] [Google Scholar]
- 52.Pornillos, O., J. E. Garrus, and W. I. Sundquist. 2002. Mechanisms of enveloped RNA virus budding. Trends Cell Biol. 12:569-579. [DOI] [PubMed] [Google Scholar]
- 53.Potgieter, A. C., A. D. Steele, and A. A. van Dijk. 2002. Cloning of complete genome sets of six dsRNA viruses using an improved cloning method for large dsRNA genes. J. Gen. Virol. 83:2215-2223. [DOI] [PubMed] [Google Scholar]
- 54.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raiborg, C., T. E. Rusten, and H. Stenmark. 2003. Protein sorting into multivesicular endosomes. Curr. Opin. Cell Biol. 15:446-455. [DOI] [PubMed] [Google Scholar]
- 56.Ramadevi, N., J. Rodriguez, and P. Roy. 1998. A leucine zipper-like domain is essential for dimerization and encapsidation of bluetongue virus nucleocapsid protein VP4. J. Virol. 72:2983-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Royer, M., S. S. Hong, B. Gay, M. Cerutti, and P. Boulanger. 1992. Expression and extracellular release of human immunodeficiency virus type 1 Gag precursors by recombinant baculovirus-infected cells. J. Virol. 66:3230-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sagata, N., T. Yasunaga, J. Tsuzuku-Kawamura, K. Ohishi, Y. Ogawa, and Y. Ikawa. 1985. Complete nucleotide sequence of the genome of bovine leukemia virus: its evolutionary relationship to other retroviruses. Proc. Natl. Acad. Sci. USA 82:677-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sakurai, A., J. Yasuda, H. Takano, Y. Tanaka, M. Hatakeyama, and H. Shida. 2004. Regulation of human T-cell leukemia virus type 1 (HTLV-1) budding by ubiquitin ligase Nedd4. Microbes Infect. 6:150-156. [DOI] [PubMed] [Google Scholar]
- 60.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 61.Schmitt, A. P., G. P. Leser, E. Morita, W. I. Sundquist, and R. A. Lamb. 2005. Evidence for a new viral late-domain core sequence, FPIV, necessary for budding of a paramyxovirus. J. Virol. 79:2988-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schwartz, D. E., R. Tizard, and W. Gilbert. 1983. Nucleotide sequence of Rous sarcoma virus. Cell 32:853-869. [DOI] [PubMed] [Google Scholar]
- 63.Stapley, B. J., and T. P. Creamer. 1999. A survey of left-handed polyproline II helices. Protein Sci. 8:587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stoltz, M. A., C. F. van der Merwe, J. Coetzee, and H. Huismans. 1996. Subcellular localization of the nonstructural protein NS3 of African horsesickness virus. Onderstepoort J. Vet. Res. 63:57-61. [PubMed] [Google Scholar]
- 65.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689-699. [DOI] [PubMed] [Google Scholar]
- 67.Strack, B., A. Calistri, and H. G. Gottlinger. 2002. Late assembly domain function can exhibit context dependence and involves ubiquitin residues implicated in endocytosis. J. Virol. 76:5472-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thayer, R. M., M. D. Power, M. L. Bryant, M. B. Gardner, P. J. Barr, and P. A. Luciw. 1987. Sequence relationships of type D retroviruses which cause simian acquired immunodeficiency syndrome. Virology 157:317-329. [DOI] [PubMed] [Google Scholar]
- 69.Timmins, J., G. Schoehn, S. Ricard-Blum, S. Scianimanico, T. Vernet, R. W. Ruigrok, and W. Weissenhorn. 2003. Ebola virus matrix protein VP40 interaction with human cellular factors Tsg101 and Nedd4. J. Mol. Biol. 326:493-502. [DOI] [PubMed] [Google Scholar]
- 70.Vana, M. L., Y. Tang, A. Chen, G. Medina, C. Carter, and J. Leis. 2004. Role of Nedd4 and ubiquitination of Rous sarcoma virus Gag in budding of virus-like particles from cells. J. Virol. 78:13943-13953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Staden, V., M. A. Stoltz, and H. Huismans. 1995. Expression of nonstructural protein NS3 of African horsesickness virus (AHSV): evidence for a cytotoxic effect of NS3 in insect cells, and characterization of the gene products in AHSV infected Vero cells. Arch. Virol. 140:289-306. [DOI] [PubMed] [Google Scholar]
- 72.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]
- 74.Wang, H., N. J. Machesky, and L. M. Mansky. 2004. Both the PPPY and PTAP motifs are involved in human T-cell leukemia virus type 1 particle release. J. Virol. 78:1503-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wechsler, S. J., L. E. McHolland, and W. J. Tabachnick. 1989. Cell lines from Culicoides variipennis (Diptera: Ceratopogonidae) support replication of bluetongue virus. J. Invertebr. Pathol. 54:385-393. [DOI] [PubMed] [Google Scholar]
- 76.Whittington, S. J., B. W. Chellgren, V. M. Hermann, and T. P. Creamer. 2005. Urea promotes polyproline II helix formation: implications for protein denatured states. Biochemistry 44:6269-6275. [DOI] [PubMed] [Google Scholar]
- 77.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu, X., S. Y. Chen, H. Iwata, R. W. Compans, and P. Roy. 1992. Multiple glycoproteins synthesized by the smallest RNA segment (S10) of bluetongue virus. J. Virol. 66:7104-7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yasuda, J., E. Hunter, M. Nakao, and H. Shida. 2002. Functional involvement of a novel Nedd4-like ubiquitin ligase on retrovirus budding. EMBO Rep. 3:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yasuda, J., M. Nakao, Y. Kawaoka, and H. Shida. 2003. Nedd4 regulates egress of Ebola virus-like particles from host cells. J. Virol. 77:9987-9992. [DOI] [PMC free article] [PubMed] [Google Scholar]