Abstract
Transport of capsids in cells is critical to alphaherpesvirus infection and pathogenesis; however, viral factors required for transport have yet to be identified. Here we provide a detailed examination of capsid dynamics during the egress phase of infection in Vero cells infected with pseudorabies virus. We demonstrate that the VP1/2 tegument protein is required for processive microtubule-based transport of capsids in the cytoplasm. A second tegument protein that binds to VP1/2, UL37, was necessary for wild-type transport but was not essential for this process. Both proteins were also required for efficient nuclear egress of capsids to the cytoplasm.
Viruses must overcome the diffusion barrier of the cytoplasm to effectively replicate in mammalian cells. This is most dramatically exemplified with neurotropic infections, such as those of the alphaherpesviruses, during which virus particles may translocate several centimeters or more between axon terminals and neuronal cell bodies. Intracellular transport of alphaherpesvirus particles to the nucleus in both neurons and non-neuronal cells is dependent on microtubules (20, 24, 27, 40).
The alphaherpesvirus virion is composed of four structural elements. The viral genome consists of a linear double-stranded DNA (ca. 120 to 230 kbp) that is housed within a proteinaceous capsid having icosahedral symmetry (∼120-nm diameter). The capsid is enclosed within a host-derived lipid envelope, and between the capsid and the envelope is a collection of viral proteins collectively referred to as the tegument. Upon entry into a cell the viral envelope fuses with the cellular plasma or endosomal membrane, depositing the capsid and tegument into the cytosol (10, 28, 29). At this phase, many tegument proteins are removed from the capsid. However, at least three tegument proteins (VP1/2, UL37, and US3) remain associated with capsids as they travel toward the nucleus (12, 23). After replication and assembly of capsids in the nucleus, progeny capsids translocate to the cytosol where they are again found associated with the VP1/2, UL37, and US3 tegument proteins (8, 13). These capsid/tegument complexes ultimately bud into a component of the secretory pathway and egress from the cell (reviewed in reference 26). The dynamics of capsid transport and assembly in the cytoplasm are poorly understood.
Although many alphaherpesvirus proteins can interact with cellular microtubule-based motors, no herpesvirus proteins are currently known to be required for capsid transport (3, 4, 6, 19, 25, 31, 47). The presence of VP1/2, UL37, and US3 on cytosolic capsids makes them prime candidates as effectors of intracellular capsid transport. Of particular interest to the present study, cells infected with viruses lacking either VP1/2 or UL37 assemble genome-containing capsids in the nucleus, and these capsids egress to the cytoplasm similar to capsids of wild-type viruses. However, once in the cytoplasm, unenveloped capsids lacking either VP1/2 or UL37 accumulate, and re-envelopment is rare or nonexistent (1, 2, 9, 16, 17). One possible explanation of this accumulation is a failure of capsids to translocate to the site of secondary envelopment within the cytoplasm. We therefore set out to characterize capsid dynamics during the egress phase of infection and to determine whether either VP1/2 or UL37 is necessary for intracellular capsid transport.
MATERIALS AND METHODS
Cells.
Derivatives of pseudorabies virus (PRV) strain Becker (PRV-GS443, PRV-GS678R, and PRV-GS993R; see below) were propagated in pig kidney epithelial cells (PK15). PK15 cells were also used to determine virus titers and for single-step growth curve analysis as previously described (41). African green monkey kidney epithelial cells (Vero) were used for live cell imaging, as they had a flatter morphology than PK15 cells. Infections of Vero cells were performed at a multiplicity of infection (MOI) of ≤0.1, which helped to keep infections consistent and minimized cell rounding. Disruption of microtubules or actin filaments in Vero cells was achieved with 10-μm nocodazole or 0.5-μm cytochalasin D, respectively. To deplete cellular ATP, Vero cells were treated with 20 mM sodium azide in glucose-free medium.
To grow the ΔUL36 virus (PRV-GS678; see below), PK15 cells were stably transfected with a fragment of the PRV genome that included the entire UL36 open reading frame (ORF) and neighboring sequences (598 nucleotides [nt] upstream and 798 nt downstream) containing portions of the UL35 and UL37 genes. The UL36-containing fragment was cloned into a neomycin phosphotransferase-encoding plasmid into which a monomeric red-fluorescent protein (mRFP1)-expression cassette was subsequently inserted, resulting in pGS901. Red-fluorescent cells were isolated by limiting dilution and subsequent expansion in the presence of 0.5 mg of G418/ml. An isolate that expressed heterogeneous levels of red fluorescence provided the best-complemented growth of PRV-GS678 and was saved as PK15-UL36 cells.
A UL37-complementing cell line was isolated to efficiently propagate the ΔUL37 virus (PRV-GS993; see below). The entire UL37 ORF and downstream 903 nt (which includes a portion of the UL36 gene) was subcloned into the cytomegalovirus immediate-early expression cassette of the pLPCX retroviral vector (Clontech), resulting in pGS1324. Cotransfection of pGS1324 and pVSV-G into a retroviral packaging cell line and subsequent isolation of retroviral particles were done as recommended by the manufacturer (Clontech). The resulting retrovirus was used to transduce PK15 cells, which were then selected with 1 μg of puromycin/ml. The resulting cell population was saved as PK15-UL37 cells.
Viruses.
Derivatives of PRV-Becker encoding either egfp or mrfp1 fused in-frame to the UL35 capsid gene, PRV-GS443 and PRV-GS847, respectively, were previously described (37, 38). Deletions of the UL36 and UL37 genes were made by replacing each respective coding sequence with a kanamycin resistance cassette flanked by Flp recombination target (FRT) sites, followed by removal of the kanamycin cassette by Flp mediated recombination.
To make the ΔUL36 virus, the pGS443 infectious clone was transformed into the EL250 E. coli strain, resulting in strain GS575 (21). A kanamycin cassette was amplified by PCR from the pUCK4 plasmid (Pharmacia) using the following primers: 5′-AAAGATTTTTCCCCCACGCGCGTGTGTTATTTCAGCCATGTAAGAAGTTCCTATACTTTCTAGAGAATAGGAACTTCCCAGTCACGACGTTGTAAAACG and 5′-AGTCCTCCCGGGTGGGCCAGACGCGGCCGAGGCCGCCCAGGAAGTTCCTATTCTCTAGAAAGTATAGGAACTTCGAAACAGCTATGACCATGATTACG. Each primer encodes a single FRT site (underlined) and 3′ homology to the pUC4K template. The 5′ portions of the primers are homologous to the PRV UL36 gene (−37 nt to +3 nt and +8911 nt to +8950 nt relative to the UL36 ORF, respectively). The first primer also encodes a stop codon (in boldface) fused downstream of the UL36 ATG initiation codon. The resulting PCR product was transformed into GS575 that was first induced to express the RED proteins (exo, beta, and gam), and recombinants were selected with 20 μg of chloramphenicol and 50 μg of kanamycin/ml (21, 48). One recombinant, GS674, was confirmed by restriction digest analysis. Arabinose-induced expression of Flp recombinase was used to remove the kanamycin cassette, resulting in GS678 (Fig. 1). Sequencing of the pGS678 infectious clone plasmid confirmed the presence of a TAA stop codon and single FRT site in place of nucleotides 4 to 8910 of the UL36 ORF. The resulting allele has a stop codon fused immediately downstream of the endogenous UL36 start codon and is deleted for the subsequent 2969 codons (the product of the UL36 ORF, VP1/2, is predicted to be a 3084-amino-acid protein). Transfection of pGS575 into PK15 cells resulted in the green fluorescent protein (GFP)-capsid virus (PRV-GS575). Transfection of pGS678 into PK15-UL36 cells resulted in the ΔUL36 GFP-capsid virus (PRV-GS678). Transfection of infectious clones by electroporation into PK15 cells was performed as previously described (23).
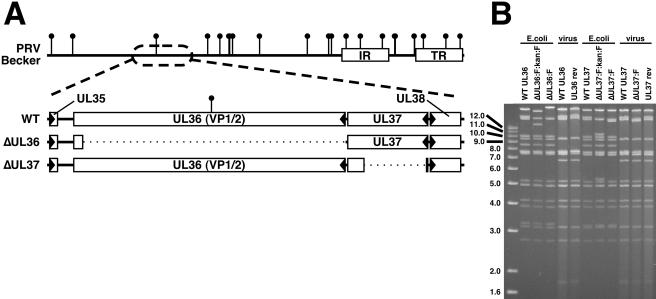
FIG. 1.
Description of VP1/2-null virus and VP1/2-complementing cell line. (A) Illustration of the PRV genome with characteristic internal and terminal repeats (IR and TR, respectively) shown as white rectangles. BamHI sites are indicated by vertical lollipops. The region of the genome relevant to this report is expanded to show the UL36 and UL37 ORFs, as well as the neighboring UL35 and UL38 ORFs (arrowheads indicate gene orientation). Below, dotted lines indicate regions deleted in the ΔUL36 (PRV-GS678) and ΔUL37 (PRV-GS993) viruses. (B) Ethidium bromide-stained agarose gel of BamHI-digested infectious clone DNAs isolated from E. coli and corresponding viral DNA isolated from nucleocapsids. The UL36 gene overlaps the two largest BamHI fragments of the viral genome, whereas the UL37 gene is exclusively on the second largest fragment. The FRT:kan:FRT insertion is ∼1.5 kbp and encodes two BamHI sites within the kanamycin resistance cassette, resulting in the truncation of the two large fragments during the ΔUL36 construction, and split of the second largest fragment into two smaller fragments during the ΔUL37 construction. Removal of the kanamycin resistance cassette and one FRT equivalent results in fusion of the remainders of the two largest fragments to produce the final ΔUL36 allele, and the reunion of the remainders of the second largest fragment to produce the final ΔUL37 allele. Additional fragment variations seen between the infectious clone plasmids and viral DNA result from loss of the E. coli vector backbone (as described in reference 36).
The ΔUL37 virus was made by the same strategy as the ΔUL36 virus described above. The pGS847 infectious clone was transformed into the EL250 Escherichia coli strain, resulting in strain GS958. The kanamycin cassette was amplified by using a pair of FRT-encoding primers (FRT sequences are underlined) with 5′ homologies to the PRV UL37 gene +6 nt to +45 nt and +2200 nt to +2239 nt relative to the UL37 ORF, respectively): 5′-GGCGCTCGTGCGCGCGCTCGAGGAGGCCGACCACGCCGTCTAAGAAGTTCCTATACTTTCTAGAGAATAGGAACTTCCCAGTCACGACGTTGTAAAACG and 5′-CCAGCGCCTCGCACTCGCGCAGCGCCTCCGTCGTCTGCGCGAAGTTCCTATTCTCTAGAAAGTATAGGAACTTCGAAACAGCTATGACCATGATTACG. The resulting PCR product was recombined into GS958 and one recombinant, GS992, was confirmed by restriction digest analysis. Arabinose-induced expression of Flp recombinase was used to remove the kanamycin cassette, resulting in GS993 (Fig. 1). The resulting allele has a stop codon fused 15 codons downstream of the endogenous UL37 start codon, and is deleted for the subsequent 718 codons (the product of the UL37 ORF is predicted to be a 919-aa protein). A deletion in UL37 that leaves the first 15 codons intact has no impact on expression of the upstream UL38 gene (17). Transfection of pGS993 into PK15-UL37 cells resulted in the ΔUL37 mRFP1-capsid virus (PRV-GS993).
Although PRV-GS678 could be propagated on PK15-UL36 cells, sequential passage of the virus ultimately resulted in repair of the ΔUL36 allele in the viral genome. PK15-UL36 cells provided less than wild-type growth kinetics of PRV-GS678, and only low titer stocks of PRV-GS678 could be isolated prior to the emergence of spontaneous revertant viruses. One spontaneous revertant virus, PRV-GS678R, was isolated from growth of PRV-GS678 on PK15 cells transfected with the UL36 gene. In contrast, we never observed spontaneous reversion of PRV-GS993 on PK15-UL37 cells, probably due to the lack of flanking homology 5′ of the transgene to the viral genome. Therefore, to make a revertant of the PRV-GS993 virus, the UL37 gene was subcloned with 800 nt of flanking sequence both upstream and downstream the UL37 ORF (pGS1522). This construct was transiently transfected into Vero cells, which were then infected with PRV-GS993 1 day posttransfection. This resulted in many large foci of viral spread which were collectively harvested 3 days postinfection. The viral stock was expanded by two subsequent rounds of infection and harvest, resulting in the PRV-GS993R stock.
DNA purification.
Infectious clone plasmids were isolated from E. coli strain EL250 by standard alkali lysis methods. Viral DNAs were isolated from nucleocapsids purified from infected PK15 cells, as previously described (35).
Immunostaining.
Coverslips were incubated for 1 h in fixation-extraction buffer (100 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid); pH 6.9], 1 mM EGTA, and 1 mM MgCl2 containing 0.1% glutaraldehyde and 1.0% Triton X-100) at room temperature. Samples were washed in phosphate-buffered saline and quenched with 1% sodium borohydride for 20 min at room temperature. Coverslips were then blocked with 1% bovine serum albumin. For microtubule staining, samples were incubated with rat monoclonal anti-tubulin antibodies (clone YL1/2, provided by J. V. Kilmartin, Laboratory of Molecular Biology, Cambridge, United Kingdom) for 30 min at 37°C, followed by incubation with anti-rat secondary antibodies (1:200) conjugated to Cy5 fluorophore (Jackson Immunoresearch Laboratories). The actin cytoskeleton was visualized by staining with phalloidin conjugated to Alexa 350 fluorophore (Molecular Probes).
Fluorescence microscopy.
Immunostained cells were observed with a Nikon Eclipse TE200 inverted microscope equipped with a ×100 oil immersion objective. Optimized Cy5 and UV filter sets (Chroma Technology Corp.) were used for Cy5 and Alexa 350 fluorescence detection, respectively. Digital images were acquired with a CH250 charged coupled device (Photometrics) controlled by Metamorph imaging software (Molecular Devices).
All other static and time-lapse images were acquired from living cells by using an inverted wide-field Nikon Eclipse TE2000-U microscope. The microscope was fitted with a Cascade:650 CCD (Roper Scientific) and was housed in a 37°C environmental box (Life Imaging Services). Infected cells were imaged in sealed chambers as previously described (37). The Metamorph software package was used for image acquisition. Fluorescent and phase contrast images of infected PK15 cells were captured by using a ×10/0.3 numerical aperture (NA) objective, and time-lapse fluorescent imaging of infected Vero cells was performed with a ×60/1.4NA objective. Cytoplasmic fluorescent punctae observed in Vero cells were consistent with emissions from individual fluorescent-capsids (37, 38). Nuclear fluorescence was significantly brighter and not diffraction limited and likely results from the formation of large nuclear capsid inclusions (7, 30).
Image analysis.
Nuclear egress in Vero cells was analyzed by acquiring both fluorescence and differential interference contrast (DIC) still images of infected cells. The DIC images were used to identify the location of nuclei. Because fluorescent capsids in proximity to the nuclear membrane cannot be spatially resolved as cytoplasmic or nuclear, cells were only scored as positive if 10 or more fluorescent punctae were considered to be outside of the nucleus.
To examine cytoplasmic capsid dynamics, time-lapse fluorescence images were captured at 5 frames/s (continuous 200-ms exposures). The fluorescent punctae of individual fluorescent capsids were subsequently tracked by using the Metamorph “Track Points” application. Because many capsids displayed little or no motion during recording, only capsids that moved at least 1 μm were included in the data sets used for subsequent analysis.
RESULTS
Isolation of fluorescent-capsid viruses lacking either the VP1/2 or UL37 tegument protein.
We have previously described infectious clones of PRV that encode the VP26 capsid protein fused to a fluorescent protein (i.e., GFP or mRFP1) and have wild-type growth properties in culture (23, 37, 38). To test the roles of VP1/2 and UL37 in intracellular capsid transport, recombination in E. coli was used to replace either nucleotides 4 to 8910 of the 9255-nt UL36 ORF or nucleotides 46 to 2199 of the 2760-nt UL37 ORF, with a TAA stop codon followed by a 34-bp FRT site (Fig. 1A). This was accomplished by RED-mediated insertion of a kanamycin cassette flanked by a pair of FRT sites, followed by Flp-mediated excision of the kanamycin cassette (Fig. 1B).
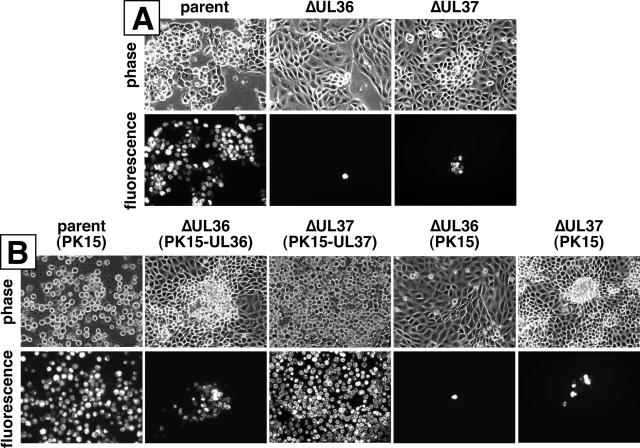
VP1/2 is essential for herpesvirus growth in cultured cells, whereas UL37 is essential for the growth of herpes simplex virus type 1 (HSV-1) and is necessary for wild-type growth of PRV (2, 9, 18, 35). To verify that the ΔUL36 (VP1/2-null) and ΔUL37 mutations introduced into the PRV infectious clones impacted viral growth as expected, each clone was transfected into PK15 cells and capsid fluorescence was monitored (Fig. 2A). A virus with intact UL36 and UL37 genes resulted in productive infections, as noted by the spread of capsid fluorescence to all cells with corresponding cytopathic effects (i.e., cell rounding). In contrast, transfection of the ΔUL36 clone resulted in fluorescent-capsid expression in single cells only. The lack of viral spread from cell to cell was consistent with a lethal mutation. The ΔUL37 clone displayed minimal spread.
FIG. 2.
Initial characterization of VP1/2- and UL37-null viruses and complementing cell lines. (A) Images of living PK15 cells 2 days posttransfection with infectious clones encoding fluorescent capsids (pGS575; “parent”) and clones carrying additional ΔUL36 (pGS678) or ΔUL37 (pGS993) mutations. (B) Images of living PK15 or complementing cells (as indicated) 2 days postinfection with viruses derived from the above three infectious clones (PRV-GS575, PRV-GS678, and PRV-GS993).
To propagate the ΔUL36 and ΔUL37 virus for further study, complementing cell lines stably expressing either VP1/2 (PK15-UL36) or UL37 (PK15-UL37) were made. PK15-UL36 cells failed to fully complement growth of the ΔUL36 virus, since VP1/2-null virus produced small foci (Fig. 2B). The inability of VP1/2-expressing cells to fully complement PRV lacking VP1/2 was previously noted by others (9). Transcomplementation of the ΔUL37 virus resulted in efficient viral spread throughout the cell culture.
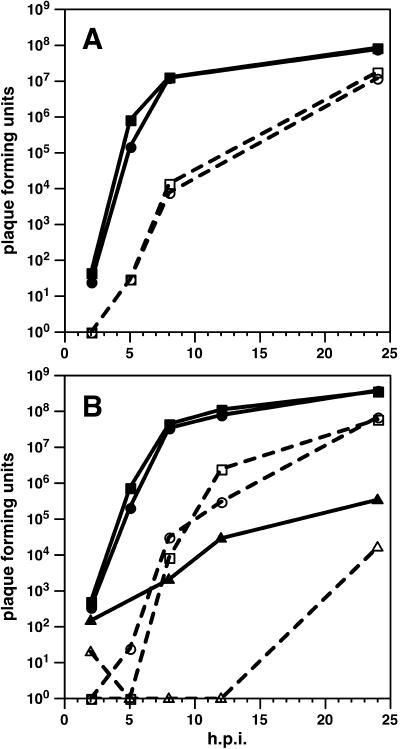
To avoid reversion of the deletion viruses by recombination with the wild-type UL36 or UL37 genes in complementing cells, complemented viral stocks were examined by infection of noncomplementing cells (Fig. 2b). In one case, a spontaneous UL36 revertant was harvested from PK15 cells and further examined (not shown). The revertant virus grew with wild-type growth kinetics and was saved for use as a control in subsequent experiments (Fig. 3A). A spontaneous UL37 revertant was never observed; however, a UL37 revertant was isolated after infection of PK15 cells transiently transfected with a full-length UL37 construct. Unlike the ΔUL37 virus, which grew poorly, the revertant virus grew with wild-type kinetics (Fig. 3B).
FIG. 3.
Single-step growth kinetics of fluorescent-capsid viruses. (A) Comparison of the growth of the parent virus (PRV-GS575; circles) with the UL36 revertant virus (PRV-GS678R; squares). The ΔUL36 virus was not viable and therefore could not be examined. (B) Comparison of the growth of the parent virus (PRV-GS958; circles) with the ΔUL37 virus (PRV-GS993; triangles) and the UL37 revertant virus (PRV-GS993R; squares). Virions were harvested from the media (dashed lines; open symbols) and cells (solid lines; filled symbols) at the indicated times.
VP1/2 and UL37 are required for efficient nuclear egress of PRV.
Cells infected with either HSV-1 or PRV harboring deletions in UL36 (VP1/2-null) are reported to accumulate unenveloped cytosolic capsids at late time points postinfection (2, 9). To our surprise, our initial observations of cells infected with the ΔUL36 virus indicated that capsids were typically restricted to the nucleus. This observation was in contrast to a previous report of HSV-1 harboring a deletion in UL36 (2).
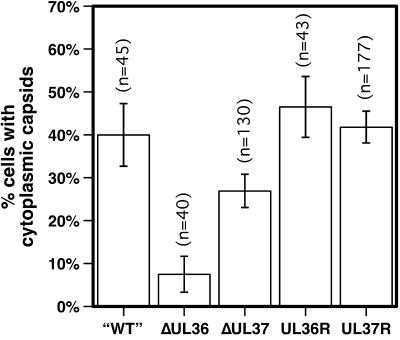
To further assess the role of VP1/2 in nuclear egress, Vero cells were infected with the fluorescent-capsid viruses harboring the wild-type UL36, ΔUL36 or the UL36-revertant alleles. Living infected cells were scored for the presence of cytoplasmic fluorescent capsids from 11 to 15 h postinfection (hpi) by fluorescence microscopy. Each virus produced capsids in the nuclei of infected cells as expected; however, only a fraction of infected cells were observed to have capsids that had translocated from the nucleus to the cytoplasm (Fig. 4). Nearly half of cells infected with wild-type and revertant UL36 viruses had clear cytoplasmic capsid fluorescence (the fraction of cells with cytoplasmic capsids increased with time; data not shown). In contrast, capsids of the ΔUL36 virus egressed from nuclei at a significantly lower frequency. Examination of the ΔUL37 virus by this assay indicated a reduction in nuclear egress that was not as dramatic as the ΔUL36 virus, while the UL37-revertant virus behaved similarly to the “wild-type” and the UL36-revertant viruses.
FIG. 4.
Nuclear egress of capsids into the cytoplasm. The percentage of living cells displaying cytoplasmic capsids 11 to 15 h postinfection is shown. Vero cells were infected with PRV-GS575, having intact UL36 and UL37 genes (“WT”), PRV-GS678 (ΔUL36), PRV-GS993 (ΔUL37), the PRV-GS678R revertant virus (UL36R), or the PRV-GS993R revertant virus (UL37R) at an MOI of ≤0.1. Because capsids near the nuclear rim could not be scored easily in this assay, cells were only counted as positive if at least 10 fluorescent-capsid punctae were observed in the cytoplasm. Error bars are standard error of the proportions (SEp). PRV-GS678 and PRV-GS993 were significantly different from each other and from each revertant virus (z < 0.01).
Microtubule-based transport of capsids requires VP1/2.
The dynamics of cytoplasmic fluorescent-capsids were examined by time-lapse microscopy in living Vero cells. Capsids of the parent virus (PRV-GS575) displayed three dynamic states: (i) static, (ii) random nonprocessive motion, and (iii) curvilinear processive motion (movie S1 in the supplemental material). The static capsids were observed around the margins of the cell and at the basal surface adjacent to the coverslip, a finding consistent with this population of capsids having exited the cell as fully assembled viral particles (data not shown). Individual moving capsids were tracked for up to 20 s or as long as they remained visible in the focal plane. An approximately equal number of PRV-GS575 capsids demonstrated processive (directed) and nonprocessive (random) movement (Fig. 5A). The randomly oriented nonprocessive motion generally did not result in capsids diffusing more than 2 μm from the initially observed starting point during the course of recording. Processively transporting capsids moved greater distances (>10 μm in at least one instance) at rates comparable to that reported in axons of neurons (1 to 5 μm/s) (37, 38).
FIG. 5.
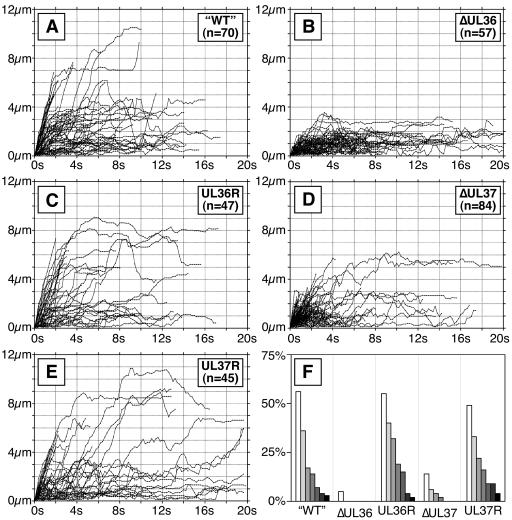
Transport of capsids in the cytoplasm. Individual moving fluorescent capsids were tracked in the cytoplasm of Vero cells at 18 hpi. Tracking was performed on time-lapse fluorescence recordings collected from a single focal plane at five frames/s. A profile of transport for each infection is shown by plots of the individual capsids tracked as distance from starting position in the first frame (y axis) versus time (x axis). Vero cells were infected with the following viruses: PRV-GS575 (A), PRV-GS678 (B), PRV-GS678R (C), PRV-GS993 (D), and PRV-GS993R (E). (F) Summary of data shown as percentage of tracked capsids that moved greater than 3, 4, 5, 6, 7, 8, and 9 μm from the origins for each infection (shown from left to right, respectively).
No curvilinear capsid transport from the ΔUL36 virus was observed; instead, motion was exclusively random and nonprocessive (Fig. 5B and F; see also movie S2 in the supplemental material). Although transport of the ΔUL37 virus was reduced from wild type, directed transport could still be observed (Fig. 5D and F). The UL36- and UL37-revertant viruses both had wild-type capsid transport dynamics (Fig. 5C, E, and F).
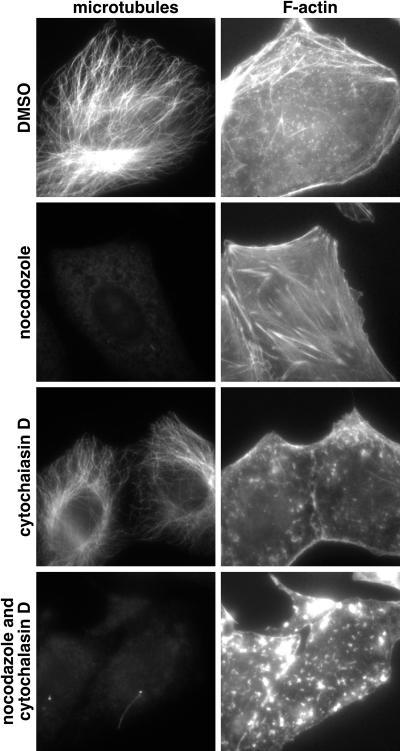
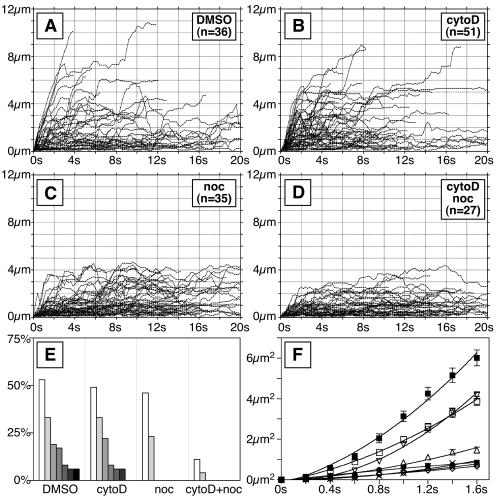
To determine whether actin filaments or microtubules were required for capsid motion, cells were treated with cytochalasin D, nocodazole, or both drugs in combination. Both drugs were effective at disrupting their respective cytoskeletal targets (Fig. 6); however, in some cells isolated nocodazole-resistant microtubules were observed (for example, see the lower left panel in Fig. 6). In the presence of nocodazole the processive curvilinear transport of capsids was lost, whereas treatment with cytochalasin D or dimethyl sulfoxide (DMSO) alone did not result in an obvious change in transport (Fig. 7A to E). Nonprocessive motion was unaffected by cytochalasin D, nocodazole, or the combination of both drugs.
FIG. 6.
Vero cell cytoskeleton susceptibility to cytochalasin D and nocodazole. Uninfected Vero cells were treated for 1 h with cytochalasin D, nocodazole, or both drugs together (or DMSO alone as control). Cells were fixed and processed for immunofluorescence imaging of filamentous actin and microtubules (image pairs).
FIG. 7.
Dependence of cytoplasmic capsid transport on cytoskeleton integrity. Infected Vero cells were treated with cytochalasin D and nocodazole (or DMSO alone as control) at 17 hpi as indicated. (A to D) Individual moving cytoplasmic fluorescent capsids were tracked at 18 hpi and analyzed as described in the legend of Fig. 5. (E) Summary of data shown as a percentage of tracked capsids that moved greater than 3, 4, 5, 6, 7, 8, and 9 μm from the origins for each infection (shown from left to right). (F) Diffusion profiles of moving cytoplasmic capsids. The mean squared displacement of the capsids from origin is shown over the first 1.6 s of the recordings. Symbols are as follows: ▪, PRV-GS575 (“wild-type”); •, PRV-GS678: ΔUL36; □, PRV-GS575 + DMSO; ▿, PRV-GS575 + cytochalasin D; ▵, PRV-GS575 + nocodazole; ○, PRV-GS575 + cytochalasin D + nocodazole; ⋄, PRV-GS678 + cytochalasin D + nocodazole; ×, PRV-GS575 + cytochalasin D + nocodazole + azide. The error shown is 95% confidence.
To further analyze the impact of cytoskeletal drugs on capsid motion, we examined the mean-square capsid displacement for each tracking experiment (Fig. 7F). This analysis allows for an examination of the overall capsid diffusion in a sample. As expected for either passive or facilitated diffusion, mean-square capsid displacement was approximately linear with time (we note that this linearity was gradually lost as capsids progressed beyond ∼5 μm, which is consistent with diffusion being caged by the plasma membrane [data not shown]). Surprisingly, nocodazole did not completely disrupt capsid transport; wild-type capsid displacement in the presence of nocodazole was greater than that of ΔUL36 capsids. Cytochalasin D alone had no effect on capsid displacement (the cytochalasin D sample was indistinguishable from the DMSO control), but when cytochalasin D was added in combination with nocodazole capsid motion was reduced to that of the ΔUL36 virus. Cotreatment of ΔUL36 infected cells with cytochalasin D and nocodazole resulted in no further reduction in capsid diffusion. Addition of azide also failed to further reduce capsid diffusion. Collectively, these results indicated that (i) directed capsid motion was dependent on VP1/2, (ii) random nonprocessive motion occurred by passive diffusion of capsids in the cytoplasm, (iii) VP1/2-mediated capsid transport was primarily dependent on microtubules, and (iv) residual capsid motion in the presence of nocodazole, relative to ΔUL36 capsids, was primarily dependent on intact actin filaments and not nocodazole-resistant microtubules.
DISCUSSION
The neurotropic properties of the alphaherpesvirus subfamily are dependent upon long distance transport of capsids along microtubules in axons (20, 27). Transport of herpesvirus capsids to the nucleus of sensory neurons and subsequent transport of progeny viral particles to distal axons occurs by bidirectional transport processes at rates consistent with dynein- and kinesin-based mechanisms (37, 38). Similar transport mechanisms likely traffic intracellular herpesvirus particles to the nucleus and cell periphery in nonpolarized cells. Herpesvirus capsids newly deposited in Vero cells travel along microtubules from the plasma membrane to the nucleus, and this transport is dependent upon the dynein-associated complex, dynactin (5, 32, 40). In contrast, transport of newly assembled herpesvirus particles to the periphery of nonpolarized cells has been less well studied. Here we report on the dynamics of herpesvirus transport during egress from Vero cells and identify the first herpesvirus protein, VP1/2, required for microtubule-based capsid transport.
The capsids of viruses with an intact UL36 gene, which encodes VP1/2, often moved in curvilinear trajectories, and this motion was sensitive to nocodazole. This transport was processive and saltatory, with similar velocities to those seen in neurons (1 to 5 μm/s) (38). Also similar to axonal transport, only a fraction of capsids moved directionally in Vero cells during the course of a recording (37). However, unlike capsids in axons, capsids in Vero cells also displayed nonprocessive motion. The random nature of the later movement and the observation that nocodazole, cytochalasin D, and azide could not impede it argues that this motion is simple diffusion. Herpesvirus capsids would not normally be expected to diffuse to any significant degree in cytoplasm (22, 39). However, we note that late in infection the cytoplasm of Vero cells becomes increasingly soluble, as indicated by increased motion of cytoplasmic structures observed by transmitted light microscopy (data not shown). This may in part be explained by virus-induced loss of actin filaments or cytopathic effects resulting in cellular distress (22, 34, 43).
In the absence of VP1/2 capsids fail to re-envelope and instead accumulate in the cytosol (2, 9). We find that these capsids do not move directionally in the cytoplasm. This lack of transport was similar to that seen when wild-type UL36 virus was treated with nocodazole, indicating that the VP1/2 tegument protein was required for microtubule-based transport of capsids. Unexpectedly, capsids of wild-type UL36 virus treated with nocodazole displayed some residual motion relative to ΔUL36 capsids. The residual motion was abrogated when infected cells were cotreated with nocodazole and cytochalasin D, indicating this motion was actin dependent. We are unclear as to whether the residual motion is a result of capsids using some aspect of the actin cytoskeleton as a motile force or if this is an indirect effect. However, we note that at least one herpesvirus protein has been reported to bind to a myosin motor (42).
There are two likely explanations of the VP1/2-null phenotype. First, VP1/2 may bind a host motor complex either directly or indirectly, possibly through another tegument protein, and deliver capsids to the site of re-envelopment (for example, the HCMV VP1/2 homologue was shown to bind the cellular p180 protein, which in turn binds kinesin, as described in references 3 and 31). In support of this model, VP1/2 remains bound to capsids during intracellular transport, making it a good candidate for a motor binding protein (23). VP1/2 is thought to bind to the capsid surface directly, while additional tegument proteins, such as UL37, require VP1/2 for capsid association (2, 9, 15, 49). Second, VP1/2 may play a direct role in the re-envelopment process, and the observed capsid transport may represent cellular vesicles containing re-enveloped capsids (i.e., virions) moving to the plasma membrane.
We expect that both explanations are true. Both PRV and HSV-1 lacking UL37 are defective for re-envelopment in the cytoplasm, resulting in accumulation of unenveloped cytosolic capsids (1, 17). This phenotype is strikingly similar to those of PRV and HSV-1 lacking UL36 (2, 9). However, in the present study, ΔUL37 capsids frequently moved directionally in the cytoplasm, albeit with reduced overall kinetics, while ΔUL36 capsids failed to do so. The capsid transport that remains intact in the absence of UL37 demonstrates that unenveloped capsids transport prior to re-envelopment, as ΔUL37 viruses rarely re-envelope (1, 17). With both VP1/2 and UL37 available, capsids of wild-type viruses would then re-envelope and become available to transport within the cellular secretory pathway, collectively resulting in wild-type transport. There is precedence for such a model with the poxvirus, vaccinia. Like herpesviruses, vaccinia intracellular mature virus particles bud into internal host membranes to form an intracellular enveloped virus, which then transports to the cell periphery and releases the virion by fusion with the plasma membrane. With vaccinia virus, host motors are used to transport intracellular mature virus particles to the site of envelopment and subsequently to move the membrane-enclosed intracellular enveloped viruses to the plasma membrane (11, 14, 33, 44-46). Development of a dual-fluorescent virus with tags on capsid and a membrane protein may allow for resolution of these possibilities.
VP1/2 also remains associated with cytosolic capsids after viral entry, and it is possible that the VP1/2 protein plays a critical role in capsid transport at this early stage of infection (12, 23). However, the dynamics of capsid transport toward the nucleus in the absence of VP1/2 could not be examined because the ΔUL36 virus does not produce extracellular virions with which to infect cells (when grown on complementing cells virions incorporate VP1/2, rendering them useless for such investigations). The role of UL37 also could not be studied during capsid transport after entry, since titers of the ΔUL37 virus recovered from noncomplementing cells were too low to effectively image using our live-cell imaging methods.
Lastly, we note that capsids of the ΔUL36 and ΔUL37 viruses were found in the cytoplasm of infected cells at a lower frequency than capsids of viruses that encoded both proteins. In agreement with our present findings, deletion of the UL37 gene from HSV-1 was previously reported to result in a partial defect in nuclear egress (1). However, in the current study, the ΔUL36 virus displayed the more dramatic nuclear egress defect, yet a previous examination of a HSV-1 ΔUL36 mutant indicated no such defect (2). The reason for these differences is not immediately clear. The mutant UL36 allele introduced into HSV-1 left ∼62% of the coding sequence intact, which could have resulted in the production of a truncated protein with partial function. Alternatively, the nuclear egress function in PRV and HSV-1 may be mediated by different proteins. The PRV ΔUL36 virus in the current study (PRV-GS678) assembled capsids in the nucleus based on transmission electron microscopy, which is consistent with the findings in the previous ΔUL36 studies in PRV and HSV (data not shown). However, we observed that deletion of UL36 in PRV resulted in a polar effect that decreased UL37 expression by ∼4-fold. This reduction in expression was unexpected since the UL36 deletion did not overlap with the predicted UL37 polyadenylation site or 3′ untranslated region but nevertheless may contribute to the difficulty in complementing the growth of the PRV ΔUL36 virus as was observed in this report and previously by others (9; data not shown). As such, we cannot rule out that the nuclear egress defect observed in the ΔUL36 virus is a synergistic defect from coincidental lowered UL37 expression.
In summary, although many herpesvirus proteins are known to bind cellular motor components, VP1/2 is the first viral protein shown to be essential for intracellular capsid transport. This role of VP1/2 is not dependent on the VP1/2 binding protein, UL37, although the latter is required for wild-type transport kinetics. These findings provide an important step toward the identification of the mechanism of herpesvirus intracellular transport and ultimately determining how alphaherpesviruses spread in the vertebrate nervous system.
Supplementary Material
Acknowledgments
We are grateful to Steven Gross, Volodya Gelfand, and Gary Borisy for helpful suggestions regarding analysis of capsid motion. We also thank Sarah Rice and Sarah Antinone for critical evaluation of the manuscript.
This study was supported by NIH grant 1RO1AI056346 to G.A.S.
Footnotes
Supplementary material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Desai, P., G. L. Sexton, J. M. McCaffery, and S. Person. 2001. A null mutation in the gene encoding the herpes simplex virus type 1 UL37 polypeptide abrogates virus maturation. J. Virol. 75:10259-10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Desai, P. J. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diefenbach, R. J., E. Diefenbach, M. W. Douglas, and A. L. Cunningham. 2004. The ribosome receptor, p180, interacts with kinesin heavy chain, KIF5B. Biochem. Biophys. Res. Commun. 319:987-992. [DOI] [PubMed] [Google Scholar]
- 4.Diefenbach, R. J., M. Miranda-Saksena, E. Diefenbach, D. J. Holland, R. A. Boadle, P. J. Armati, and A. L. Cunningham. 2002. Herpes simplex virus tegument protein US11 interacts with conventional kinesin heavy chain. J. Virol. 76:3282-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dohner, K., A. Wolfstein, U. Prank, C. Echeverri, D. Dujardin, R. Vallee, and B. Sodeik. 2002. Function of Dynein and dynactin in herpes simplex virus capsid transport. Mol. Biol. Cell 13:2795-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Douglas, M. W., R. J. Diefenbach, F. L. Homa, M. Miranda-Saksena, F. J. Rixon, V. Vittone, K. Byth, and A. L. Cunningham. 2004. Herpes simplex virus type 1 capsid protein VP26 interacts with dynein light chains RP3 and Tctex1 and plays a role in retrograde cellular transport. J. Biol. Chem. 279:28522-28530. [DOI] [PubMed] [Google Scholar]
- 7.Epstein, M. A. 1962. Observations of the mode of release of herpesvirus from infected HeLa cells. J. Cell Biol. 12:589-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuchs, W., H. Granzow, B. G. Klupp, M. Kopp, and T. C. Mettenleiter. 2002. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 76:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 2004. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J. Virol. 78:11879-11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuller, A. O., and P. G. Spear. 1987. Anti-glycoprotein D antibodies that permit adsorption but block infection by herpes simplex virus 1 prevent virion-cell fusion at the cell surface. Proc. Natl. Acad. Sci. USA 84:5454-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geada, M. M., I. Galindo, M. M. Lorenzo, B. Perdiguero, and R. Blasco. 2001. Movements of vaccinia virus intracellular enveloped virions with GFP tagged to the F13L envelope protein. J. Gen. Virol. 82:2747-2760. [DOI] [PubMed] [Google Scholar]
- 12.Granzow, H., B. G. Klupp, and T. C. Mettenleiter. 2005. Entry of pseudorabies virus: an immunogold-labeling study. J. Virol. 79:3200-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Granzow, H., B. G. Klupp, and T. C. Mettenleiter. 2004. The pseudorabies virus US3 protein is a component of primary and of mature virions. J. Virol. 78:1314-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hollinshead, M., G. Rodger, H. Van Eijl, M. Law, R. Hollinshead, D. J. Vaux, and G. L. Smith. 2001. Vaccinia virus utilizes microtubules for movement to the cell surface. J. Cell Biol. 154:389-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klupp, B. G., W. Fuchs, H. Granzow, R. Nixdorf, and T. C. Mettenleiter. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 76:3065-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2001. Effect of the pseudorabies virus US3 protein on nuclear membrane localization of the UL34 protein and virus egress from the nucleus. J. Gen. Virol. 82:2363-2371. [DOI] [PubMed] [Google Scholar]
- 17.Klupp, B. G., H. Granzow, E. Mundt, and T. C. Mettenleiter. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 75:8927-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knipe, D. M., W. Batterson, C. Nosal, B. Roizman, and A. Buchan. 1981. Molecular genetics of herpes simplex virus. VI. Characterization of a temperature-sensitive mutant defective in the expression of all early viral gene products. J. Virol. 38:539-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koshizuka, T., Y. Kawaguchi, and Y. Nishiyama. 2005. Herpes simplex virus type 2 membrane protein UL56 associates with the kinesin motor protein KIF1A. J. Gen. Virol. 86:527-533. [DOI] [PubMed] [Google Scholar]
- 20.Kristensson, K., E. Lycke, M. Röyttä, B. Svennerholm, and A. Vahlne. 1986. Neuritic transport of herpes simplex virus in rat sensory neurons in vitro. Effects of substances interacting with microtubular function and axonal flow [nocodazole, taxol and erythro-9-3-(2-hydroxynonyl)adenine]. J. Gen. Virol. 67:2023-2028. [DOI] [PubMed] [Google Scholar]
- 21.Lee, E. C., D. Yu, J. Martinez de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56-65. [DOI] [PubMed] [Google Scholar]
- 22.Luby-Phelps, K. 2000. Cytoarchitecture and physical properties of cytoplasm: volume, viscosity, diffusion, intracellular surface area. Int. Rev. Cytol. 192:189-221. [DOI] [PubMed] [Google Scholar]
- 23.Luxton, G. W., S. Haverlock, K. E. Coller, S. E. Antinone, A. Pincetic, and G. A. Smith. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. USA 102:5832-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mabit, H., M. Y. Nakano, U. Prank, B. Saam, K. Dohner, B. Sodeik, and U. F. Greber. 2002. Intact microtubules support adenovirus and herpes simplex virus infections. J. Virol. 76:9962-9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez-Moreno, M., I. Navarro-Lerida, F. Roncal, J. P. Albar, C. Alonso, F. Gavilanes, and I. Rodriguez-Crespo. 2003. Recognition of novel viral sequences that associate with the dynein light chain LC8 identified through a pepscan technique. FEBS Lett. 544:262-267. [DOI] [PubMed] [Google Scholar]
- 26.Mettenleiter, T. C. 2004. Budding events in herpesvirus morphogenesis. Virus Res. 106:167-180. [DOI] [PubMed] [Google Scholar]
- 27.Miranda-Saksena, M., P. Armati, R. A. Boadle, D. J. Holland, and A. L. Cunningham. 2000. Anterograde transport of herpes simplex virus type 1 in cultured, dissociated human and rat dorsal root ganglion neurons. J. Virol. 74:1827-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicola, A. V., J. Hou, E. O. Major, and S. E. Straus. 2005. Herpes simplex virus type 1 enters human epidermal keratinocytes, but not neurons, via a pH-dependent endocytic pathway. J. Virol. 79:7609-7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicola, A. V., A. M. McEvoy, and S. E. Straus. 2003. Roles for endocytosis and low pH in herpes simplex virus entry into HeLa and Chinese hamster ovary cells. J. Virol. 77:5324-5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nii, S., C. Morgan, and H. M. Rose. 1968. Electron microscopy of herpes simplex virus. II. Sequence of development. J. Virol. 2:517-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogawa-Goto, K., S. Irie, A. Omori, Y. Miura, H. Katano, H. Hasegawa, T. Kurata, T. Sata, and Y. Arao. 2002. An endoplasmic reticulum protein, p180, is highly expressed in human cytomegalovirus-permissive cells and interacts with the tegument protein encoded by UL48. J. Virol. 76:2350-2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogawa-Goto, K., K. Tanaka, W. Gibson, E. Moriishi, Y. Miura, T. Kurata, S. Irie, and T. Sata. 2003. Microtubule network facilitates nuclear targeting of human cytomegalovirus capsid. J. Virol. 77:8541-8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rietdorf, J., A. Ploubidou, I. Reckmann, A. Holmstrom, F. Frischknecht, M. Zettl, T. Zimmermann, and M. Way. 2001. Kinesin-dependent movement on microtubules precedes actin-based motility of vaccinia virus. Nat. Cell Biol. 3:992-1000. [DOI] [PubMed] [Google Scholar]
- 34.Schumacher, D., B. K. Tischer, S. Trapp, and N. Osterrieder. 2005. The protein encoded by the US3 orthologue of Marek's disease virus is required for efficient de-envelopment of perinuclear virions and involved in actin stress fiber breakdown. J. Virol. 79:3987-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, G. A., and L. W. Enquist. 2000. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc. Natl. Acad. Sci. USA 97:4873-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith, G. A., S. P. Gross, and L. W. Enquist. 2001. Herpesviruses use bidirectional fast-axonal transport to spread in sensory neurons. Proc. Natl. Acad. Sci. 98:3466-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, G. A., L. Pomeranz, S. P. Gross, and L. W. Enquist. 2004. Local modulation of plus-end transport targets herpesvirus entry and egress in sensory axons. Proc. Natl. Acad. Sci. USA 101:16034-16039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sodeik, B. 2000. Mechanisms of viral transport in the cytoplasm. Trends Microbiol. 8:465-472. [DOI] [PubMed] [Google Scholar]
- 40.Sodeik, B., M. W. Ebersold, and A. Helenius. 1997. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 136:1007-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tirabassi, R. S., and L. W. Enquist. 1998. Role of envelope protein gE endocytosis in the pseudorabies virus life cycle. J. Virol. 72:4571-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Leeuwen, H., G. Elliott, and P. O'Hare. 2002. Evidence of a role for nonmuscle myosin II in herpes simplex virus type 1 egress. J. Virol. 76:3471-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Minnebruggen, G., H. W. Favoreel, L. Jacobs, and H. J. Nauwynck. 2003. Pseudorabies virus US3 protein kinase mediates actin stress fiber breakdown. J. Virol. 77:9074-9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ward, B. M. 2005. Visualization and characterization of the intracellular movement of vaccinia virus intracellular mature virions. J. Virol. 79:4755-4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ward, B. M., and B. Moss. 2001. Vaccinia virus intracellular movement is associated with microtubules and independent of actin tails. J. Virol. 75:11651-11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ward, B. M., and B. Moss. 2001. Visualization of intracellular movement of vaccinia virus virions containing a green fluorescent protein-B5R membrane protein chimera. J. Virol. 75:4802-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye, G. J., K. T. Vaughan, R. B. Vallee, and B. Roizman. 2000. The herpes simplex virus 1 U(L)34 protein interacts with a cytoplasmic dynein intermediate chain and targets nuclear membrane. J. Virol. 74:1355-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou, Z. H., D. H. Chen, J. Jakana, F. J. Rixon, and W. Chiu. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 73:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.