Abstract
Enteroviruses are members of the family Picornaviridae that cause widespread infections in human and other mammalian populations. Enteroviruses are genetically and antigenically highly variable, and recombination within and between serotypes contributes to their genetic diversity. To investigate the dynamics of the recombination process, sequence phylogenies between three regions of the genome (VP4, VP1, and 3Dpol) were compared among species A and B enterovirus variants detected in a human population-based survey in Scotland between 2000 and 2001, along with contemporary virus isolates collected in the same geographical region. This analysis used novel bioinformatic methods to quantify phylogenetic compatibility and correlations with serotype assignments of evolutionary trees constructed for different regions of the enterovirus genome. Species B enteroviruses showed much more frequent, time-correlated recombination events than those found for species A, despite the equivalence in population sampling, concordant with a linkage analysis of previously characterized enterovirus sequences obtained over longer collection periods. An analysis of recombination among complete genome sequences by computation of a phylogenetic compatibility matrix (PCM) demonstrated sharply defined boundaries between the VP2/VP3/VP1 block and sequences to either side in phylogenetic compatibility. The PCM also revealed equivalent or frequently greater degrees of incompatibility between different parts within the nonstructural region (2A-3D), indicating the occurrence of extensive recombination events in the past evolution of this part of the genome. Together, these findings provide new insights into the dynamics of species A and B enterovirus recombination and evolution and into the contribution of structured sampling to documenting reservoirs, emergence, and spread of novel recombinant forms in human populations.
The genus Enterovirus in the family Picornaviridae is a group of nonenveloped RNA viruses that cause a wide range of diseases in humans and other mammals. The enterovirus genome is a single strand of positive-sense RNA of approximately 7,500 nucleotides (nt) comprising a long open reading frame flanked 5′ and 3′ by untranslated regions (UTRs). The encoded polyprotein is subsequently cleaved to produce structural (capsid proteins VP1 to VP4) and nonstructural (2A and 3D) proteins. Primary infection with an enterovirus leads to viral replication in the tissue around the gastrointestinal tract, followed by a transient viremia and sometimes migration into other tissues (2, 34). Although infection in immunocompetent individuals is often asymptomatic or causes mild febrile illness, a persistent or widely disseminated, systemic infection associated with severe disease outcomes is observed in highly immunosuppressed individuals and neonates (6, 13, 18).
Enteroviruses were originally classified as polioviruses, coxsackie type A or B viruses (CVA and CVB), or echoviruses (enteric cytopathic human orphan viruses), depending on the infectious properties of the virus, such as its pathogenicity in mice. From the 1960s on, enteroviruses within these groups were further differentiated into serotypes by the use of panels of specific neutralizing antisera, leading to the current total of 68 recognized human enterovirus (HEV) serotypes (polioviruses 1 to 3; CVA 1 to 22 and 24; CVB 1 to 6; echoviruses 1 to 7, 9, 11 to 21, 24 to 27, and 29 to 33; and enteroviruses (EV) 68 to 71, 73, 74 to 78, and 89 to 91). These fall into four main species by phylogenetic analysis (species A to D) (19), differing from each other by >40% nucleotide sequence divergence.
At any one time point, a range of different enterovirus serotypes circulate in human populations. The evolution of enteroviruses occurs through genetic drift, where particularly rapid sequence changes occur in the capsid region (3, 8, 25, 41) but also recombine between the capsid and nonstructural coding parts of the genome and the 5′ UTR (1, 7, 9, 14, 27, 29, 31, 36, 37). To date, documented examples of recombination have been limited to members of the same species (e.g., between species B serotypes), with the possible exception of the 5′ UTR, where only two genetic groups can be identified, each of which is associated with more than one species (37). In vitro, intraspecies recombinants are usually viable, although differences in pathogenicity between recombinant and parental viruses have been observed in newborn and adult mouse models (15, 16). The evolution of novel circulating recombinants of enteroviruses may therefore potentially lead to changes in disease associations or severity in naturally occurring enterovirus infections in humans.
In this study, we have compared the frequency and nature of recombination occurring within species A and B human enteroviruses circulating in a single geographical area over comparable sample collection periods. In addition to determining the dynamics of naturally occurring enterovirus recombination, we have developed a series of novel phylogenetic methods to map favored sites of recombination points among species A, B, and C complete genome sequences. These combined analyses provide new information on the tempo and mode of enterovirus evolution in vivo.
MATERIALS AND METHODS
Samples.
Scottish blood donor samples were obtained in a previous study, in which a highly sensitive reverse transcriptase PCR (RT-PCR) amplification method using highly conserved primers specific for the 5′ UTR was used to screen approximately 330,000 blood donation samples in pools of 95 between 2000 and 2001 (44). Viremic donors were identified by testing cross pools and individual donations; through the use of this procedure, no pools were found that contained more than one PCR-positive component donation (44). Samples found to be positive using the 5′ UTR RT-PCR assay were further investigated by amplification and sequencing of regions of the capsid (VP1) for serotype identification as previously described (44). The sequence of the 251-nt region of VP1 (positions 3090 to 3341, numbered relative to the poliovirus P3/Leon/37 strain; accession number K01392) was capable of reliably identifying the enterovirus serotype in each of the blood donor samples. Pairwise Jukes-Cantor-corrected distances between previously published sequences of different species A and B serotypes were always >0.31 and >0.295, respectively, while distances within serotypes were invariably below these thresholds. This distance-based method for serotype classification was used in addition to phylogenetic analysis to identify serotypes in blood donor samples and led to the identification of the following enterovirus serotypes: species A, CVA2 (n = 1), CVA5 (n = 2), CVA10 (n = 1), CVA16 (n = 5), and enterovirus 71 (n = 3); and species B, E9 (n = 1), E11 (n = 3), E13 (n = 1), E18 (n = 1), E30 (n = 4), CVB2 (n = 1), CVB3 (n = 1), CVB4 (n = 1), and CVB5 (n = 1).
A total of 32 clinical isolates of common enterovirus serotypes collected during the observation periods of 1997 and 2004 were obtained from Edinburgh (Regional Virus Laboratory, The Royal Infirmary of Edinburgh; n = 8), Glasgow (West of Scotland Specialist Virology Centre, Gartnavel General Hospital; n = 16), and Epsom (The Virology Laboratory, West Park Hospital, Epsom and St. Helier University Hospitals NHS Trust; n = 8), Scotland. Most isolates were serotyped by standard serological methods, and their designations were confirmed by nucleotide sequencing of the VP1 region as described above (Fig. 1 and 2, middle panels) (44).
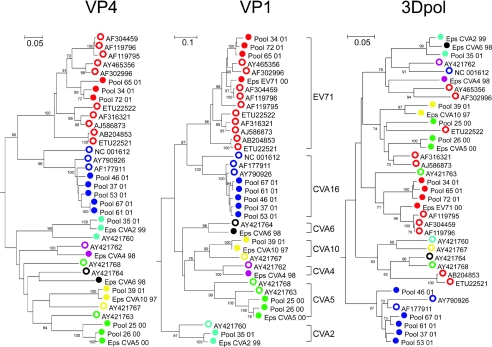
FIG. 1.
Phylogenetic analysis of sequences of VP4 (positions 744 to 962, numbered relative to the poliovirus P3/Leon/37 strain; accession number K01392), VP1 (positions 3046 to 3335), and 3Dpol (positions 6968 to 7151) for enterovirus A variants detected in survey samples (filled circles) and comparison with available complete genome sequences and matched VP4, VP1, and 3Dpol sequences (unfilled circles; accession numbers are shown). The names of blood donor samples are prefixed by “Pool,” followed by a unique number and sample year to differentiate them from clinical isolates (prefixed with “Edin,” “Glas,” and “Eps” [Edinburgh, Glasgow, and Epsom, respectively] to identify referral sources). Different colors were used to label serotypes. The tree was constructed by neighbor joining of 100 resampled bootstrap sets of Jukes-Cantor-corrected pairwise distances; branches present in ≥70% of trees are labeled with their bootstrap values.
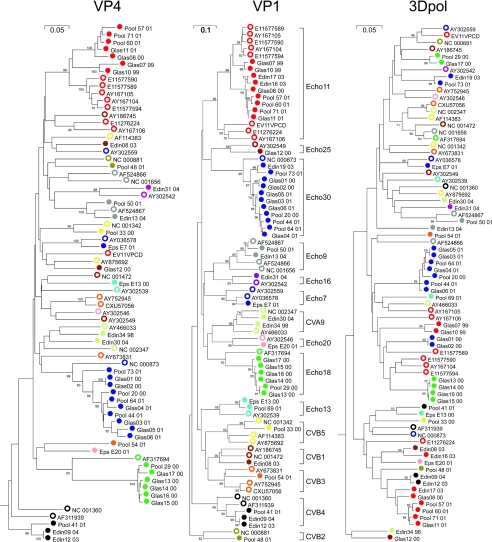
FIG. 2.
Phylogenetic analysis of species B variants in VP4, VP1, and 3Dpol regions (nucleotide positions are given in the legend to Fig. 1). Survey and published sequences were labeled and the tree was constructed as described in the legend to Fig. 1.
Amplification of 3Dpol region.
Pan-enterovirus primers were designed based on conserved sequences within the 3Dpol gene of HEV types A to D. Due to the proximity of the 3Dpol gene to the 3′ terminus of the enterovirus genome, reverse transcription was performed using (dT)18 or (dT)20V as a primer. First-round PCR was carried out with 3D outer sense and 3D outer antisense primers with the sequences TWG CHT TTG AYT ACW CNG GNT ATG A and TAY TCB TSY TCN CCR TTR TGC CA, respectively, for 30 cycles of 30 s at 94°C, 30 s at 50°C, and 50 s at 72°C. A second round of PCR was performed using inner sense (ATG ATH GCH TAT GGD GAY GAY GT) and antisense (TCR TGD ATB TCY TTC ATK GGC AT) primers with the same buffer and thermal cycling conditions. All 61 samples that were amplified and sequenced in VP1 were amplifiable in the 3Dpol region.
Amplification of VP4.
Pan-enterovirus primers were designed based on conserved sequences within the 5′ UTR and VP2 regions. Reverse transcription was performed using primer RT (CCR TCR TAR AA). First-round PCR was carried out with VP4 outer sense and VP4 antisense primers with the sequences GCG GAA CCG ACT ACT TTG GGT G and GGN ARC TTC CAC CAC CAN CC, respectively, using a “touchdown” thermal cycling program of 30 s at 94°C, 45 s at the annealing temperature, and 50 s at 72°C. The annealing temperatures used were 53°C for 5 cycles, 50°C for 5 cycles, 47°C for 5 cycles, and 42° for 25 cycles. Initial studies with a single degenerate antisense primer showed that a simple heminested PCR was not sensitive enough for amplification of this region. Therefore, several different inner antisense primers were designed, each targeted to a different group of human enteroviruses, as follows: for HEV-Ai, TCR CTR TAN CCA CAY GCT TC; for HEV-Aii, TCR CTR TAN CCA CAY GCC TC; for HEV-Bi, ACI TCN GGY TGI GTI GGY TGI TC; for HEV-Bii, ACH ACR TTI GCR CAY TCI TG; for HEV-Biii, TGI GTN GTI ATN GTI GAI TTN CC; for HEV-Ci, ACY CTR TCR CTI TAN CCI CAH GCC TC; and for HEV-Cii, ACY CTR TCR CTI TAN CCI CAH GCT TC. Each individual secondary PCR used 2 μl of primary product as a template and was done with the same buffer and thermal cycling conditions as the first round of PCR. Of the 61 samples that were amplified and sequenced in VP1, only 5 were not amplifiable in the VP4 region (Eps_EV71 00, Edin_16, Edin_17, Edin_19, and Pool_69).
Nucleotide sequencing.
Amplified DNAs from RT-PCR-positive pools were directly sequenced using a Thermo Sequenase cycle sequencing kit (USB) or a BigDye kit (ABI) and the inner sense or antisense primer used for amplification. Thermo Sequenase reactions incorporated 33P-labeled dideoxynucleoside triphosphates (Sigma), and reaction products were resolved by electrophoresis in 12% acrylamide gels (Sequagel; National Diagnostics), followed by autoradiography. BigDye reactions were sequenced by the University of Edinburgh automated sequencing service.
Sequence listings and phylogenetic analysis.
A full listing of the study samples, serotypes, and sampling/isolation dates and the accession numbers/loci of complete genome sequences of the reference isolates analyzed in Fig. 1 and 2 is available upon request. Complete genome sequences and partial sequences (i.e., paired VP1 and 3Dpol sequences that matched the regions sequenced in the current study) used for analyses of recombination frequencies (see Fig. 3 and 4) and complete genome sequences of enterovirus species A, B, and C investigated for phylogenetic compatibility (see Fig. 5 and 6) are also available upon request. For this analysis, all sequences selected were ≥2% divergent from each other. The outgroup for phylogenetic analysis was a majority (50%) consensus sequence generated from pairs of representative sequences of each species.
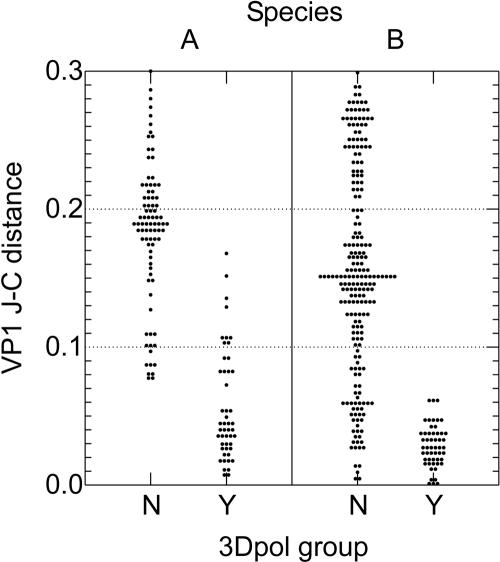
FIG. 3.
Relationship between 3Dpol recombination and divergence in VP1. Jukes-Cantor (J-C) distances were calculated by pairwise comparisons of VP1 sequences (y axis) of enterovirus A and B isolates of the same serotype and categorized into those with 3Dpol sequences in the same (Y) and different (N) genetic groups (x axis).
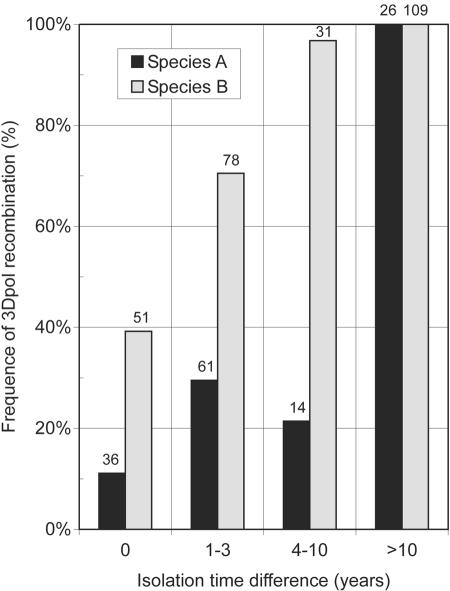
FIG. 4.
Timescale of 3Dpol recombination. The frequency of discordant 3Dpol genetic groupings (y axis) was compared to the time elapsed between the collection or isolation of samples or isolates of enterovirus species A and B (black and gray bars, respectively). Numbers above bars indicate the numbers of comparisons from which the percentages were calculated.
FIG. 5.
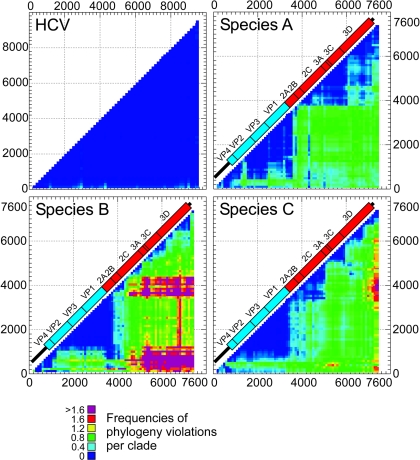
Phylogenetic compatibility matrices of sequence alignments of species A, B, and C enteroviruses and of HCV genotypes 1 to 6 (nonrecombinant control). The numbers of phylogeny violations required to order the trees generated from each combination of fragments (x and y axes) are recorded in color (see the legend), with phylogenetically compatible regions shown in deep blue. An enterovirus genome diagram drawn to scale has been superimposed to enable regions of phylogenetic compatibility to be matched to specific genome regions. Sequence fragments were numbered relative to the poliovirus P3/Leon/37 strain (accession number K01392).
FIG. 6.
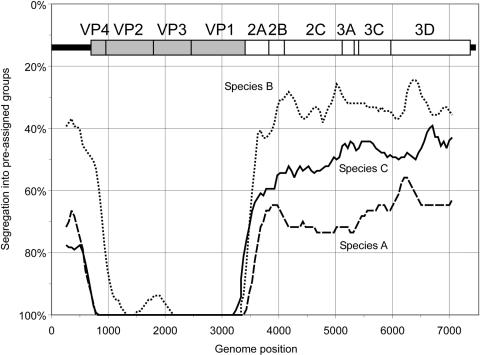
Correspondence between the phylogeny of sequential fragments throughout the enterovirus genome (midpoint recorded on x axis) and the assignment of species A, B, and C variants into serotypes. Correspondence between phylogeny and serotype is indicated by the concordance of the serotype assignment with phylogenetic ordering (see the text); a y axis value of 100% corresponds to trees where serotypes invariably segregated, while 0% corresponds to phylogenetic trees where serotypes were randomly distributed throughout the tree. An enterovirus genome diagram drawn to scale has been added to indicate positions where changes in segregation occurred. Sequence positions were numbered relative to the poliovirus P3/Leon/37 strain (accession number K01392).
The enterovirus nucleotide sequences obtained in this study were edited using the Simmonic 2005 version 1.4 sequence package (39; http://www.polio.vir.gla.ac.uk/software). Complete genome sequences were aligned by CLUSTALW with standard settings (5); for the coding region, amino acid sequence alignments were used to preserved codon boundaries in the nucleotide sequences. The phylogenetic trees shown in Fig. 1 and 2 were calculated from 2,000 repetitions of the Jukes-Cantor algorithm in the MEGA (molecular evolution genetic analysis) software package (21), with pairwise deletion for missing data. Pairwise P- and Jukes-Cantor-corrected distances between sequences and groups were calculated in the Simmonic package.
Statistical analysis.
All graphs (except that in Fig. 6, which was produced in Excel) were produced using Systat, version 9 (Systat Software Inc.).
Nucleotide sequence accession numbers.
The sequences obtained in this study have been submitted to GenBank and have been assigned the accession numbers DQ251288 to DQ251439.
RESULTS
Sequence relationships between VP4, VP1, and 3Dpol in enterovirus survey samples.
To investigate the frequency of recombination in species A and species B enterovirus genomes, we obtained nucleotide sequences from the VP4 and VP1 capsid regions and compared their phylogenies with those obtained from sequences collected at the other end of the genome for 3Dpol (Fig. 1 and 2). This analysis included all available published sequences matching the regions sequenced in the survey samples.
For each species A survey sample, the phylogenetic groupings into serotypes observed for VP1 were preserved for VP4 and 3Dpol (Fig. 1, filled circles), with the exception of the separate grouping of the CVA5 isolate Pool 25 (green) from Pool 26 and bCVA5 in the 3Dpol tree. However, in the case of the CVA6 (turquoise) and CVA2 (black) survey samples, which were genetically distinct in VP1, all three grouped together in the 3Dpol tree, which is evidence for a further recombination event. Far more frequent recombination was apparent among previously published sequences (unfilled circles). For example, published sequences of CVA16 (blue), EV71 (red), and CVA5 (green) each split into two or more phylogenetic groupings in the 3Dpol region and became interspersed with 3Dpol sequences from other serotypes.
Species B variants in the survey samples showed evidence of more frequent recombination events than species A samples (Fig. 2), not only between VP1 and 3Dpol but also between VP1 and VP4, for both published and survey isolates. As observed for species A viruses, the breakdown in VP1 sequence groupings occurred most commonly with published sequences, with some obvious examples being the scattering of Echo11 and CBV5 sequences (red and yellow unfilled circles) in the 3Dpol tree and some splitting of these serotype-associated groups in the VP4 tree. For the survey samples, there were several examples of some (but not all) VP1 sequence groups breaking down in 3Dpol, with examples including Echo11 variants (red filled circles) and Echo30 variants (blue filled circles).
The occurrence of these inferred recombination events showed a relationship with the degree of divergence of the VP1 sequence; thus, for Echo11, survey samples Gla07 and Glas10, which formed a separate subgroup within the Echo11 clade, were the ones that were distinct in the 3Dpol tree; similarly, the Echo30 outliers, Edin19/Pool 73 and Glas01/Glas02, were separate from each other and from the main group for 3Dpol. If sequence differences in VP1 are related to the time of divergence, then the occurrence of recombination may be a time-related phenomenon. This possibility is investigated below.
VP1 sequence divergence and time of isolation with recombination.
To formally test for an association between sequence divergence in VP1 and within-serotype recombination, comparisons of the survey samples and all published sequences of human enterovirus species A and B for which matched VP1 and 3Dpol sequences and dates of isolation were available (a full list is available upon request) were first classified according to their phylogenetic groupings within the 3Dpol tree. Pairwise Jukes-Cantor-corrected distances between 3Dpol sequences within species A and B serotypes fell into two distributions. Those with distances of <0.14 (species B) or <0.15 (species A) from each other invariably grouped together into one of the bootstrap-supported clades (examples shown in Fig. 1), while those with distances greater than these values grouped separately. The mean and range of pairwise distance values for the second distribution of these species A and B variants (0.26 ± 0.04 [mean ± 1 standard deviation] and 0.24 ± 0.04, respectively) were equivalent to those obtained by comparing 3Dpol sequences from different serotypes (0.24 ± 0.04 and 0.24 ± 0.04, respectively), which almost invariably showed distinct 3Dpol sequences from each other.
Comparisons of species B enterovirus variants with 3Dpol sequences from the same genetic group invariably showed very closely similar VP1 sequences (Fig. 3) (mean Jukes-Cantor distance, 0.029; range, 0 to 0.062), while those with different 3Dpol sequences (representing recombinant viruses) were usually, but not invariably, considerably more divergent in VP1 (mean distance, 0.156; range, 0.005 to 0.299). A different relationship between recombination and VP1 sequence divergence was found among species A sequences. There were several examples of comparisons between variants in the same 3Dpol genetic group that were nevertheless divergent in VP1, in many cases with distances of >0.1, well into the distribution of VP1 divergence values of recombinant viruses. Since the timing of collection of isolates was similar in this and previous studies, these observations provide evidence of different dynamics of recombination of species A and B enteroviruses.
This finding was consistent with a comparison of times of sample collection or isolation dates with frequencies of recombination (Fig. 4). For species B, approximately 40% of isolates isolated in the same calendar year showed different 3Dpol sequences. This value rose to 70% for enteroviruses isolated 2 to 3 years apart, while virtually all species B variants contained different 3Dpol sequences when the period was longer than 3 years. Recombination over time was noticeably lower for species A (Fig. 3). Only 10% of variants isolated in the same year were recombinant, and no more than 30% of variants were recombinant when isolated up to 10 years apart. Only when prototype isolates were compared with contemporary samples (>25 years of separation) was there evidence of universal recombination.
Mapping recombination by phylogenetic compatibility.
Mapping of the positions of recombination was carried out by phylogeny tree scanning, using a set of new computational methods that are based on recording the order of each variant in an alignment in a phylogenetic tree and the positions in the alignment where phylogenetic relationships change. For this analysis, complete genome sequences of human enteroviruses A and B were aligned (n = 28 and 61, respectively) (the sequences are available upon request). For comparison, a further set of poliovirus and nonpoliovirus species C complete genome sequences (n = 51) was included in the analysis. Boot-strapped phylogenetic trees were generated by the programs SEQBOOT, DNADIST, NEIGHBOR, and CONSENSE in the PHYLIP v3.62 package, produced by successively generated sequence fragments from alignments of complete genome sequences of each species.
To observe changes in phylogenetic relationships, the ordering of sequences in trees generated for each fragment was compared with those for all the other fragments to produce a phylogenetic compatibility matrix using the program TreeOrderScan in the Simmonic 2005 version 1.4 package (http://www.vir.gla.ac.uk/software). This program first produces optimally ordered trees in which the branching order is as closely matched as possible to that of the tree generated from a different fragment. Reordering of trees is achieved by the rotation of branches and through the movement of sequences or groups of sequences, provided that this occurs within groupings below the specified bootstrap threshold value (70% for all analyses shown in this study).
The compatibility of one tree with another is then computed by measurement of the number of times the phylogeny of one tree has to be violated (i.e., transfer of a sequence or group of sequences between different bootstrap-supported clades) to match the tree orders. For the analysis described in this study, a bootstrap value of 70% was used as the threshold for scoring phylogeny violations, as this is frequently used for assigning “robust” support for phylogenetic groups (17). Sequences can be assigned to predefined groups during the analysis (serotypes in the current study), and phylogenetic compatibility is thus computed separately for phylogeny violations between and within groups.
Different fragments in enterovirus genomes show different degrees of phylogenetic separation, and for the analysis shown, data were normalized by dividing the number of phylogeny violations by the smallest number of clades between the two trees. In extreme cases, where one or another tree being compared contains no bootstrap-supported clades, no phylogeny violation can be computed. Since trees for every fragment are matched to trees for every other fragment, inter- or intraserotype phylogeny violations can be displayed as a half-diagonal matrix, in which the values at each x and y coordinate record the number and genome positions of phylogeny violations for each pairwise comparison.
As a negative control, an alignment of hepatitis C virus (HCV) genotype 1 to 6 complete genome sequences was used, for which inter- and intragenotype recombination events are known to be extremely rare (38). The numbers of phylogeny violations between trees upon pairwise comparisons of sequence fragments of 300 bp with a bootstrap threshold of 70% were zero or low throughout the coding part of the genome (shaded dark blue in Fig. 5), while low degrees of phylogenetic incompatibility were occasionally observed between the relatively invariant 5′ UTR and the coding region.
Comparable analyses of alignments of enteroviruses were subsequently carried out on 76 sequentially generated fragments of aligned species A, B, and C sequences (300 bases in length, at increments of 100 bases; 70% bootstrap threshold value). For each species, the region of the genome encoding the capsid proteins VP2, VP3, and VP1 contained sequence fragments that produced trees that were phylogenetically compatible with each other (shaded blue) but incompatible with nonstructural genes and the 5′ UTR. These observations are consistent with the observation of discordant phylogenies between the VP1 and 3Dpol regions for species A and B (Fig. 1 and 2). There was also evidence of a lesser degree of incompatibility between VP2/VP3/VP1 and VP4 phylogenies for species B and C serotypes (shaded pale blue), again predicted from the specific examples of recombination in species B observed in Fig. 2.
The compatibility matrix provided evidence of extensive, major incompatibility between different regions within the nonstructural regions of all three species, indicating that recombination must occur frequently throughout this part of the genome. The greatest incompatibility scores were observed for species B, with >1.6 phylogeny violations per clade upon comparison of the 5′ UTR/VP4 and 2A/2B regions with 3A-3D. Species B also showed no compatibility between fragments separated by more than 300 to 400 bases, except for the VP2/VP3/VP1 block described above. The frequency of recombination in species B was shown to be higher than that in species C or A, as these last viruses showed much lower mean incompatibility scores and frequent phylogenetic compatibility between regions outside VP2/VP3/VP1 (such as within 3A/3B/3C for species C and 2A-3C for species A).
The observed differences in frequencies of phylogeny violations between species were unrelated to the degree of diversity between sequences. Mean pairwise distances between serotypes within species A, B, and C were comparable for both structural regions and nonstructural regions (for one structural region [positions 744 to 3377], the distances were 0.346, 0.340, and 0.340 for species A, B, and C, respectively; for one nonstructural region [positions 3378 to 7361], the distances were 0.225, 0.223, and 0.193, respectively). These distances are, in turn, similar to those between HCV genotypes (0.31 between genotypes) for the control sequences, where consistently low or zero frequencies of phylogeny violations were observed.
Phylogenetically informative regions in enterovirus genomes.
Regions in the genome where nucleotide sequence phylogeny was congruent with the neutralization characteristics of different enterovirus serotypes were determined through application of a related tree-scoring technique. Enterovirus variants were uniquely labeled according to their designated serotypes, and phylogenetic trees were constructed from sequentially generated fragments as described above. The correspondence in the order of sequences in the tree with their serotype designations was measured by counting the number of serotype label transitions between sequences in the list order for the tree. Expected values for a perfectly segregated tree where sequences grouped by serotype would therefore correspond to one less than the number of serotypes in the data set, while the number for a tree where there was no relationship between serotype and phylogeny (i.e., serotypes randomly distributed in the tree) would lead to a much larger number of label transitions, depending on and computable from the number of assigned groups and the number of members within each. Actual segregation values from the species A, B, and C datasets can be plotted as values from 0% to 100% within the scale corresponding to these opposed outcomes (Fig. 6).
For the region from nucleotide positions 800 to 1000 to approximately 3500 in the alignment, sequences almost invariably segregated according to their assigned serotype, with segregation scores at or close to 100%. For either side of this structural gene region, there were sharp transitions into almost completely random or serotype-incompatible tree orders within each species, leading to the observed low segregation values. The 5′ transition point in species B sequences was approximately 300 bases downstream of that for species A and C and likely reflects frequent recombination events between VP4 and VP2 that break the linkage of VP4 with the variant's serotype designation. All three species showed similar transition points at the 3′ end of the structural region (nt 3300 to 3500). In general, transition points corresponded closely to the positions in the compatibility matrices where phylogenies changed from compatibility to incompatibility on either side of the capsid region (Fig. 5).
DISCUSSION
Species-associated differences in recombination frequency.
For this study, we used a population-based sampling method to identify and characterize enterovirus infections in the adult community in the United Kingdom; infections identified in blood donors through large-scale screening were obtained independently of any clinical symptoms and/or medical referral. In general, the common species A and B enteroviruses amplified from blood donors were closely similar to and phylogenetically interspersed with those from clinical isolates in the VP4, VP1, and 3Dpol genomic regions (Fig. 1 and 2). Thus, there is no evidence that enterovirus variants infecting blood donors are systematically different genetically from those associated with symptomatic infections. However, while similar numbers of species A and B infections (n = 12 and 14, respectively) were found in blood donors throughout the study period, species B viruses were predominant among typed isolations reported centrally (Scottish Centre for Infection and Environmental Health [43]) and therefore were unrepresentative numerically of enterovirus infections in the community.
Despite the equivalent sampling times and populations studied, sequence comparisons of different genomic regions of species A and B viruses, along with those of contemporary U.K. isolates matching serotypes found in blood donors, showed substantial differences in the frequencies of discordant phylogenetic relationships (Fig. 1 and 2). Whereas genetic linkage between VP4, VP1, and 3Dpol was preserved in almost all species A variants in our survey samples, there were frequent examples of recombination between VP1, 3Dpol, and VP4 in species B variants.
The existence of differences in the frequency of recombination between species was examined by an extended comparison of sequences that included all available complete genome sequences of species A and B serotypes along with all available paired VP1 and 3Dpol sequences available from public databases that matched the regions sequenced in this study. For species A variants, there was an almost entirely separate distribution of VP1 pairwise distances between viruses of the same serotype with shared 3Dpol sequences and those in which recombination had occurred (Fig. 3). Using the rate of sequence change in VP1 of 1.35 × 10−2 per site per year determined for EV71 (3) (comparable to rates measured for other human enterovirus species [25, 41]), the 0.10 to 0.12 threshold that divides most recombinant and nonrecombinant viruses corresponds to a divergence time of approximately 6 to 8 years, over which period recombination would have occurred in one or both descendants. The apparent long-term stability of circulating species A serotypes is consistent with the low frequency of recombination between variants isolated within 10 years of each other (11% to 29%).
For species B viruses, rates of recombination for the extended data set differed substantially, with no examples of virus pairs with >0.065 sequence divergence being nonrecombinant between VP1 and 3Dpol. Indeed, there were examples of recombination events occurring in variants with VP1 distances as low as 0.005 to 0.015, with divergence times therefore measurable in terms of weeks or months rather than years. Similarly, using dates of isolation (Fig. 4), approximately 80% of isolates obtained 1 to 3 years apart were recombinant, rising to close to 100% between years 4 and 10.
Although the phylogenetic compatibility matrix (PCM; Fig. 5) was developed to map positions of recombination, the computed output (phylogeny violations per clade) provides an independent, absolute value with which to compare rates of recombination between data sets. Marked differences in frequencies of violations of phylogeny were observed between species, consistent with the previously described analyses of paired VP1 and 3Dpol sequences. While species C variants appeared intermediate in recombination frequency between species A and B, this figure may be biased by sampling differences; the interruption in the circulation of wild-type polioviruses through global immunization has led to several biases in the representation of sequences that potentially bias the analysis of recombination frequency. These include an overrepresentation of vaccine-derived isolates among available complete genome sequences of polioviruses (sequences are available upon request) and differences in isolation times between wild-type and vaccine-related polioviruses and other species C serotypes.
Sites of recombination.
By exhaustive comparisons of fragment sets generated from alignments of species A, B, and C complete genome sequences, it was possible to map regions of phylogenetic incongruity and thus infer sites of favored recombination using the PCM. This is a more global approach to previously used methods to map recombination, such as SIMPLOT, in which phylogenetic groupings or sequence similarities of individual sequences to a set of reference sequences are determined (e.g., see references 1 and 27). Use of the PCM is particularly of value for data sets where the identities of the parental sequences involved in recombination are unknown (i.e., for each phylogenetic incompatibility, it is generally unknown which enterovirus sequence is the recombinant and which is nonrecombinant).
Analyses of species A, B, and C by the PCM identified a large phylogenetically compatible region between nucleotide positions 1000 to 1200 and 3600 to 3800 in each, with zero or close to zero frequencies of phylogeny violations. This observation is consistent with previous analyses demonstrating a virtual absence of recombination in P1 or the VP2/VP3/VP1 section of the capsid-encoding region (9, 30, 36, 37). While the incompatibility of the capsid region with the nonstructural region and the 5′ UTR was expected from this and previous analyses of recombination in enteroviruses (1, 9, 22, 23, 30, 36, 37), the high degree of phylogenetic incompatibility throughout the nonstructural regions of all three species revealed by the PCM analysis (Fig. 5) indicates a much more extensive series of recombination events than previously suspected. Indeed, among species B sequences, the 2C-3D 3′ end of the genome shows greater incompatibility with 2A/2B and the 5′ UTR/VP4 regions than with the VP2/VP3/VP1 block.
Parallel analyses of species C and A viruses showed larger areas of compatibility within the nonstructural region, particularly in species A, where a block from 2B to 3C showed little evidence of intraregion recombination. While it is possible that these viruses differ functionally from species B in the compatibility constraints operating within and between proteins, it is also possible (and more feasible biologically) that the appearance of these phylogenetically congruent regions is simply the consequence of the relatively fewer recombination events occurring among the available complete genome sequences used in the analysis. As exemplified by the species B data, each nonstructural protein or part of a protein may be functionally interchangeable with any other variant within each species. The findings from the PCM analysis provide little support for the existence of specifically favored sites of recombination in enteroviruses. Sites of RNA secondary structure formation have been previously shown to promote recombination in copy-choice mechanisms of RNA recombination (20, 35, 42), in contrast to premature termination (32) and nonreplicative models (10, 11). However, there is no evidence from the PCM that recombination was specifically favored at sites of RNA structure, such as the conserved cis-replicating element (alignment positions 4439 to 4493; Fig. 5) (12).
The complete breakdown of genetic linkage throughout the nonstructural region (even between fragments separated by as little 300 to 400 bases for species B) is more compatible with and accounts for the previously observed variability in the positions of mapped breakpoints in comparisons of individual recombinant viruses. Examples of previously determined recombination points include 2C in Echo 9/18 (1), 2B in CVB4 (22), 2A in EV11/EV19 (24), and in larger analyses, several positions in P2 and P3 among naturally occurring enteroviruses and poliovirus vaccine strains (9, 37). Although the nonstructural regions of enteroviruses show less sequence diversity than the capsid gene, the existence of such extensive recombination supports the idea that this section of the genome may be evolutionarily much older, but more constrained in sequence space, than genes encoding structural proteins.
Alternatively, it is possible that recombination within the capsid region, particularly between serotypes with different receptor use, is less likely to generate viable offspring than recombination elsewhere in the genome. The poor or absent replication ability of artificially generated recombinant polioviruses with mosaic capsids derived from different serotypes (4, 26, 40) provides some evidence that functional constraints limit recombination in this region to a greater extent than elsewhere in the genome (where artificially generated recombinants are generally viable). These may arise through structural incompatibilities between capsid proteins from different serotypes in protein-protein interactions during virus assembly or maturation. For enterovirus serotypes that use different cellular receptors, recombinant viruses in the capsid region may produce mosaic capsids that fail to create correct binding structures to allow cell attachment and/or entry.
Sources of enterovirus diversity.
One of the peculiar aspects of enterovirus recombination is the asymmetry between structural and nonstructural coding regions. A relatively constant range of serotypes has continued to circulate in human populations, often episodically, throughout the world over the past 40 years. However, the accumulated set of distinct genetic lineages in the nonstructural region (particularly well documented for 3Dpol) is far greater than the number of serotypes (several hundred in the case of species B), and their occurrence in human enterovirus populations is far more transitory. For example, none of the 3Dpol sequences of the prototype enterovirus strains isolated in the 1960s and 1970s have ever since reappeared in subsequent enterovirus isolates, either in association with the original or in association with other serotypes within the species (23, 29, 30, 37). With some documented exceptions (e.g. see reference 29), the evolution of enterovirus serotypes over time generally comprises gradual drift, diversification of the capsid-encoding region, and episodic exchange of the entire downstream region with an apparently unending series of phylogenetically distinct, novel nonstructural sequence variants.
Since the original differentiation of nonstructural region sequences into the large number of clades observed for 3Dpol likely occurred over an equivalent or greater timescale than the evolution of different serotypes, the observation of continuous stepwise changes in 3Dpol sequences over time within each serotype indicates that the viral reservoir where these recombination events occur is much more extensive and diverse than the pool of human enteroviruses sampled to date. In focusing on other potential sources of enterovirus infections, it is intriguing that the recently discovered species A serotypes EV76, EV89, EV90, and EV91, isolated from Bangladesh, are closely related to a series of enteroviruses isolated from macaques (28, 33). It was indeed speculated that contact between humans and this species of Asian monkey in Bangladesh (and potentially elsewhere in the macaque range) facilitated the spread of these simian viruses into humans (28).
Given this occurrence, it is therefore tempting to imagine other reservoirs for species A serotypes, as well as for other enterovirus species where large-scale asymmetric recombination has been shown to occur. The markedly greater frequency of recombination of species B enteroviruses observed in this study may indeed originate through more frequent exchange with external genetic reservoirs of diversity. In future work, it will be of value to carry out more structured sampling of enterovirus variants in human populations over defined sampling windows in several different geographical regions in parallel. Documenting the dynamics and geographical extent of the spread of individual recombinant forms would provide valuable new information on the sources of enterovirus gene diversity and the relationship between the emergence of new forms and changes in pathogenicity.
Acknowledgments
Clinical isolates were kindly provided by Kirstine Eastick (Specialist Virology Centre, Edinburgh Royal Infirmary), Bill Carman and Ann Smith (West of Scotland Specialist Virology Centre, Gartnavel Hospital, Glasgow, Scotland), and Justin Bendig (Virology Laboratory, West Park Hospital, Epsom, Scotland, and St. Helier University Hospitals NHS Trust).
J.B.W. was supported by a grant from Baxter Healthcare.
REFERENCES
- 1.Andersson, P., K. Edman, and A. M. Lindberg. 2002. Molecular analysis of the echovirus 18 prototype: evidence of interserotypic recombination with echovirus 9. Virus Res. 85:71-83. [DOI] [PubMed] [Google Scholar]
- 2.Bodian, M. D. and D. M. Horstmann. 1965. Polioviruses, p. 430-473. In F. L. Horsfall and I. Tamm (ed.), Vital and rickettsial infections of man, 4th ed. Pitman Lippincott, London, United Kingdom.
- 3.Brown, B. A., M. S. Oberste, J. P. Alexander, Jr., M. L. Kennett, and M. A. Pallansch. 1999. Molecular epidemiology and evolution of enterovirus 71 strains isolated from 1970 to 1998. J. Virol. 73:9969-9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burke, K. L., G. Dunn, M. Ferguson, P. D. Minor, and J. W. Almond. 1988. Antigen chimaeras of poliovirus as potential new vaccines. Nature 332:81-82. [DOI] [PubMed] [Google Scholar]
- 5.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherry, J. D. 1998. Enteroviruses: coxsackieviruses, echoviruses, and polioviruses, p. 1787-1839. In R. D. Feigin and J. D. Cherry (ed.), Textbook of pediatric infectious diseases, 2nd ed. Saunders, London, United Kingdom.
- 7.Chevaliez, S., A. Szendroi, V. Caro, J. Balanant, S. Guillot, G. Berencsi, and F. Delpeyroux. 2004. Molecular comparison of echovirus 11 strains circulating in Europe during an epidemic of multisystem hemorrhagic disease of infants indicates that evolution generally occurs by recombination. Virology 325:56-70. [DOI] [PubMed] [Google Scholar]
- 8.Chua, B. H., P. C. McMinn, S. K. Lam, and K. B. Chua. 2001. Comparison of the complete nucleotide sequences of echovirus 7 strain UMMC and the prototype (Wallace) strain demonstrates significant genetic drift over time. J. Gen. Virol. 82:2629-2639. [DOI] [PubMed] [Google Scholar]
- 9.Cuervo, N. S., S. Guillot, N. Romanenkova, M. Combiescu, A. Aubert-Combiescu, M. Seghier, V. Caro, R. Crainic, and F. Delpeyroux. 2001. Genomic features of intertypic recombinant Sabin poliovirus strains excreted by primary vaccinees. J. Virol. 75:5740-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gmyl, A. P., E. V. Belousov, S. V. Maslova, E. V. Khitrina, A. B. Chetverin, and V. I. Agol. 1999. Nonreplicative RNA recombination in poliovirus. J. Virol. 73:8958-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gmyl, A. P., S. A. Korshenko, E. V. Belousov, E. V. Khitrina, and V. I. Agol. 2003. Nonreplicative homologous RNA recombination: promiscuous joining of RNA pieces? RNA 9:1221-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodfellow, I., Y. Chaudhry, A. Richardson, J. Meredith, J. W. Almond, W. Barclay, and D. J. Evans. 2000. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 74:4590-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grist, N. R., E. J. Bell, and F. Assaad. 1978. Enteroviruses in human disease. Prog. Med. Virol. 24:114-157. [PubMed] [Google Scholar]
- 14.Guillot, S., V. Caro, N. Cuervo, E. Korotkova, M. Combiescu, A. Persu, A. Aubert-Combiescu, F. Delpeyroux, and R. Crainic. 2000. Natural genetic exchanges between vaccine and wild poliovirus strains in humans. J. Virol. 74:8434-8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harvala, H., H. Kalimo, J. Bergelson, G. Stanway, and T. Hyypia. 2005. Tissue tropism of recombinant coxsackieviruses in an adult mouse model. J. Gen. Virol. 86:1897-1907. [DOI] [PubMed] [Google Scholar]
- 16.Harvala, H., H. Kalimo, L. Dahllund, J. Santti, P. Hughes, T. Hyypia, and G. Stanway. 2002. Mapping of tissue tropism determinants in coxsackievirus genomes. J. Gen. Virol. 83:1697-1706. [DOI] [PubMed] [Google Scholar]
- 17.Hillis, D. M., and J. J. Bull. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 42:182-192. [Google Scholar]
- 18.Kaplan, M. H., S. W. Klein, J. McPhee, and R. G. Harper. 1983. Group B coxsackievirus infections in infants younger than three months of age: a serious childhood illness. Rev. Infect. Dis. 5:1019-1032. [DOI] [PubMed] [Google Scholar]
- 19.King, A. M. Q., F. Brown, P. Christian, T. Hovi, T. Hyypiä, N. J. Knowles, S. M. Lemon, P. D. Minor, A. C. Palmenberg, T. Skern, and G. Stanway. 2000. Picornaviridae, p. 657-683. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, D. J. McGeoch, J. Maniloff, M. A. Mayo, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses, 7th report of the International Committee on Taxonomy of Viruses. San Diego Academic Press, San Diego, Calif.
- 20.Kirkegaard, K., and D. Baltimore. 1986. The mechanism of RNA recombination in poliovirus. Cell 47:433-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 22.Lindberg, A. M., P. Andersson, C. Savolainen, M. N. Mulders, and T. Hovi. 2003. Evolution of the genome of human enterovirus B: incongruence between phylogenies of the VP1 and 3CD regions indicates frequent recombination within the species. J. Gen. Virol. 84:1223-1235. [DOI] [PubMed] [Google Scholar]
- 23.Lukashev, A. N., V. A. Lashkevich, O. E. Ivanova, G. A. Koroleva, A. E. Hinkkanen, and J. Ilonen. 2003. Recombination in circulating enteroviruses. J. Virol. 77:10423-10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lukashev, A. N., V. A. Lashkevich, G. A. Koroleva, J. Ilonen, and A. E. Hinkkanen. 2004. Recombination in uveitis-causing enterovirus strains. J. Gen. Virol. 85:463-470. [DOI] [PubMed] [Google Scholar]
- 25.Martin, J., G. Dunn, R. Hull, V. Patel, and P. D. Minor. 2000. Evolution of the Sabin strain of type 3 poliovirus in an immunodeficient patient during the entire 637-day period of virus excretion. J. Virol. 74:3001-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murdin, A. D., H. H. Lu, M. G. Murray, and E. Wimmer. 1992. Poliovirus antigenic hybrids simultaneously expressing antigenic determinants from all three serotypes. J. Gen. Virol. 73:607-611. [DOI] [PubMed] [Google Scholar]
- 27.Norder, H., L. Bjerregaard, and L. O. Magnius. 2002. Open reading frame sequence of an Asian enterovirus 73 strain reveals that the prototype from California is recombinant. J. Gen. Virol. 83:1721-1728. [DOI] [PubMed] [Google Scholar]
- 28.Oberste, M. S., K. Maher, and M. A. Pallansch. 2002. Molecular phylogeny and proposed classification of the simian picornaviruses. J. Virol. 76:1244-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oberste, M. S., S. Penaranda, K. Maher, and M. A. Pallansch. 2004. Complete genome sequences of all members of the species human enterovirus A. J. Gen. Virol. 85:1597-1607. [DOI] [PubMed] [Google Scholar]
- 30.Oberste, M. S., S. Penaranda, and M. A. Pallansch. 2004. RNA recombination plays a major role in genomic change during circulation of coxsackie B viruses. J. Virol. 78:2948-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oprisan, G., M. Combiescu, S. Guillot, V. Caro, A. Combiescu, F. Delpeyroux, and R. Crainic. 2002. Natural genetic recombination between co-circulating heterotypic enteroviruses. J. Gen. Virol. 83:2193-2200. [DOI] [PubMed] [Google Scholar]
- 32.Pierangeli, A., M. Bucci, M. Forzan, P. Pagnotti, M. Equestre, and B. R. Perez. 1998. Identification of an alternative open reading frame (“hidden gene?”) stringently required for infectivity of poliovirus cDNA clones. New Microbiol. 21:309-320. [PubMed] [Google Scholar]
- 33.Poyry, T., L. Kinnunen, T. Hovi, and T. Hyypia. 1999. Relationships between simian and human enteroviruses. J. Gen. Virol. 80:635-638. [DOI] [PubMed] [Google Scholar]
- 34.Reetoo, K. N., S. A. Osman, S. J. Illavia, C. L. Cameron-Wilson, J. E. Banatvala, and P. Muir. 2000. Quantitative analysis of viral RNA kinetics in coxsackievirus B3-induced murine myocarditis: biphasic pattern of clearance following acute infection, with persistence of residual viral RNA throughout and beyond the inflammatory phase of disease. J. Gen. Virol. 81:2755-2762. [DOI] [PubMed] [Google Scholar]
- 35.Romanova, L. I., V. M. Blinov, E. A. Tolskaya, E. G. Viktorova, M. S. Kolesnikova, E. A. Guseva, and V. I. Agol. 1986. The primary structure of crossover regions of intertypic poliovirus recombinants: a model of recombination between RNA genomes. Virology 155:202-213. [DOI] [PubMed] [Google Scholar]
- 36.Santti, J., H. Harvala, L. Kinnunen, and T. Hyypia. 2000. Molecular epidemiology and evolution of coxsackievirus A9. J. Gen. Virol. 81:1361-1372. [DOI] [PubMed] [Google Scholar]
- 37.Santti, J., T. Hyypia, L. Kinnunen, and M. Salminen. 1999. Evidence of recombination among enteroviruses. J. Virol. 73:8741-8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simmonds, P. 2004. Genetic diversity and evolution of hepatitis C virus—15 years on. J. Gen. Virol. 85:3173-3188. [DOI] [PubMed] [Google Scholar]
- 39.Simmonds, P., and D. B. Smith. 1999. Structural constraints on RNA virus evolution. J. Virol. 73:5787-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stanway, G., P. J. Hughes, G. D. Westrop, D. M. Evans, G. Dunn, P. D. Minor, G. C. Schild, and J. W. Almond. 1986. Construction of poliovirus intertypic recombinants by use of cDNA. J. Virol. 57:1187-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takeda, N., M. Tanimura, and K. Miyamura. 1994. Molecular evolution of the major capsid protein VP1 of enterovirus 70. J. Virol. 68:854-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tolskaya, E. A., L. I. Romanova, V. M. Blinov, E. G. Viktorova, A. N. Sinyakov, M. S. Kolesnikova, and V. I. Agol. 1987. Studies on the recombination between RNA genomes of poliovirus: the primary structure and nonrandom distribution of crossover regions in the genomes of intertypic poliovirus recombinants. Virology 161:54-61. [DOI] [PubMed] [Google Scholar]
- 43.Welch, J. B., K. Maclaran, T. Jordan, and P. N. Simmonds. 2003. Frequency, viral loads, and serotype identification of enterovirus infections in Scottish blood donors. Transfusion 43:1060-1066. [DOI] [PubMed] [Google Scholar]
- 44.Welch, J. B., K. McGowan, B. Searle, J. Gillon, L. M. Jarvis, and P. Simmonds. 2001. Detection of enterovirus viraemia in blood donors. Vox Sang. 80:211-215. [DOI] [PubMed] [Google Scholar]