Abstract
The aim of the present study was to identify and functionally characterize the equine herpesvirus 1 (EHV-1) UL20 protein (UL20p). Using a specific antiserum, UL20p was shown to be associated with membranes of infected cells, as well as with envelopes of purified virions. By Western blot analysis, UL20p was detected in two main forms exhibiting Mrs of 25,000 and 75,000. Both moieties did not enter the separating gel after heating of protein samples to 99°C. The slower-migrating form of UL20p contains N-linked carbohydrates, and its presence is dependent of that of other viral proteins. Infection of cells that either constitutively express UL20p or a gK-green fluorescent protein (GFP) fusion protein with various EHV-1 deletion mutants revealed a relatively stable hetero-oligomer containing gK and UL20p with an apparent Mr of 75,000. As demonstrated by confocal microscopy, UL20p distribution in Rk13 cells changed from a diffuse granular or netlike appearance to a pattern confined to the Golgi network when gK was coexpressed. Analysis of a UL20 deletion mutant of EHV-1 strain RacL11 indicated an involvement of UL20p in cell-to-cell spread, as well as in very late events in virus egress. Based on these and electron microscopic studies we suggest that the EHV-1 UL20 protein might be necessary to avoid fusion of mature virions with membranes of their transport vesicles.
Equid herpesvirus 1 (equine herpesvirus 1 [EHV-1]) is one of the most important viral pathogens of horses and classifies as a Varicellovirus within the subfamily Alphaherpesvirinae (21).
Alphaherpesviruses express relatively few proteins with the potential to span membranes several times. Accordingly, only four such open reading frames (ORFs) have been identified in the EHV-1 sequence, and they are homologous to the herpes simplex virus type 1 (HSV-1) ORFs UL10, UL20, UL43, and UL53 (31). Of these, the EHV-1 UL10 product glycoprotein M (gM), has been studied most intensively, and it has been shown that gM is involved in virus egress, more precisely in secondary envelopment, and plays a role in cell-to-cell spread (23, 28). The protein encoded by UL53, gK, is less dispensable for virus replication in Rk13 cells than gM. However, like gM, gK influences a late step in virus egress, direct cell-to-cell spread, and the efficiency of virus penetration (20). EHV-1 gK and gM share additional characteristics such as N glycosylation, early-late expression kinetics, and the fact that they both depend on a viral complex partner for complete processing (20, 25). In the present study we sought to characterize the UL20 product to further investigate the specific roles of multiply hydrophobic proteins in EHV-1 replication and to address the question whether the special structural features might be related to common functional properties.
In contrast to the EHV-1 UL20 protein, which has not been studied before, the respective homologs of HSV-1 and pseudorabies virus (PRV) are well characterized. The structure of the PRV UL20p is still unknown, since no specific antibodies are available, but the HSV-1 protein is an unglycosylated, early-late structural protein which localizes to nuclear membranes and the Golgi apparatus (9, 32). The respective roles of the HSV-1 and PRV UL20 homologs overall are similar, but in detail they are clearly distinct. A UL20-negative PRV displayed a marked reduction in plaque sizes and replicated to decreased virus titers. Interestingly, the extent of these defects varied with the cell line used and was especially prominent on Vero cells. Electron microscopy of again Vero cells infected in the absence of UL20p revealed the accumulation of complete, enveloped particles in cytoplasmic vesicles, whereas all other stages of replication appeared normal (10).
Similarly, HSV-1 UL20p is important for final stages of virus replication (3, 8). However, whereas the UL20 product of PRV is suggested to support intracellular transport of mature virions to the cell surface (10), the HSV-1 UL20 protein is proposed to regulate membrane fusion involved in virion assembly (8). HSV-1 UL20p is thought to either support secondary envelopment or to prevent another putative de-envelopment step, i.e., the fusion of viral envelopes with membranes of their transport vesicles or with viral envelopes of other mature virions within these vesicles. These conclusions were drawn because the number of unenveloped capsids increased within the cytoplasm of Vero cells infected with various UL20 deletion mutants, and relatively few enveloped virions were observed. Also, deleting UL20 from HSV-1 syncytial strains resulted in accumulation of unenveloped particles in vesicles surrounded by two membranes (8). Taken together, both the PRV and HSV UL20 homologs function in a late step of virus egress in Vero cells, but the precise mechanisms seem to differ.
The primary aim of the present study was to assess whether UL20p might play a role in EHV-1 egress and whether a putative overlap in function with other multiply hydrophobic proteins like gK or gM would be detectable. An interaction of UL20p with gK was likely because (i) processing and localization of EHV-1 gK has been previously demonstrated to depend on the presence of (an)other as-yet-unidentified viral component(s) (20) and (ii) expression of the respective UL20 proteins is necessary for complete processing of gK in HSV-1 and PRV. Furthermore, the HSV-1 proteins have been shown to be interdependent for intracellular localization (6, 7, 9). Nevertheless, a putative physical interaction has not yet been proven, and the functional relevance of these findings is not yet understood.
To determine the precise function of the UL20 homolog in EHV-1 replication in Rk13 cells, an antiserum was raised allowing direct identification and in-depth analysis of the UL20 gene product. Physical and functional interactions were investigated by Western blot analysis and confocal laser scanning microscopy, and the presence of a stable complex dependent on gK and UL20p expression for formation and positive for UL20p and gK components was demonstrated. Interestingly, the defect of UL20 negative EHV-1 seems to affect even a later step in virus replication than that of the respective gK deleted mutant, suggesting that UL20p functions not only in complex with gK but also independently of the glycoprotein.
MATERIALS AND METHODS
Plasmids.
Plasmid ptL20 contains a 2.8-kb PstI fragment of EHV-1 strain RacL11 within vector pTZ18R and encompasses UL20 encoding, as well as upstream and downstream sequences (Fig. 1B). To either express a truncated UL20-mycHIS fusion protein in Escherichia coli or the full-length UL20 polypeptide in eukaryotic cells, either bp 1 to 208 or the complete 719-bp UL20 ORF (gene 41 [31]) was amplified by PCR. The respective primer sequences (5′-ATAGAATTCATGCCACAGGTATTAATG-3′,5′-ATATCTAGATGGGCAGAA AATGATGGG-3′, 5′-ATGGATCCAAGCTACGCTTAACGGAG-3′) included restriction enzyme recognition sites for insertion into either vector pBADgIIIB (pBAD20t; Invitrogen) or vector pcDNAI/Amp (pc20; Invitrogen). Correct amplification was confirmed by custom sequence analysis (MWG Biotech). Plasmid ptDp carries the 6.6-kb PstI-fragment of RacL11 DNA containing gene 71 and adjacent sequences (vector pTZ18R). Plasmids pcgM, pcgK, and pcgKGFP were designed to express the respective proteins and have been described elsewhere (20, 23).
FIG. 1.
The UL20-negative virus, L11Δ20, was constructed by mutagenesis of the EHV-1 BAC pRacL11 (24). (A) The BamHI map of pRacL11 is illustrated, and sequences of mini F plasmid pHA2 are highlighted as a black box. (B) The position of the UL20 ORF (gene 41) is given within the PstI fragment contained in plasmid ptL20, which was used to repair UL20 sequences in L11Δ20R. (C and D) The short linear DNA fragment depicted in panel B served for recombination in Escherichia coli, thereby replacing UL20 sequences with a FRT (FLP recognition target) site flanked kanamycin resistance gene (kan) (C), which was then removed (D). Binding sites of primers used for control sequencing are given. (E) A Kyte-Doolittle hydrophilicity plot (husar; GCG Software Package, Heidelberg, Germany) of the predicted amino acid sequence of EHV-1 UL20 (31) is shown. Arrows point to two N-glycosylation consensus sites, whereas the bar marks the region of the polypeptide used to generate an antiserum.
Cells and viruses.
Rabbit kidney cell line Rk13 was maintained as previously described (18). Cell line SC20 constitutively expresses the EHV-1 UL20 gene product and was isolated after cotransfection of Rk13 cells with plasmids pc20 and pSV2neo (18). UL20p expression of Geneticin-resistant cell clones was confirmed by immunofluorescence analysis using the UL20p-specific antiserum. EHV-1 strains RacL11 and RacH (11), including the recombinant viruses generated in the present study, were propagated on Rk13 or SC20 cells as indicated in the text. EHV-1 mutants deleted in gM (HΔgM-GFP), gK (HΔgK), or gp2 (L11Δgp2) have been described previously (20, 24, 29).
Generation of recombinant viruses.
The complete UL20 sequence was deleted in the bacterial artificial chromosome (BAC) clone of EHV-1 strain RacL11 (pRacL11 [24]) by using Red mutagenesis as described by Datsenko and Wanner (4). A linear DNA fragment was generated by PCR amplification of the kanamycin resistance (kanr) gene and flanking FRT-sites (FLP recognition target) of plasmid pKD13 (4) (Fig. 1B). Besides kanr recognition sequences, the respective primers included sequences complementary to those up- and downstream of UL20 (primer 1, 5′-GAC-TAG-TCC-ATA-CCC-TGC-ACC-GCT-CGC-AGG-CTG-CCA-GAA-ATA-TTT-CTC-TCC-GAA-TTT-TTG-AGG-GTT-GGA- GGT-GTA-GGC-TGG-AGC-TGC-TTC-3′; primer 2, 5′-TCT-CTT-CTC-ACG-TTT-TAT-ACT-GTC-AGT-CGG-CCG-CTA-CCC-CAG-TGG-TGC-CTC- CCA-CGG-AGG-CAC-CCC-TAA-TTC-CGG-GGA-TCC-GTC-GAC-CT-3′). After transformation of the PCR product into E. coli cells harboring pRacL11, colonies containing an insertion of the kanr gene were selected on kanamycin-containing agar plates. The inserted kanr sequences were then removed by conditionally expressing the FLP recombinase in these cells, and a UL20-deleted pRacL11, pRacL11Δ20, resulted that carried an insertion of 82 bp only (Fig. 1D). The final UL20 negative virus, L11Δ20, was plaque purified after cotransfection into SC20 cells DNA of pRacL11Δ20 and of plasmid ptDp, which contains the RacL11 sequences necessary to replace the mini F plasmid pHA2 and to repair gene 71 (24). The UL20-negative but gp2-positive (gene 71 repaired) L11Δ20 virus was used for generating a UL20 revertant virus, L11Δ20R, by homologous recombination of L11Δ20 with plasmid ptL20 in Rk13 cells. DNA of all viruses constructed in the present study was examined by PCR amplification (primer, 5′-CGGTTATGTAGAACACAA-3′ and 5′-GTACCGGAATTTTGAACG-3′) and nucleotide sequencing, by restriction enzyme fragment analysis and Southern blotting with a digoxigenin-labeled PstI fragment contained in ptL20 as a probe (22).
Western blotting.
Cells were infected at a multiplicity of infection (MOI) of 2 (unless stated otherwise) with the respective viruses and lysed at the indicated times postinfection (p.i.). After addition of sample buffer containing sodium dodecyl sulfate (SDS) and 2-mercaptoethanol (26), samples were usually either heated to 99°C for 5 min or kept on ice throughout relative to the tendency of the protein of interest to aggregate upon boiling. Proteins were separated by SDS-12% polyacrylamide gel electrophoresis (PAGE) and detected by Western blotting as previously described (22). Apparent Mrs were exactly calculated in relation to a precision protein standard (Bio-Rad), but in most experiments another protein marker (Biolabs) was used running slightly different. Extracellular virions were purified from supernatants of infected Rk13 cells (MOI = 2; 16 h p.i.) by repeated centrifugation through a 30% sucrose cushion (22).
Purified virions were split into two fractions by incubation at 45°C in buffer containing Triton X-100 (1%; 20 min) and pelleting of capsids and attached proteins at 45,000 × g for 30 min. Supernatants that contained envelopes and solubilized tegument proteins were carefully separated from pellets. After another washing step, the pellet was resuspended in lysis buffer.
To remove N-linked carbohydrates, virion preparations (5 μg) were incubated in the presence or absence of 1 U of PNGase-F (16 h, 37°C; Roche Molecular Biochemicals) in deglycosylation buffer (100 μl [13]). O-linked carbohydrates were digested after removal of sialic acid side chains. To do this, virions (5 μg) were first incubated with 1 U of neuraminidase (1 h, 37°C; Roche Molecular Biochemicals) in a buffer (100 μl) containing sodium acetate (pH 5.2, 50 mM) and calcium chloride (4 mM) and then pelleted and suspended in a Tris-phosphate buffer (20 mM, pH 7.4, 100 μl). These preparations were then incubated in the presence or absence of O-glycosidase (2 U, 16 h, 37°C; Roche Molecular Biochemicals).
Indirect immunofluorescence.
For indirect immunofluorescence studies, cells grown on glass coverslips were either directly used, infected as indicated, or transfected with DNA of the given plasmids. At the stated times postinfection or transfection cells were fixed with 2% paraformaldehyde (15 min), permeabilized with Triton X-100 (0.1%; 10 min) unless stated otherwise, and processed by standard procedures (22). Concanavalin A (ConA) directly labeled with Alexa Fluor 594 was used to track the endoplasmic reticulum (2 μg/ml, 30 min). Fluorescence of samples was digitally documented by using either conventional fluorescence microscopy (Zeiss Axioskop; Spot Advanced Software, Diagnostic Instruments) or an LSM510 laser scanning microscope (LSM510 Software; Zeiss). Pictures were adjusted in Adobe Photoshop 7.0, and figures were assembled by using CorelDraw 7.
Antibodies.
To generate EHV-1 UL20 specific antibodies, amino acids 1 to 69 of the UL20 polypeptide were expressed in E. coli as a mycHis fusion protein using the pBADgIII expression system (Invitrogen) and purified by affinity chromatography according to the manufacturer's instructions. Two rabbits were immunized five times in 3-week intervals with 50 to 100 μg of the purified protein mixed with Freund adjuvant.
The monoclonal anti-major capsid protein antibody ZB4 (kindly provided by G. B. Caughman), anti-gB monoclonal antibody (MAb) 3F6 (1), anti-gB MAb 4B6 (17), anti-gM MAb F6 (5), and anti-gp2 MAb 3B12 (17), as well as rabbit polyclonal antisera directed against gM (28), UL34p (19), and UL45p (22), were used in the present study. Antibodies directed against green fluorescent protein (GFP; rabbit serum) and Alexa Fluor 546- or 488-conjugated anti-rabbit or anti-mouse immunoglobulin G (IgG) secondary antibodies were purchased from Molecular Probes; antibodies to γ-adaptin (clone 100/3) and peroxidase-conjugated anti-rabbit or anti-mouse IgG secondary antibodies were from Sigma.
Virus growth kinetics, penetration assays, and plaque size measurements.
Rk13 or SC20 cells in six-well plates were infected with the indicated viruses (MOI = 5). After a 60-min period of adsorption at 4°C, inocula were removed, and attached viruses were allowed to penetrate for another 60 min at 37°C, followed by inactivation of remaining extracellular virus with a citrate buffer (40 mM citric acid, 10 mM KCl, 135 mM NaCl; pH 3.0). At the indicated time points p.i., supernatants and infected cells, which were again treated with low pH to achieve inactivation of contaminating extracellular infectivity, were collected separately. Virus titers were determined independently by plaque titration on Rk13 or SC20 cells.
Penetration assays were performed as previously described (18). The efficiency of penetration was determined by the number of plaques present after citrate buffer treatment relative to the number of plaques present after control treatment (phosphate-buffered saline) at various times after the temperature was shifted from 4°C (adsorption) to 37°C.
Sizes of the virus plaques were examined on Rk13 or SC20 cells (50 PFU/well) at day 3 under a methocellulose overlay. Plaque areas were measured by using the SPOTadvanced software (Diagnostic instruments), and mean areas of respective plaques were set in relation to mean areas of parental RacL11 plaques (100%). Representative plaques were immunofluorescently labeled for gB and digitally documented by using a Zeiss Axioskop.
Electron microscopy.
Rk13 cells were infected with the indicated viruses (MOI of 2) and prepared for electron microscopic investigations at 16 h p.i. as previously described (19). Briefly, cells were fixed in 5% glutaraldehyde and 4% formaldehyde (pH 7.4, 2 h) and then washed with 0.1 M sodium phosphate buffer (Ph 7.4) before they were postfixed with 1% OsO4-0.8% K3Fe(CN)6 in 0.1 M sodium phosphate buffer (2 h). All samples were stained with 2% aqueous uranyl acetate for 90 min, dehydrated in graded ethanol, and finally embedded in ERL 4206. Thin sections were stained with uranyl acetate and lead citrate and examined with a transmission electron microscope EM 10C/CR (Zeiss, Oberkochen, Germany) at 60 kV. Pictures were digitally scanned and assembled by using Adobe Photoshop 7.0.
RESULTS
Construction of an EHV-1 UL20 deletion mutant.
To investigate the function of the UL20 product in EHV-1 replication, a UL20-deleted RacL11 was generated by using BAC-mutagenesis. In a first step, the complete ORF was removed from the pRacL11 BAC clone in E. coli (24), resulting in recombinant clone pRacL11Δ20 (Fig. 1D). In pRacL11, the mini F plasmid pHA2, which contains a GFP cassette, replaces gene 71 encoding for gp2 (Fig. 1A) (24). To, in a second step, reconstitute in eukaryotic cells a virus carrying the UL20-deletion only, UL20p-expressing SC20 cells were cotransfected with DNA of pRacL11Δ20 and of plasmid ptDp. GFP-negative progeny virus was purified to homogeneity, and gp2 expression of the isolated L11Δ20 was confirmed by immunofluorescence analysis with a gp2-specific antibody (17; data not shown). In addition, L11Δ20R, the respective UL20 repair virus, was isolated in Rk13 cells. The generated recombinant viruses were genetically characterized as explained in Materials and Methods to confirm their genotypes and to exclude spurious modifications (data not shown) before they were functionally assessed in the course of the present study.
The EHV-1 UL20 protein is involved in a late step of virus replication.
To gain basic information on the replication of the generated recombinant viruses, one-step growth kinetics of RacL11, L11Δ20, and L11Δ20R were compared on Rk13 cells and on SC20 cells (Fig. 2). The L11Δ20 virus stocks used in these experiments were propagated on complementing SC20 cells. Virus titers in supernatants (extracellular infectivity) and of cells (cell-associated infectivity) were determined at various times p.i. On Rk13 cells (Fig. 2A) the growth properties of parental RacL11 virus and the UL20 repaired virus were virtually undistinguishable at all times p.i. and were similar to those of L11Δ20 until 8 h p.i. At later times, cell-associated infectivity of L11Δ20 remained almost unaffected and was reduced by <5-fold compared to parental and revertant virus (at24 h p.i., 1.8-fold for L11Δ20 versus RacL11). In contrast, the production of extracellular infectious progeny was inhibited drastically. The relative reduction in virus titers in supernatants increased over time, and mean titers were 415-fold lower compared to those of parental virus at 12 h p.i., and 1,527-fold reduced at 24 h p.i. (L11Δ20 versus RacL11). Taken together, although L11Δ20 produced significantly less extracellular infectivity, a compensatory increase in intracellular infectious virus was not observed. Parallel analysis on SC20 cells (Fig. 2B), however, demonstrated significant in trans complementation for virus growth by constitutive expression of UL20p. On these cells the maximum observed difference in extracellular L11Δ20 to RacL11 titers at 24 h p.i. was 3.6-fold, allowing the conclusion that the observed phenotype was specific for the deletion of the UL20 gene. The data suggested a function of the EHV-1 UL20 homologous protein in a very late step of the viral replication cycle—after completion but without accumulation of intracellular infectious virus progeny.
FIG. 2.
One-step growth kinetics of RacL11 (▪), L11Δ20 ( ), or L11Δ20R (○). Rk13 (A) or SC20 cells (B) were infected at an MOI of 5, and extracellular and cell-associated virus titers were determined independently at 0, 4, 8, 12, and 24 h p.i. Shown are means of three individual experiments. Standard deviations are given above symbols as error bars.
), or L11Δ20R (○). Rk13 (A) or SC20 cells (B) were infected at an MOI of 5, and extracellular and cell-associated virus titers were determined independently at 0, 4, 8, 12, and 24 h p.i. Shown are means of three individual experiments. Standard deviations are given above symbols as error bars.
L11Δ20 plaques are markedly reduced in size.
The ability to form large plaques in cell culture is generally accepted as a marker for efficient cell-to-cell spread of infectivity, a mechanism which is not yet fully understood and which seems to be affected by many viral components (12). To address the implications of UL20p on this viral phenotype, the sizes of plaques induced by RacL11, L11Δ20, or L11Δ20R were determined on Rk13 or SC20 cells.
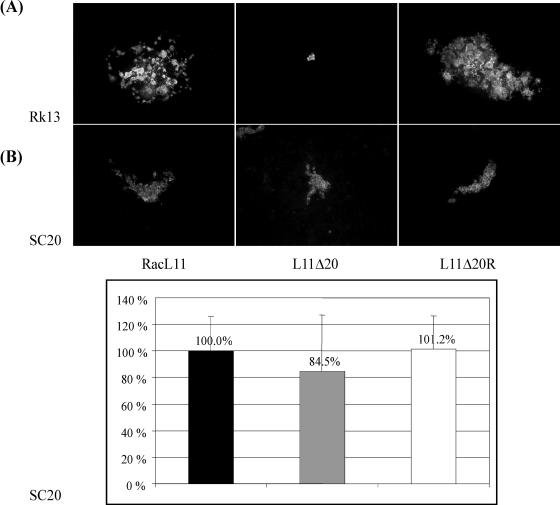
Representative plaques visualized by gB-specific immunofluorescence are depicted in Fig. 3. Interestingly, although UL20p obviously functions in a final egress step, the deletion also substantially affected cell-to-cell spread as indicated by the formation of minute plaques. Plaque formation was complemented, albeit not completely, on SC20 cells and restored by L11Δ20R, confirming the direct link between UL20p expression and plaque sizes. Plaque areas of all viruses were markedly smaller on SC20 cells than on Rk13 cells. This observation could be related to a specific function of UL20p as discussed below but could also be a consequence of an intrinsic property to the selected cell clone. In conclusion, the deletion of the UL20 protein resulted in a substantial decrease in cell-to-cell spread capability.
FIG. 3.
Plaque phenotype of RacL11, L11Δ20, or L11Δ20R. Rk13 (A) or SC20 cells (B) in six-well plates were infected with 50 PFU of the respective viruses per well. At day 3 p.i. plaques were immunofluorescently labeled (anti-gB MAb 4B6), and photographs were taken. Representative pictures of individual plaques are shown. Plaque areas were measured on SC20 cells with the help of the SPOTadvanced software. Mean plaque areas of RacL11 were set to 100%. Standard deviations are depicted above bars.
L11Δ20 penetrates as efficiently into Rk13 cells as wild-type virus.
As other multiply hydrophobic proteins of EHV-1, gK and gM, facilitate virus penetration (20, 23), the influence of deleting UL20 sequences on this specific step was assessed. For these assays all virus stocks were produced of Rk13 cells. Surprisingly, and in contrast to the viruses with either gK or gM deleted, the UL20-negative mutant was protected from extracellular acid treatment as efficiently as wild-type virus (data not shown). It was therefore concluded that virus penetration is not affected by deleting UL20.
L11Δ20 capsids accumulate within the cytoplasm of infected cells.
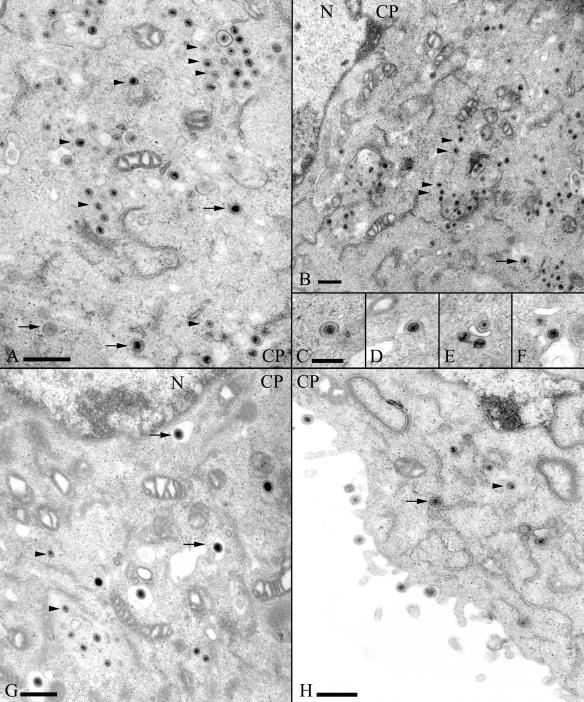
Analysis of L11Δ20 growth in Rk13 cells indicated a defect in a very late step in virus egress. To more precisely define the role of UL20p in virus assembly, Rk13 cells infected with RacL11, L11Δ20, or L11Δ20R were processed for electron microscopic analysis at 16 h p.i. This time point was chosen according to the onset of the marked decrease in extracellular infectivity observed in growth kinetics (Fig. 2A). Ultrastructurally, the repaired virus behaved in a way that was indistinguishable from parental RacL11 (data not shown; Fig. 4G and H). In contrast, several irregularities were noted in Rk13 cells infected with L11Δ20. (i) Extracellular particles were completely absent (data not shown), corroborating the suggested block in egress. (ii) Within the cytoplasm, enveloped particles in vesicles (arrows) were observed, rarely more than one particle per vesicle, and the number of unenveloped cytoplasmic capsids (arrow heads) was clearly increased. (iii) Upon examination at higher magnifications, particles appeared to be stalled during budding steps at cytoplasmic vesicles (Fig. 4A to F).
FIG. 4.
Electron micrographs of Rk13 cells infected with L11Δ20 (A to F) or with L11Δ20R (G to H) (both at 16 h p.i.). Different kinds of particles are demonstrated within the cytoplasm of cells (CP), and some enveloped particles are designated by arrows, whereas arrowheads point to examples of unenveloped cytoplasmic capsids. Inlays C to F depict particles during “budding,” as frequently observed in cells infected with L11Δ20. Bars represent 500 nm in A, B, G, and H and 250 nm in C.
To confirm these observations by quantitative data, virus particles within 20 random thin sections of cells infected with L11Δ20 or L11Δ20R were counted. The direct comparison of particle counts was slightly hampered by the striking difference in numbers of polynucleated cells. Whereas cells frequently harbored more than one nuclear section after infection with L11Δ20R or RacL11 (data not shown), such cells were completely absent after infection with L11Δ20. Nevertheless, some differences were obvious, and the results are summarized in Table 1. Particles clearly accumulated within the cytoplasm of L11Δ20-infected cells and were unable to exit. In addition, a statistically significant increase in particles captured in the process of fusion with vesicles was observed. Based on the images, directionality of processes cannot be assessed and, in the absence of UL20p, there either is a defect in secondary envelopment, i.e., the budding of particles into vesicles, or a defect of egress thereafter, i.e., the refusion of viral envelopes with membranes of vesicles.
TABLE 1.
Mean particle numbers in sections of cells infected with L11Δ20R or L11Δ20
| Virus | Mean particle no. (%)a
|
|||||
|---|---|---|---|---|---|---|
| Capsids in nuclei | Capsids between nuclear membranes | Capsids in cytoplasm | Budding particles | Virions in vesicles | Free virions | |
| L11Δ20R | 40.1 (79.6) | 0.5 (1.0) | 3.65* (7.2) | 0.75* (1.5) | 2.15 (4.3) | 3.2* (6.4) |
| L11Δ20 | 33.4 (55.2) | 1.35 (2.2) | 16.4* (27.1) | 7* (13.5) | 1.2 (2.0) | 0.0* (0.0) |
Values are mean particle numbers; relative numbers of particles are given in parentheses as a percentage. *, Differences in mean numbers were significant (P < 0.05) as assessed by pairwise testing according to the Student's t test.
Identification of the EHV-1 UL20 protein.
The EHV-1 UL20 polypeptide sequence contains several hydrophobic stretches and is predicted to span membranes up to four times. To identify the UL20 gene product, an antiserum was raised in rabbits against the relatively hydrophilic amino-terminal portion (amino acids 1 to 69) of the polypeptide (Fig. 1E). In Western blot analysis of cells infected with RacL11, the generated serum reacted mainly at three different molecular sizes; two of these reactivities disappeared upon denaturation at 99°C (Fig. 5A and B), suggesting instability or aggregation of hydrophobic proteins. Since these protein species at Mrs of 25,000 and 75,000 were not detected in cell lysates of the UL20-negative L11Δ20 (Fig. 5C), they were considered specific for UL20 expression. In contrast, the Mr 175,000 reactivity was still reactive with the anti-UL20 serum in L11Δ20-infected cells and therefore appeared to be unspecific. To further investigate the properties of the observed proteins, samples were prepared under various conditions before SDS-PAGE and Western blot analysis. These conditions included (i) exposure to different temperatures, (ii) the use of sample buffer with or without addition of 2-mercaptoethanol, and (iii) cycles of freezing and thawing before the addition of sample buffer and immediate analysis. The latter treatment was included to determine cell lysate stability at −20°C. The results are summarized in Fig. 5. The reactivity of the Mr 25,000 protein was quite robust and only sensitive to heat denaturation at 99°C, whereas the Mr 75,000 protein appeared less stable. Although its detection was independent of the presence of 2-mercaptoethanol, it disappeared after heat denaturation and also after repeated freezing and thawing of samples. Samples intended for the investigation of UL20p were therefore kept on ice throughout in the following experiments, and freezing was avoided. Taken together, UL20p specific reactivity was identified in two relatively stable forms of 25 and 75 kDa in EHV-1-infected cells by Western blot analysis. The exact nature of these proteins seemed puzzling and was further characterized and is discussed below.
FIG. 5.
(A and B) Western blot analysis of Rk13 cells, uninfected (Rk) or infected with RacL11 for 14 h (L), that were assayed for UL20p after various treatments. Lysates of cells were either resuspended in the usual sample buffer containing 2-mercaptoethanol (A) or in buffer without the reducing agent (B) and either kept on ice (4°C), heated to 56°C for 2 min, or heated to 99°C for 5 min. Finally, lysates were either analyzed immediately after harvest (dir) or after having been subjected to one (1x) or two (2x) cycles of freezing and thawing before addition of sample buffer. (C) Rk13 cells, uninfected (Rk) or infected with RacL11 (L), L11Δ20 (Δ), or L11Δ20R (R), were lysed at 14 h p.i. and kept on ice for detection of either gM or the UL20 protein. In contrast, samples intended for analysis of the mcp (major capsid protein), UL45p, UL34p, or gB were denatured by heating. Parallel blots were probed with antibodies against the given proteins. Sizes of a prestained molecular weight marker (Biolabs) are given in thousands.
Beyond the lack of UL20 protein expression demonstrated in lysates of Rk13 cells infected with L11Δ20, the Western blot analyses (Fig. 5C) also revealed complete restoration of UL20p expression in the repaired L11Δ20R virus. In addition, the correct synthesis and processing of several other EHV-1 proteins by L11Δ20 was confirmed in parallel blots probing for the major capsid protein, which is encoded by the adjacent UL19 ORF, for UL34p, gB, gM, and the true-late, glycosylated UL45p. It could therefore be concluded that deleting UL20 sequences did not appreciably influence synthesis and processing of a representative number of early- and true-late EHV-1 proteins.
The EHV-1 UL20 protein is incorporated into the viral envelope.
Both forms of UL20p were detected in virions collected from supernatants of RacL11-infected cells by Western blotting. The purity of these preparations was demonstrated by the absence of nonstructural proteins such as UL34p or UL45p and the exclusive presence of the mature form of gB (19, 22, 30) (Fig. 6). Virion proteins were divided into two fractions relative to their Triton X-100 solubility (45°C). One fraction represented soluble components of the viral envelope and tegument, while the other fraction contained proteins resistant to detergent dissociation from capsids. Similar to gB and in contrast to the major capsid protein, UL20p was found to be completely solubilized, indicating its association with viral envelopes. UL11p, a component of the tegument (27), was detected in both fractions (Fig. 6B). However, after separation of virions into fractions significantly less UL20p seemed to be reactive. This loss of reactivity could be consequence of heating the samples to 45°C and partial aggregation of UL20p.
FIG. 6.
(A) RacL11-infected cells (L-ic) or virions (L-v) purified from supernatants of infected cells were subjected to SDS-12% PAGE and analyzed for the presence of gB, UL34p, or UL45p. (B) The same virion preparations (L-v) were separated into soluble envelope proteins (s) or pelleted capsid associated proteins (p), and parallel blots were probed for UL20p, the envelope protein gB, the major capsid protein (mcp), or the tegument protein UL11p (SDS-15% PAGE). (C) Purified virions (L-v) were subjected to deglycosylation with either N-glycosidase F (-NGF) or O-glycosidase (-OG). Parallel blots were incubated with the anti-UL20-serum (UL20p) or the anti-gB MAb 3F6 (gB), as indicated. Molecular weight marker sizes (Biolabs) are given in thousands.
For the 239-amino-acid UL20 polypeptide an Mr of about 26,500 is predicted (31). This corresponds nicely to the Mr 25,000 form detected in Western blots. The slower-migrating form could result from either processing or represent a UL20p oligomer. Although UL20p contains two putative N glycosylation consensus sites, no cleavable signal sequence was detected (Fig. 1E). In addition, the corresponding, amino-terminal portion of the HSV-1 UL20 homolog is known to protrude into the cytoplasm and the protein not to be glycosylated (9, 32). To address the possible glycosylation of UL20p, several deglycosylation experiments were performed, and the results are summarized in Fig. 6C. Whereas the faster-migrating UL20p species was not affected, removal of N-linked carbohydrates resulted in an increased electrophoretic mobility of the Mr of 75,000 reactivity, shifting to an Mr of 60,000. Removal of O-linked sugar moieties had no detectable effect, although the expected shifts of the electrophoretic mobility of gB were observed by either treatment.
These data could therefore either indicate N glycosylation of the UL20 polypeptide and leave the remaining difference in migration of these protein species unexplained or could indicate formation of a heterooligomer between the UL20 protein and another glycosylated protein.
UL20p is present in a complex containing gK.
To further assess potential hetero-oligomerization of UL20p with (an)other viral protein(s), expression of UL20p in the absence of selected viral proteins was assessed. SC20 cells that constitutively express UL20p were infected with different EHV-1 deletion mutants, and the UL20p reactivity was analyzed by Western blotting (Fig. 7). UL20p expressed by uninfected SC20 cells was generally hardly detectable, and the high-molecular-weight UL20p specific reactivity was not observed at all, not even after prolonged exposure (data not shown), whereas infection with L11Δ20, parental RacL11, or a gM-deleted EHV-1 (HΔgM-GFP [29]) resulted in its formation. In contrast, after infection with a gK-negative virus (HΔgK [20]), the Mr of 75,000 form was clearly absent and was replaced by an Mr of about 105,000 after infection with HgKGFP. The latter virus expresses a gK-GFP fusion protein instead of authentic gK (20). Interestingly, the Mr 25,000 UL20p observed after infection with HΔgK was weaker than that after infection with the other viruses, although other proteins were expressed at apparently normal levels (Fig. 7A) (20). This observation and the difficulty to detect UL20p in uninfected SC20 cells might indicate instability of UL20p in the absence of gK and will be addressed in future experiments. Finally, in SC20 cells another UL20p specific reactivity was also noted that ran only slightly slower than the usual form at 25,000. This additional reactivity was usually even dominant in lysates of SC20 cells infected with LΔ20 (Fig. 7A, right blot). Currently, we do not know what might cause this additional shift in the apparent molecular weight, but alterations either in further modifications such as phosphorylation or in protein unfolding, caused by an imbalance between the complex forming proteins, might serve as putative explanations.
FIG. 7.
(A) SC20 cells were either lysed directly or after infection (16 h) with L11Δ20, RacL11, HΔgK, HΔgM, or HgKGFP and probed either for UL20p or gM. Two independent blots of lysates of SC20 cells and of SC20 cells infected with L11Δ20, respectively, are shown. (B) Accordingly, TCgKGFP cells were either lysed directly or after infection (16 h) with L11Δ20, RacL11, HΔgK, HΔgM, or HgKGFP, and blots were incubated with either the anti-GFP or the anti-gM antibody. (C) Virion preparations of RacH (H-v) or HgKGFP (HgKGFP-v) were analyzed for gK-GFP, UL20p, or gB using the anti-GFP polyclonal, the anti-gB monoclonal 3F6, or the anti-UL20 serum. Molecular weight marker sizes (Biolabs) are given in thousands.
In another series of experiments the reciprocal approach was chosen. As no antiserum is available to directly detect EHV-1 gK, the GFP-tagged version was used. The gK-GFP fusion protein can complement for all functions of EHV-1 gK, and its complete processing and TGN localization depends on the presence of one or more other viral components (20). The fusion protein is constitutively expressed by cell line TCgKGFP. After TCgKGFP cells were infected with parental RacL11 or with viruses deleted in gK, gM, or UL20p, it was shown that gK-GFP required UL20p not only to fully mature to a glycoprotein of Mr 55,000 to 65,000 (20) but also to form the hetero-oligomer (Fig. 7B; at an Mr of about 105,000 in SDS-12% PAGE).
Because the Mr 105,000 protein band was relatively hard to distinguish from the superimposing unspecific signal at the top of the gel in cells infected with HgKGFP (Fig. 7A) and because the putative hetero-oligomeric form seemed to be preferentially incorporated into virions (Fig. 6B), the data were corroborated by analyzing purified virions. Extracellular RacH and HgKGFP virions were tested for the presence of several EHV-1 proteins by Western blot analysis. As shown in Fig. 7C, the formation of a complex containing both UL20p and gK was confirmed by the fact that the Mr 75,000 UL20p-containing form disappeared and was replaced by the slower-migrating form in HgKGFP particles (Mr 105,000). In a parallel blot, a protein reacted at exactly the same position on HgKGFP virions within the gel when probed for GFP. Since RacH virions do not contain any GFP or GFP fusion protein, no reaction was observed in this lane. Together, these findings show that for formation of the Mr 75,000 UL20p-containing protein not only viral infection but also specifically the presence of EHV-1 gK was necessary, and we concluded that the Mr 75.000 protein represents a hetero-oligomeric complex containing UL20p and gK. Since UL20p alone runs at an apparent Mr of 25,000, the additional Mr of about 50,000 might represent mature gK and therefore suggests a one-plus-one stoichiometry of the hetero-oligomeric complexes components. Nevertheless, these calculations have to be interpreted very carefully, since the multiple hydrophobic proteins of EHV-1 are very sensitive to variations in conditions. It is conceivable that the components are denatured differently and thus appear at different relative Mrs, when detected in an oligomer as opposed to when detected separately.
The EHV-1 UL20 protein is regulated with early-late kinetics and associates with cellular membranes.
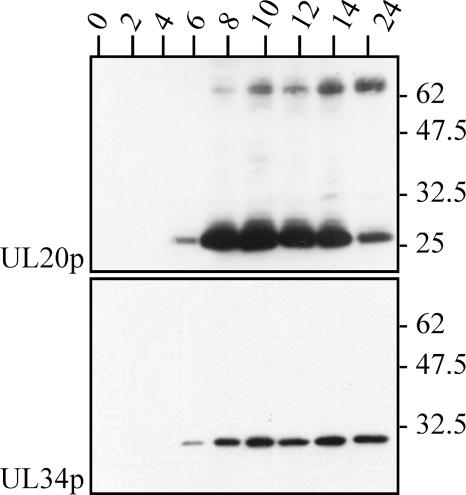
Western blot analysis of time course experiments demonstrated that expression kinetics corresponded well to those of UL34p (Fig. 8). The Mr 25,000 UL20p moiety was noted as early as 6 h p.i., whereas the Mr 75,000 form of UL20p was first detectable at 8 h p.i. and its intensity seemed to increase with time. No intermediate form appeared. These findings suggested that expression of UL20p occurred with early-late kinetics, which was confirmed in experiments with phosphonoacetic acid, an inhibitor of de novo viral DNA synthesis (data not shown).
FIG. 8.
Western blot analysis of Rk13 cells that had been infected with RacL11 at an MOI of 2. Cells were lysed at the time points p.i. (indicated in hours), frozen, and thawed immediately before use. Blots were probed with antisera specific for UL20p or UL34p. Sizes of the molecular weight marker (Biolabs) are indicated in thousands.
To address the subcellular localization of the UL20 protein, indirect immunofluorescence analyses were performed. Unfortunately, direct analysis of EHV-1-infected cells for UL20p was inconclusive because at least one additional protein was shown to be strongly reactive with the anti-UL20-serum, as determined with cells infected with L11Δ20 (data not shown). To avoid any cross-reactivity, Rk13 cells were transfected with plasmid pc20 and immunofluorescently labeled by using the anti-UL20-serum. Specific fluorescence was distributed throughout the cytoplasm in a bright granular or netlike pattern that mostly colocalized with the signal of the ConA, which was used as an endoplasmic reticulum (ER) marker, but not with a trans-Golgi network (TGN) marker protein, γ-adaptin (Fig. 9A).
FIG. 9.
Rk13 cells were transfected, individually (A) or in different combinations (B), with DNA of plasmids expressing the proteins indicated at the left border of the respective pictures. At 48 h after transfection cells were fixed as explained in the text. gK-GFP was detected by its autofluorescence (green), while pUL20, gM, or γ-adaptin were immunofluorescently stained with the respective antibodies (anti-UL20-serum, anti-gM-MAb F6, anti-γ-adaptin MAb) and reacted with Alexa Fluor 546 (red)- or 488 (green)-conjugated anti-rabbit or anti-mouse IgG secondary antibodies. To mark the ER region, directly labeled (Alexa Fluor 594, red) ConA was used. Slides were analyzed and digitally documented by using a confocal LSM510 microscope and the respective software (Zeiss). Representative examples of planes through single cells are given.
The subcellular localization of UL20p depends on expression of gK.
To address a putative influence of the interaction of UL20p with gK on the subcellular localization, Rk13 cells were cotransfected with plasmids encoding for UL20p and either gM, gK, or the gK-GFP fusion protein. Partial colocalization but no changes in the UL20p fluorescence pattern were observed upon coexpression of gM, whereas a clear redistribution of UL20p-specific fluorescence was noted when either gK or gK-GFP was cotransfected. gK-GFP was detected by its autofluorescence, but expression of authentic gK could not directly be confirmed, since no specific anti-EHV-1 gK-sera are available. For this reason only the data obtained with the gK-GFP fusion protein are given. In cells positive for gK-GFP and UL20p, an accumulation of both specific fluorescence signals was noted in a confined area, reminiscent of the Golgi apparatus (Fig. 9B). To demonstrate that this area indeed represented parts of the Golgi network, cells expressing UL20p and gK-GFP were first judged for gK-GFP expression by using an Axiovert microscope, fixed with acetone to destroy GFP fluorescence, and then stained for UL20p and γ-adaptin. Changing the fixation procedure (acetone versus paraformaldehyde-Triton) did not appreciably alter the distribution of UL20p fluorescence (compare lines 2, 3, and 4 in Fig. 9B), but coexpression of gK-GFP clearly redistributed the UL20p signal to the TGN. From these data we concluded that (i) coexpression of gM with UL20p had no impact on the localization of UL20p, (ii) expression of UL20p did not influence the ER-retention of gM expressed in the absence of its complex partner pUL49.5 (25), and (iii) the specific localization of EHV-1 gK (gK-GFP) and UL20p to the trans-Golgi network required coexpression of the proteins and therefore suggested protein interaction.
DISCUSSION
The present study was designed to structurally and functionally examine the EHV-1 UL20 protein. The salient findings were (i) that UL20p and gK are present in a hetero-oligomeric complex, (ii) that both proteins are interdependent on each other for proper localization within transfected cells and that UL20p is necessary for complete processing of the glycoprotein, and (iii) that the presence of EHV-1 UL20p is important for late events in virus egress.
The role of UL20p in EHV-1 replication in vitro was investigated by characterizing properties of a virus with UL20 deleted. Low extracellular virus titers observed after infection with the L11Δ20 virus were morphologically reflected by the absence of free extracellular virions, while “normal” intracellular titers were reflected by the presence of enveloped virions in cytoplasmic vesicles. The apparent loss of infectious virus particles correlated with an accumulation of naked capsids within the cytoplasm of L11Δ20-infected cells. Based on the findings of relatively high cell-associated infectivity but large numbers of nonenveloped particles in the cytoplasm of infected cells, we concluded that UL20p-negative particles fail to execute proper exit that occurs after secondary envelopment. The observed increase in particles captured during fusion with vesicles could theoretically be interpreted in several ways, but based on the observations just mentioned it may indicate that EHV-1 UL20p is required to prevent refusion of mature virions with transport vesicles in Rk13 cells.
Moreover, the inefficient egress of L11Δ20 virions seemed to even be reduced with time of infection, likely by the gradual loss of cellular transport dynamics. Two dynamic processes could theoretically determine the outcome of virion transport in the absence of UL20p, the velocity of de-envelopment versus that of particle transport. If transport was faster than refusion could occur, UL20p would be unimportant. We speculate that the speed of transport in Rk13 cells decelerates as infection proceeds, allowing more and more particles to “reversely penetrate” from transport vesicles into the cytoplasm, while fewer and fewer particles are able to exit cells. Similar dynamics could also explain the apparent cell-dependent differences in the importance of the HSV-1 or PRV UL20 proteins (3, 10). Foster et al. (8) already stated that HSV-1 UL20p either functions in secondary envelopment or prevents de-envelopment of virions in cytoplasmic vesicles and reported an ultrastructural phenotype of UL20-negative HSV-1 (KOS/UL20-null) in Vero cells that is very similar to that shown for EHV-1 in the present study. Our data finally allow suggesting a possible role for UL20p in preventing de-envelopment, at least in EHV-1-infected cells. In contrast, the function of UL20p by itself in the case of PRV seems to differ as no unusual fusion events were observed (10). The transport of cytoplasmic vesicles containing enveloped particles was inhibited during infection with UL20-negative viruses. However, PRV gK is suggested to prevent fusion very late in infection, i.e., the reentry of mature particles into the producing cell (13). In the light of these and our findings, the role of UL20p cannot be reasonably discussed any further without taking gK also into account.
Using a UL20p specific antiserum, two main UL20p moieties were specifically detected in lysates of EHV-1-infected cells. Whereas the Mr 25,000 moiety was in good agreement with the predicted size of the polypeptide, the origin of the slower-migrating form of Mr 75,000 seemed enigmatic at first but was then clearly demonstrated to represent a physical complex containing EHV-1 gK and UL20p. We have not formally proven the stoichiometry of the complexing partners, but intermediate forms were never noted and the size of the observed hetero-oligomer seems to indicate that it contains gK and UL20p in a one to one ratio.
In another series of experiments the subcellular localization of UL20p in Rk13 cells was assessed and shown to change upon coexpression of gK. Again, a defined ratio of gK to UL20p seemed necessary to effectively localize both fluorescence signals to the Golgi network (data not shown). The physical interaction between gK and UL20p appears to be important for correct localization of UL20p within cells and for complete processing and localization of gK (20). Similar functional dependence between the two protein homologs has been shown in PRV and HSV-1 (6, 9).
In transcomplementation studies we observed that defects in L11Δ20 replication were repaired by UL20p expressed by SC20 cells, which confirmed that no other ORF was appreciably affected by the mutagenesis procedures. However, defects were not completely restored on SC20 cells, likely caused by the fact that not all cells of this cell line express UL20p as assessed by immunofluorescence analysis (data not shown). In addition, the relative levels of UL20p expression might play a role, because they could influence complex formation between UL20p and gK. Interestingly, plaques were generally smaller on SC20 cells even with parental viruses, and fusion of cells was rarely observed (Fig. 3). This could indicate an inhibition of fusion by UL20p in processes other than vesicle transport, when overexpressed or when present during early infection (HCMV-IE promoter). These observations are supported by the findings that UL20p of HSV-1 and PRV are both able to inhibit fusion of cells in transient fusion assay, albeit to lesser extents than when expressed in combination with gK (2, 14).
Taken together, these findings indicate that the regulation of fusion seems to be the overall function of the gK-UL20p complex, not only by negatively regulating but also by promoting fusion events, since both gK and UL20p seem to be required for syncytium formation and as gK facilitates virus penetration (20; this study). The effect of both homologous proteins on syncytia formation has been addressed and discussed in several HSV-1 studies (9, 16).
Another interesting question was how the defect in direct cell-to-cell spread in the absence of UL20p could be explained. This defect was clearly unrelated to the efficiency of penetration, and the virus with UL20 deleted penetrated as fast as wild-type viruses. This observation was intriguing by itself because the interaction partner of UL20p, gK, was shown to be involved in virus penetration (20). Incorporation of gK into virions is apparently independent of UL20p, which is supported by the fact that Dietz et al. (6) were able to detect gK, albeit incompletely processed, in UL20-negative PRV virions. To confirm virion incorporation of EHV-1 gK in the absence of UL20p, a UL20-negative recombinant virus that expresses a tagged version of gK will be generated and analyzed in future experiments. At the moment, it could even be speculated further that EHV-1 gK function does not depend on the presence of UL20p at all, since deletion of gK resulted in a phenotype affecting replication at an earlier step of virus replication (20). On the other hand, UL20p might be very dependent on the expression of gK since initial experiments indicated reduced amounts of UL20p in cells infected with the gK negative EHV-1. UL20p might thus be less stable in the absence of gK. If UL20p was less stable in the absence of gK, this could also account for the difficulties in detecting UL20p in uninfected SC20 cells by Western blotting, whereas the protein was readily observed after infection with L11Δ20 (Fig. 7).
Direct cell-to-cell spread is known to at least partially require other viral proteins than egress of virions, followed by infection of new cells, and is proposed to involve sorting of particles to basolateral membranes and tight junctions. It is conceivable that UL20p is as necessary in inhibiting refusion in vesicles targeted to basolateral membranes as in those to apical membranes, although Melancon et al. (15) identified different structural domains of the HSV-1 UL20 protein involved either in formation of syncytial plaques or in virus assembly. Future experiments will address this and other potential roles of UL20p to further dissect the role of UL20p alone or in complex with gK.
Acknowledgments
We thank Christine Brandmüller for excellent technical assistance. We also thank Jens von Einem and Klaus Osterrieder for supplying pRacL11 and HΔgM-GFP, for advice on BAC mutagenesis, and for critical reading of the manuscript. We are indebted to Barry L. Wanner for providing the E. coli strain and the plasmids used for Red mutagenesis, to George P. Allen for MAb 3F6, to Gretchen B. Caughman for MAb ZB4, and to Lindsay Day and Dick Killington for the anti- gM MAb.
This study was supported by DFG grant NE653/4-3 to A.N.
REFERENCES
- 1.Allen, G. P., and M. R. Yeargan. 1987. Use of lambda gt11 and monoclonal antibodies to map the genes for the six major glycoproteins of equine herpesvirus 1. J. Virol. 61:2454-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avitabile, E., G. Lombardi, T. Gianni, M. Capri, and G. Campadelli-Fiume. 2004. Coexpression of UL20p and gK inhibits cell-cell fusion mediated by herpes simplex virus glycoproteins gD, gH-gL, and wild-type gB or an endocytosis-defective gB mutant and downmodulates their cell surface expression. J. Virol. 78:8015-8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baines, J. D., P. L. Ward, G. Campadelli-Fiume, and B. Roizman. 1991. The UL20 gene of herpes simplex virus 1 encodes a function necessary for viral egress. J. Virol. 65:6414-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Day, L. 1999. Ph.D. thesis. Characterisation of selected glycoproteins of equine herpesvirus-1: immune responses in the murine model. University of Leeds, Leeds, United Kingdom.
- 6.Dietz, P., B. G. Klupp, W. Fuchs, B. Kollner, E. Weiland, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein K requires the UL20 gene product for processing. J. Virol. 74:5083-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foster, T. P., X. Alvarez, and K. G. Kousoulas. 2003. Plasma membrane topology of syncytial domains of herpes simplex virus type 1 glycoprotein K (gK): the UL20 protein enables cell surface localization of gK but not gK-mediated cell-to-cell fusion. J. Virol. 77:499-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foster, T. P., J. M. Melancon, J. D. Baines, and K. G. Kousoulas. 2004. The herpes simplex virus type 1 UL20 protein modulates membrane fusion events during cytoplasmic virion morphogenesis and virus-induced cell fusion. J. Virol. 78:5347-5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foster, T. P., J. M. Melancon, T. L. Olivier, and K. G. Kousoulas. 2004. Herpes simplex virus type 1 glycoprotein K and the UL20 protein are interdependent for intracellular trafficking and trans-Golgi network localization. J. Virol. 78:13262-13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 1997. The UL20 gene product of pseudorabies virus functions in virus egress. J. Virol. 71:5639-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hübert, P. H., S. Birkenmaier, H. J. Rziha, and N. Osterrieder. 1996. Alterations in the equine herpesvirus type-1 (EHV-1) strain RacH during attenuation. J. Vet. Med. B 43:1-14. [DOI] [PubMed] [Google Scholar]
- 12.Johnson, D. C., and M. T. Huber. 2002. Directed egress of animal viruses promotes cell-to-cell spread. J. Virol. 76:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klupp, B. G., J. Baumeister, P. Dietz, H. Granzow, and T. C. Mettenleiter. 1998. Pseudorabies virus glycoprotein gK is a virion structural component involved in virus release but is not required for entry. J. Virol. 72:1949-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klupp, B. G., J. Altenschmidt, H. Granzow, W. Fuchs, and T. C. Mettenleiter. 2005. Identification and characterization of the pseudorabies virus UL43 protein. Virology 334:224-233. [DOI] [PubMed] [Google Scholar]
- 15.Melancon, J. M., T. P. Foster, and K. G. Kousoulas. 2004. Genetic analysis of the herpes simplex virus type 1 UL20 protein domains involved in cytoplasmic virion envelopment and virus-induced cell fusion. J. Virol. 78:7329-7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melancon, J. M., R. L. Luna, T. P. Foster, and K. G. Kousoulas. 2005. Herpes simplex type 1 gK is required for gB-mediated virus-induced cell fusion, while neither g and gK nor gB and UL20p function redundantly in virion de-envelopment. J. Virol. 79:299-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer, H., and P. H. Hübert. 1988. Isolation and characterization of monoclonal antibodies against an attenuated vaccine strain of equine herpesvirus type 1 (EHV-1). Vet. Microbiology 21:95-101. [DOI] [PubMed] [Google Scholar]
- 18.Neubauer, A., B. Braun, C. Brandmüller, O. R. Kaaden, and N. Osterrieder. 1997. Analysis of the contributions of the equine herpesvirus 1 glycoprotein gB homolog to virus entry and direct cell-to-cell spread. Virology 227:281-294. [DOI] [PubMed] [Google Scholar]
- 19.Neubauer, A., J. Rudolph, C. Brandmüller, F. T. Just, and N. Osterrieder. 2002. The equine herpesvirus 1 UL34 gene product is involved in an early step in virus egress and can be efficiently replaced by a UL34-GFP fusion protein. Virology 300:189-204. [DOI] [PubMed] [Google Scholar]
- 20.Neubauer, A., and N. Osterrieder. 2004. Equine herpesvirus type 1 (EHV-1) glycoprotein K is required for efficient cell-to-cell spread and virus egress. Virology 329:18-32. [DOI] [PubMed] [Google Scholar]
- 21.O'Callaghan, D. J., and N. Osterrieder. 1999. The equine herpesviruses, p.508-515. In R. G. Webster and A. Granoff (ed.), Encyclopedia of virology. Academic Press/Harcourt Brace & Company, San, Diego, Calif.
- 22.Oettler, D., O. R. Kaaden, and A. Neubauer. 2001. The equine herpesvirus 1 UL45 homolog encodes a glycosylated type II transmembrane protein and is involved in virus egress. Virology 279:302-312. [DOI] [PubMed] [Google Scholar]
- 23.Osterrieder, N., A. Neubauer, C. Brandmüller, B. Braun, O. R. Kaaden, and J. D. Baines. 1996. The equine herpesvirus 1 glycoprotein gp21/22a, the herpes simplex virus type 1 gM homolog, is involved in virus penetration and cell-to-cell spread of virions. J. Virol. 70:4110-4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rudolph, J., D. J. O'Callaghan, and N. Osterrieder. 2002. Cloning of the genomes of equine herpesvirus type 1 (EHV-1) strains KyA and RacL11 as bacterial artificial chromosomes (BAC). J. Vet. Med. B 49:31-36. [DOI] [PubMed] [Google Scholar]
- 25.Rudolph, J., C. Seyboldt, H. Granzow, and N. Osterrieder. 2002. The gene 10 (UL49.5) product of equine herpesvirus 1 is necessary and sufficient for functional processing of glycoprotein M. J. Virol. 76:2952-2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook, J., D. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Schimmer, C., and A. Neubauer. 2003. The equine herpesvirus 1 UL11 gene product localizes to the trans-golgi network and is involved in cell-to-cell spread. Virology 308:23-36. [DOI] [PubMed] [Google Scholar]
- 28.Seyboldt, C., H. Granzow, and N. Osterrieder. 2000. Equine herpesvirus 1 (EHV-1) glycoprotein M: effect of deletions of transmembrane domains. Virology 278:477-489. [DOI] [PubMed] [Google Scholar]
- 29.Seyboldt, C. 2000. Structural and functional analysis of the equine herpesvirus type 1 glycoprotein M. Ph.D. thesis. Ludwig-Maximilians-Universität München, Munich, Germany.
- 30.Sullivan, D. C., G. P. Allen, and D. J. O'Callaghan. 1989. Synthesis and processing of equine herpesvirus type 1 glycoprotein 14. Virology 173:638-646. [DOI] [PubMed] [Google Scholar]
- 31.Telford, E. A., M. S. Watson, K. McBride, and A. J. Davison. 1992. The DNA sequence of equine herpesvirus-1. Virology 189:304-316. [DOI] [PubMed] [Google Scholar]
- 32.Ward, P. L., G. Campadelli-Fiume, E. Avitabile, and B. Roizman. 1994. Localization and putative function of the UL20 membrane protein in cells infected with herpes simplex virus 1. J. Virol. 68:7406-7417. [DOI] [PMC free article] [PubMed] [Google Scholar]