Abstract
Crimean-Congo hemorrhagic fever virus (genus Nairovirus, family Bunyaviridae) genome M segment encodes an unusually large (in comparison to members of other genera) polyprotein (1,684 amino acids in length) containing the two major structural glycoproteins, Gn and Gc, that are posttranslationally processed from precursors PreGn and PreGc by SKI-1 and SKI-1-like proteases, respectively. The characteristics of the N-terminal 519 amino acids located upstream of the mature Gn are unknown. A highly conserved furin/proprotein convertase (PC) cleavage site motif (RSKR247) is located between the variable N-terminal region that is predicted to have mucin-like properties and the rest of PreGn. Mutational analysis of the RSKR247 motif and use of a specific furin/PC inhibitor and brefeldin A demonstrate that furin/PC cleavage occurs at the RSKR247 motif of PreGn as the protein transits the trans Golgi network and generates a novel glycoprotein designated GP38. Immunoprecipitation analysis identified two additional proteins, GP85 and GP160, which contain both mucin and GP38 domain regions, and whose generation does not involve furin/PC cleavage. Consistent with glycosylation predictions, heavy O-linked glycosylation and moderate levels of N-glycans were detected in the GP85 and GP160 proteins, both of which contain the mucin domain. GP38, GP85, and GP160 are likely soluble proteins based on the lack of predicted transmembrane domains, their detection in virus-infected cell supernatants, and the apparent absence from virions. Analogy with soluble glycoproteins and mucin-like proteins encoded by other hemorrhagic fever-associated RNA viruses suggests these proteins could play an important role in viral pathogenesis.
Crimean-Congo hemorrhagic fever (CCHF) virus is a tick-borne virus that causes a severe hemorrhagic disease in humans with a case fatality rate of approximately 30%. It is a member of the Nairovirus genus of the family Bunyaviridae (22). Members of the five genera of the Bunyaviridae family possess tripartite negative-sense genomes composed of S, M, and L segments. The M segment generally encodes two structural glycoproteins, while an additional encoded nonstructural protein, NSM, has been described for members of the Orthobunyavirus, Phlebovirus, and Tospovirus genera. Mature glycoproteins encoded by the M segment are generally processed by cotranslational cleavage (26). The glycoprotein-processing events for the Nairoviruses, particularly CCHF virus, appear to be unique in that further posttranslational cleavage of the glycoprotein precursor products is required for production of the mature glycoproteins (18, 24, 34). The IbAr10200 strain of CCHF virus generates two primary precursor proteins, the 140-kDa PreGn and the 85-kDa PreGc, which give rise to the two mature structural proteins Gn (37 kDa) and Gc (75 kDa), respectively (24, 34). The Gn glycoprotein is processed early in the secretory pathway at its N terminus following the consensus motif RRLL519 by the protease SKI-1, which cleaves at hydrophobic residues within protein substrates as they transit through the secretory pathway (24, 27, 28, 34). SKI-1 is also involved in the proteolytic processing of the precursor glycoprotein of two arenaviruses, Lassa virus and lymphocytic choriomeningitis virus (4, 17). Although the N terminus of Gc possesses a SKI-1-like consensus motif (RKPL1040), it does not appear to be processed by SKI-1 but is likely cleaved by a related protease (34).
In addition to the Gn- and Gc-coding regions, the open-reading frame (ORF) of CCHF virus strain IbAr10200 encodes 519 amino acids (aa) upstream of the N terminus of mature Gn. This region includes the mucin-like variable domain (aa 1 to 247, including the predicted signal peptide) and a second region comprised of aa 248 to 519, previously alluded to with the tentative name P35 or connector (3, 24, 34). An interesting and distinct feature observed at the junction of the highly variable mucin-like domain and the adjacent region is the presence of a predicted furin-like cleavage recognition motif RSKR at aa 244 to 247. Many viruses utilize furin or related proprotein convertases (PCs) to process their glycoprotein precursors into mature proteins (2, 30, 31, 35, 38, 39). The summary of research involving many cellular and viral proteins led to the identification of a consensus sequence R-S-(K/R)-R in furin/PC substrates (1, 16, 20). Such a predicted furin-like cleavage site was completely conserved in the glycoproteins of all CCHF virus isolates (V. Deyde and S. Nichol, unpublished results).
In the current study, we tested the hypothesis that furin-like proteases in addition to SKI-1-like proteases contribute to the posttranslational cleavage of the CCHF virus glycoprotein precursor and the generation of additional processed proteins. Our study indicates that a novel 38-kDa protein (designated as GP38) is processed as a result of furin/PC-mediated processing at RSKR247 and a C-terminal cleavage by SKI-1 following RRLL519. The processing of GP38 is abrogated by treatment of glycoprotein-expressing cells with brefeldin A or furin/PC inhibitor decanoyl-RVKR-chloromethylketone (dec-RVKR-CMK), demonstrating the need for proteins to reach the late Golgi compartments and the involvement of furin/PCs, respectively. Consistent with cleavage site properties of other furin/PC substrates, mutation of the RSKR247 motif abrogated the processing of GP38.
MATERIALS AND METHODS
Viruses, cell lines, and antibodies.
In the present study, we used CCHF virus strain IbAr10200, which was originally isolated from Hyalomma excavatum ticks from Sokoto, Nigeria. Virus propagation and stocks were done in either Vero E6 or SW-13 cells. Experiments involving CCHF virus infection (at an approximate multiplicity of infection of 0.1 in SW-13 cells) were conducted in the biosafety level 4 laboratory at the Centers for Disease Control and Prevention, Atlanta, GA. Samples lysed in detergent buffers were gamma-irradiated for 2 × 106 rads, and culture supernatants not treated with detergents were gamma irradiated for 5 × 106 rads. For transient expression of the virus glycoproteins (wild type or mutants), we used SW-13 cells routinely maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum and antibiotic-antimycotic or penicillin-streptomycin. Recombinant vaccinia virus (vvTF7-3) expressing bacteriophage T7 RNA polymerase was propagated in HeLa cells, had titers determined in CV-1 cells, and was used in transient transfection experiments.
A hyperimmune mouse ascitic fluid (HMAF) raised against the proteins of CCHF virus strain IbAr10200, a polyclonal antibody raised to the mucin region of CCHF virus strain IbAr10200, and a peptide antibody against aa 540 to 551 in the Gn-coding region (referred to here as Gn/540-551) have been described previously (24). We also generated a panel of antibodies against selected antigenic peptides within the CCHF virus strain IbAr10200 glycoprotein ORF under contract (Research Genetics). These included antibodies to aa 294 to 309 (DCDDTFFQKRIEEFFI, referred to as GP38/294-309), aa 379 to 392 (RHSTRIVDTPGPKI, referred to as GP38/379-392), and aa 491 to 503 (EVRKGQSVLRQYK, referred to as GP38/491-503), all within the region between mucin and Gn. Monoclonal antibodies (MAbs) 6C11 and 7A7 were kindly provided by J. Smith (formerly of the U.S. Army Medical Research Institute for Infectious Diseases [USAMRIID], Fort Detrick, MD).
Preparation and analysis of proteins from virus infection.
SW-13 cells were infected with CCHF virus for approximately 24 h, and proteins were radiolabeled using 75 to 100 μCi/ml of [35S]cysteine (Perkin Elmer Life Sciences) for various labeling and chase periods as indicated in the figure legends. CCHF virus-specific proteins present in cells or media were analyzed by immunoprecipitation using specific antibodies. First, cell monolayers were lysed with 1% Triton X-100 in TNE buffer (24) supplemented with Complete protease inhibitor cocktail tablets (Roche). Supernatants were harvested and analyzed directly. Before the addition of antibodies, the lysates or supernatants were subjected to gamma irradiation to inactivate the virus. The immunoprecipitation protocol was previously described in detail (24). Samples were resolved on a NuPAGE gel system (Invitrogen) and analyzed by autoradiography.
For Western blot analysis, SW-13 cells were infected with CCHF virus, and supernatants were harvested on the second day. Supernatants were concentrated using a combination of a Centriplus YM-10 centrifugal filter unit (Millipore) and vacuum-dry centrifugation or were concentrated using vacuum-dry centrifugation alone. Samples were loaded onto a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) system containing 5 M urea in the stacking gel and 2.5 M urea in the separating gel as previously described (11) or a NuPAGE gel system, both under reducing conditions. Resolved proteins were transferred onto nitrocellulose membranes using a semidry blotting system (Sigma) and then probed with specific antibodies as indicated in the figure legends. Peroxidase-labeled goat anti-rabbit and anti-mouse immunoglobulin G (Kirkegaard and Perry Laboratories) and 3,3′-diaminobenzidine were used for detection of the proteins.
For preparation of partially purified virions, SW-13 cells were infected with CCHF virus. Approximately 21 h after infection, cells were labeled with 120 μCi/ml [3H]glucosamine (NEN) and then harvested the next day. Supernatants were clarified and loaded onto a 20% sucrose cushion in SW40 or SW41 tubes. Virus was pelleted at 30,000 rpm at 4°C for 3.5 h. Virus pellets were resuspended in nonreducing sample buffer. β-mercaptoethanol was added to the samples before loading onto an SDS-PAGE system containing urea (11).
Generation of CCHF virus-mutated glycoprotein expression constructs.
The cloning strategy employed for the M segment ORF of CCHF virus strain IbAr10200 WT (wild type) has been described previously (24). For the introduction of a stop codon following RRLL519, we used primers CCHF Topo forward, 5′-CACCATGGAAGTAAGTAAC-3′, and reverse, 5′-CACTGGGTTCTTCTTAAAGCAATCTTCTAGA-3′. Since the forward primer had a CACC included before the ATG of the gene, it could be directly cloned into the pcDNA TOPO-3.2 directional cloning vector (Invitrogen). For the RSKR247 motif mutants, we employed the QuikChange II XL site-directed mutagenesis kit (Stratagene) and followed the methods outlined by the manufacturer. We used the following primer pairs: RSKR247→ISKR247 (forward, 5′-AGTCCAACGAACATTTCTAAAAGAAAC-3′, and reverse, 5′-GTTTCTTTTAGAAATGTTCGTTGGACT-3′) and RSKR247→RSII247 (forward, 5′-CCCAGTCCAACGAACAGGTCTATAATAAACCTTAAGATGGAAATAATC-3′, and reverse, 5′-GATTATTTCCATCTTAAGGTTTATTATAGACCTGTTCGTTGGACTGGG-3′), where bold type indicates a change in that position. PCR was performed using pcDNA3.2 WT (24) as the template following cycling conditions recommended by the manufacturer. Resulting PCR products were treated with Dpn1 to digest the template DNA, and the DNA with the introduced mutations was transformed into XL10-Gold ultracompetent cells by procedures recommended by the manufacturer (Stratagene). All mutations were confirmed by complete sequencing of the ORF.
Preparation and analysis of proteins from transfections.
Transfection experiments of CCHF virus clones were conducted as described previously (24). Samples were pulse-labeled with 100 μCi/ml of [35S]cysteine and chased for the time periods indicated in the figure legends. Lysed cells and supernatants were immunoprecipitated using antibodies indicated in the figure legends. Proteins were resolved on Nu-PAGE gels under reducing conditions and visualized by autoradiography.
Brefeldin A and dec-RVKR-CMK treatment of samples from CCHF virus-infected or M segment gene-transfected cells.
Brefeldin A (Sigma) was reconstituted in ethanol and used in experiments at a final concentration of 10 μg/ml. Furin inhibitor dec-RVKR-CMK (BioMol) was reconstituted in methanol and used at the concentrations indicated in the figure legends. Both inhibitors were added during starvation, pulse-labeling, and chase periods.
Treatment of immunoprecipitated CCHF virus glycoproteins with glycosidase enzymes.
In order to investigate the nature of glycosidic modifications on the CCHF virus glycoproteins, enzymatic digestion of immunoprecipitated proteins was performed using an enzymatic deglycosylation kit with prO-LINK extender (Prozyme) per the manufacturer's protocol. Proteins were incubated at 37°C overnight with various combinations of the following enzymes: PNGase F, O-glycanase (endo-α-N-acetylgalactosaminidase), sialidase, β(1-4)-galactosidase, and β-N-acetylglucosaminidase. The digested proteins were resolved on Nu-PAGE gels under reducing conditions and analyzed by autoradiography.
RESULTS
Identification of novel proteins generated from the CCHF virus glycoprotein precursor.
Comparison of deduced CCHF virus glycoprotein amino acid sequences revealed the conservation of an RS(K/R)R motif (aa 244 to 247 for strain IbAr10200) at the junction of the mucin domain and the remainder of the glycoprotein precursor. Such a high degree of conservation suggested a biological significance and the possibility of a cleavage mediated by furin or furin-like PCs. In order to characterize any additional processed glycoproteins of CCHF virus, we generated antibodies targeted against distinct antigenic epitopes in the glycoprotein sequences upstream of Gn. SW-13 cells were infected by CCHF virus, labeled with [35S]cysteine for 30 min, and chased for 4 h. The supernatants were subjected to immunoprecipitation using an antipeptide antibody (GP38/379-392) raised against aa 379 to 392 located in the region between mucin and mature Gn (see Fig. 1B for orientation), and the proteins were resolved on a 3 to 8% gel (Fig. 1A, lane 1). The antibody GP38/379-392 reacted with three CCHF virus glycoprotein-specific proteins with apparent molecular masses of 160 kDa, 85 kDa, and 38 kDa. These data suggest that each of these proteins contained the protein domain including aa 379 to 392, or one or two of them were coimmunoprecipitated.
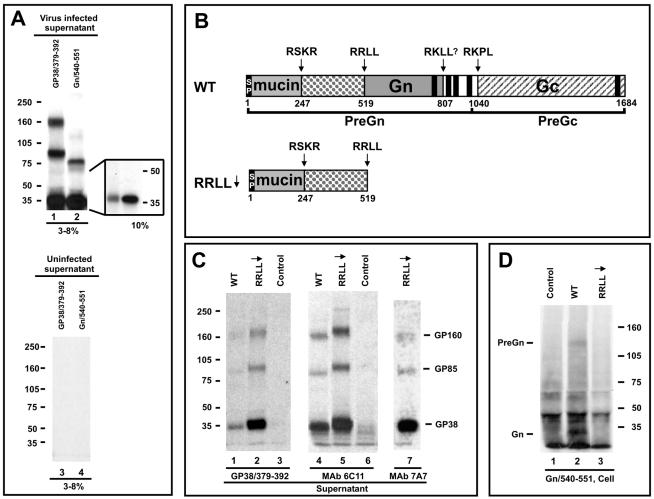
FIG. 1.
Identification of novel CCHF viral glycoproteins expressed from the N-terminal region of the CCHF virus glycoprotein ORF. (A) Autoradiograph of immunoprecipitated viral proteins from the supernatants of CCHF virus-infected SW-13 cells. At 24 h after infection, proteins were pulse-labeled with [35S]cysteine for 30 min and chased for 4 h. Immunoprecipitated proteins were run on a 3 to 8% NuPAGE gel. Proteins were immunoprecipitated with the antibodies GP38/379-392 (lanes 1 and 3) and Gn/540-551 (lanes 2 and 4). Inset shows samples from lanes 1 and 2 run on a 10% NuPAGE gel. (B) Schematic of the WT construct of CCHF virus and a truncation mutant generated that ends at the motif RRLL519 (RRLL↓). (C) Autoradiograph of viral proteins immunoprecipitated from the supernatants of SW-13 cells transfected with WT and RRLL↓ constructs with the indicated antibodies and run on a 3 to 8% NuPAGE gel. WT, RRLL↓, and untransfected (control) samples were pulse-labeled with [35S]cysteine for 30 min and were either chased for 3 h and precipitated with the antibody GP38/379-392 (lanes 1 to 3) or chased for 5 h and precipitated with MAb 6C11 (lanes 4 to 6). Proteins expressed from the RRLL↓ construct were pulse-labeled with [35S]cysteine for 30 min, chased for 3.5 h, and precipitated with MAb 7A7 (lane 7). (D) Lanes 1 to 3, WT and RRLL↓ constructs and untransfected control expressed in SW-13 cells. Proteins were pulse-labeled with [35S]cysteine for 45 min and chased for 3 h. Lysed cell monolayers were immunoprecipitated with Gn/540-551 antibody and run on a 3 to 8% NuPAGE gel.
Given their similarity in protein migration, it was important to demonstrate that the 38-kDa protein that was reactive to the GP38/379-392 antibody was distinct from the 37-kDa CCHF virus mature Gn protein previously described. Virus-infected cell supernatants were generated as above but were immunoprecipitated using a Gn-specific antibody generated against aa 540 to 551 (24), referred to here as Gn/540-551. As expected, this antibody showed strong reactivity to the 37-kDa mature Gn protein (Fig. 1A, lane 2). In addition, a 75-kDa protein was also detected along with mature Gn (probably a dimer of Gn or coprecipitated Gc). In order to more precisely determine the size of the 38-kDa protein, we resolved the proteins on a 10% gel (see inset, Fig. 1A). The 38-kDa protein appeared to resolve slightly higher than mature Gn. To further confirm that the 38-kDa protein was distinct from mature Gn, a truncated expression construct termed RRLL↓ was generated in which only the coding sequences encompassing aa 1 to 519 were preserved and all additional sequences of the M segment (including all of Gn and PreGc/Gc) were deleted (Fig. 1B). If the 38-kDa protein was still immunoprecipitated by antibody GP38/379-392 in the absence of any Gn coding sequence, this would distinguish the 38-kDa protein from Gn and confirm its unique identity. Vaccinia virus T-7 RNA polymerase-driven plasmids were transfected into SW-13 cells, and the resulting proteins were labeled with [35S]cysteine. The supernatant proteins were immunoprecipitated with antibody GP38/379-392 and resolved on a 3 to 8% gel (Fig. 1C, lanes 1 to 3). From the cells expressing the WT glycoprotein, three proteins—similar to those observed in supernatants of CCHF virus-infected cells—were resolved at 160, 85, and 38 kDa (compare Fig. 1A, lane 1 and C, lane 1). All three proteins (160, 85, and 38 kDa) were also present in the supernatant from the RRLL↓-expressing cells (Fig. 1C, lane 2), further suggesting that these proteins are expressed from sequences upstream of Gn. We observed that the 38-, 85-, and 160-kDa proteins appear slightly larger when generated from the RRLL↓ construct. This phenomenon is due to minor glycosylation differences, as the above-mentioned proteins from WT or RRLL↓-expressing cells resolved with similar mobility upon PNGase treatment (data not shown).
During this investigation, we also used two MAbs (6C11 and 7A7) that had been provided to us as Gn-specific antibodies (kindly provided by J. Smith, formerly of USAMRIID). MAb 6C11 was also recently reported as being Gn-specific (3). However, when these antibodies were used in immunoprecipitation experiments involving supernatants of WT- and RRLL↓ (lacking mature Gn)- expressing cells labeled with [35S]cysteine, the 38-, 85-, and 160-kDA proteins were detected (Fig. 1C, lanes 4, 5, and 7). These findings are similar to those observed with the GP38/379-392 antibody. To confirm the specificity of the MAbs (as well as antibody GP38/379-392) relative to the RRLL↓ clone-expressed proteins, we utilized the Gn-specific antibody Gn/540-551 to confirm that no nonspecific protein could be detected from this construct. As expected, no protein of approximately 38 kDa was visualized from RRLL↓-expressing cells relative to the WT (Fig. 1D, compare lanes 2 and 3). These results indicate that both MAbs 6C11 and 7A7 are reactive with epitopes within the 38-, 85-, and 160-kDa proteins and not reactive to those within mature Gn, contrary to previous reports (3). We therefore utilized one of the MAbs, 6C11, in further investigations designed to better understand the processed protein subunits representing the N terminus of PreGn.
The above data indicate that the 160-, 85-, and 38-kDa proteins are all generated by the amino acid sequences located N-terminally to mature Gn. We previously suggested that the 160-kDa protein found in supernatants is a more glycosylated form of the 140-kDa PreGn, in which Gn is an integral part (24, 34). This suggestion was based on the reactivity of the 160-kDa protein to antibodies against (i) the mucin domain, (ii) the region between the mucin domain and mature Gn, and (iii) erroneously designated Gn-specific MAbs (discussed above). However, we now know this must be incorrect, as the Gn-specific peptide antibody does not react with the 160-kDa protein in the medium (Fig. 1A, lane 2) and this protein is observed even in the absence of Gn-encoding sequences (Fig. 1C, lanes 2, 5, and 7).
GP38 and mucin domains are contained within GP85 and GP160.
Western blot analysis of CCHF virus-infected cell supernatants was performed to address whether the multiple protein bands (38-, 85-, and 160-kDa) that were observed on gels following immunoprecipitation using antibody GP38/379-392 and MAbs 6C11 and 7A7 were due to coimmunoprecipitation. Results obtained using antibody GP38/379-392 clearly confirm that the reactivity to the 38-, 85-, and 160-kDa proteins was not due to coimmunoprecipitation (Fig. 2A, lane 1). Furthermore, two additional antibodies raised against distinct epitopes (GP38/294-309 and GP38/491-503) of the protein located between RSKR247 and the N terminus of Gn confirmed the same results (data not shown).
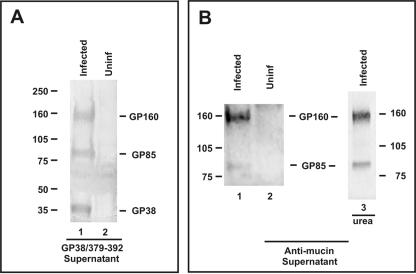
FIG. 2.
Western blot analyses of novel CCHF virus glycoproteins. SW-13 cells were infected with CCHF virus. Supernatants were harvested and concentrated. (A) Supernatants were run on a 3 to 8% NuPAGE gel, and antibody GP38/379-392 against aa 379 to 392 within the GP38-coding region of the CCHF virus glycoprotein precursor was used to probe the proteins. (B) Proteins were probed with a polyclonal monospecific antibody to the mucin region of CCHF virus strain IbAr10200. Lanes 1 and 2, supernatants were run on a 3 to 8% NuPAGE gel. Lane 3, supernatant was run on a 5% SDS-PAGE gel containing urea.
Together, these results indicate that the 38-kDa protein, which hereafter will be referred to as GP38, is a unique viral glycoprotein distinct from Gn. Based on the reactivity of the specific antibodies GP38/294-309, GP38/379-392, and GP38/491-503 to GP38 and considering the predicted size of a potential protein generated by cleavage at the proposed furin-like cleavage site RSKR247 and the previously confirmed cleavage site RRLL519 (24, 34), we hypothesized that GP38 is most likely a processed protein comprised of aa 248 to 519 (see Fig. 1B). The identity of the 85- and 160-kDa proteins (referred to hereafter as GP85 and GP160) was unclear at this point. The proposed furin-like cleavage at RSKR247 would further suggest that a discrete mucin-like protein would also be generated (see Fig. 1B). A monospecific polyclonal antibody raised against the mucin region of the CCHF virus strain IbAr10200 was previously shown in Western blot analyses to react with a 160-kDa protein in infected supernatants similar to the GP160 protein reactive to GP38-specific antibodies (24). Improved experimental conditions have allowed us to observe both 160- and 85-kDa proteins in infected supernatants using the mucin-specific antibody in Western blot analyses (Fig. 2B, lane 1), consistent with GP160 and GP85 containing the mucin protein domain in addition to the GP38 domain. Furthermore, a Western blot analysis in which proteins from infected supernatants were resolved on a gel containing urea (as well as the usual reducing conditions) and reacted to the mucin-specific antibody again showed significant amounts of both GP160 and GP85 (Fig. 2B, lane 3), suggesting that GP160 may not be a dimer of GP85. Neither protein is likely to contain mucin domain alone since GP38-specific antibodies also react with both proteins. In addition, the mucin domain (aa 1 to 247 minus the predicted signal peptide) does not contain cysteine residues, so proteins containing only this domain would not be labeled with [35S]cysteine and detected by immunoprecipitation. Several approaches to identifying the cleaved mucin protein were used, including labeling the CCHF virus-infected or M segment gene-transfected cells using [3H]leucine, [3H]threonine, or [3H]glucosamine, followed by immunoprecipitation employing the mucin-specific polyclonal antibody as well as a panel of antibodies generated to different peptide regions of the mucin domain. Unfortunately, we could not obtain clear data showing an N-terminal cleavage product consisting of the mucin domain alone.
Glycosylation of proteins containing the mucin and GP38 domains (aa 1 to 519).
As discussed previously, the mucin domain has the potential for extensive O-linked glycosylation, while it was predicted that the GP38 region contains only a minor amount of O-glycans (24). In addition, the mucin domain of the CCHF virus strain IbAr10200 contains five potential N-linked sites and GP38 contains two possible N-glycans. In order to investigate the contribution of O- and N-glycans to the overall molecular mass of the proteins expressed from aa 1 to 519 (containing only mucin and GP38 domains), the RRLL↓ construct was expressed in SW-13 cells and labeled with [35S]cysteine and the supernatant proteins were immunoprecipitated with CCHF virus HMAF polyclonal antibody. The immunoprecipitates were digested with different combinations of enzymes and run on a 3 to 8% gel (Fig. 3A). When the immunoprecipitates were not treated with any enzymes, GP160, GP85, and GP38 were visualized (Fig. 3A, lane 1), while PNGase F treatment increased the mobility of the proteins to approximately 135, 75, and 32 kDa, respectively, demonstrating the presence of N-glycans on all three proteins (Fig. 3A, lane 2). Removal of the O-glycans from the three proteins with a combination of O-glycanase (endo-α-N-acetylgalactosaminidase) and sialidase revealed a shift in mobility to approximately 105, 65, and 35 kDa, respectively (Fig. 3A, lane 3). The enzymes β(1-4)-galactosidase and β-N-acetylglucosaminidase remove additional O-glycan modifications, and when added to O-glycanase and sialidase, the three proteins shifted further to approximately 90, 60, and 34 kDa (Fig. 3A, lane 4). These data demonstrate that GP160 and GP85 possess high levels of O-glycosylation, further suggesting the presence of the mucin domain within these two proteins. When PNGase F was used in addition to the four O-glycan enzymes, the mobility of the proteins was observed at approximately 78, 50, and 30 kDa (Fig. 3A, lane 5), indicating the extent of overall glycosylation. Although PNGase F is likely to remove all N-glycans from the proteins, it is more difficult to determine if all O-glycans are removed with the enzymes used. Thus, we do not know if all O-glycans were completely removed from the proteins in our experiments.
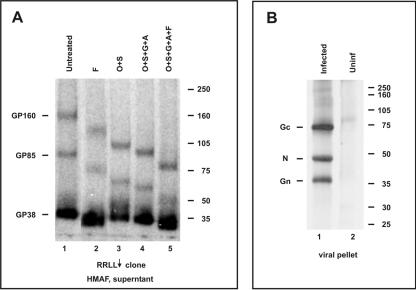
FIG. 3.
Characterization of the newly identified CCHF virus glycoproteins. (A) After SW-13 cells were transfected with the RRLL↓ truncation construct, proteins were pulse-labeled with [35S]cysteine for 30 min and chased for 3 h, and the supernatant proteins were immunoprecipitated with hyperimmune mouse ascitic fluid generated against CCHF virus. Proteins were then treated with various combinations of enzymes. Lane 1, untreated; lane 2, PNGase F; lane 3, O-glycanase (endo-α-N-acetylgalactosaminidase) and sialidase; lane 4, O-glycanase, sialidase, β(1-4)-galactosidase, and β-N-acetylglucosaminidase; lane 5, O-glycanase, sialidase, β(1-4)-galactosidase, β-N-acetylglucosaminidase, and PNGase F. (B) SW-13 cells were infected with CCHF virus and proteins were labeled overnight with [3H]glucosamine. Virions were partially purified by pelleting through a 20% sucrose cushion and then run on a 10% SDS-PAGE gel containing urea.
Together, these data indicate that GP38 is a mature processed glycoprotein that is likely generated from the predicted furin-like cleavage at the site RSKR247 at its N terminus and the confirmed SKI-1 cleavage site RRLL519 at its C terminus. As for the biological significance of the newly identified viral proteins, there is no evidence so far that GP160, GP85, and GP38 are major structural elements of the CCHF virion. In order to label all the virus glycoprotein products, including a possible mucin protein (which otherwise would not be labeled by [35S]cysteine due to lack of cysteines in that domain), we employed [3H]glucosamine labeling. Analysis of a partially purified virion preparation of CCHF virus strain IbAr10200 showed the presence of only three predominant structural proteins, Gc, N, and Gn (Fig. 3B, lane 1), which was similar to results presented previously for the Matin strain of CCHF virus (24). It is interesting to note that the N protein was also labeled with [3H]glucosamine, and this probably was facilitated by nucleocytoplamic O-glycosylation machinery (8). Similar O-glycosylation was reported for the nucleoprotein of Ebola virus (15).
Generation of GP38 is sensitive to brefeldin A and dec-RVKR-CMK treatments.
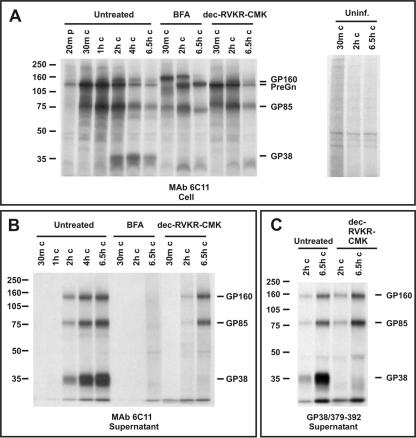
After confirming that GP38 is a unique cleavage product, we performed an extensive pulse-chase analysis using 35[S]cysteine to monitor its appearance in CCHF virus-infected cells and supernatants. Due to reactivity of the antipeptide antibodies to nonspecific proteins resolving around 35 to 38 kDa within cell lysates, we selected MAb 6C11 to study the processing of GP38 as well as GP85 and GP160 in cells and supernatants. In CCHF virus-infected SW-13 cells, PreGn can be seen at 20-min of pulse and can be seen to increase through the 1-h chase period before beginning to decrease (Fig. 4A, untreated lanes), which is similar to previous findings with polyclonal CCHF virus HMAF (24). The reactivity of PreGn to MAb 6C11 verifies that GP38 is indeed part of PreGn. Previous experiments have already confirmed the presence of the mucin domain and Gn in the PreGn precursor protein (24). At 30 min of chase, a protein of approximately 85 kDa begins to appear in the cell, which is likely GP85. At 2 h of chase, GP38 is clearly visible within the cell and GP160 begins to appear. Over the course of early chase periods, GP38, GP85, and GP160 appear to increase in the cell followed by decreasing levels during late chase periods. The intracellular decrease of these proteins corresponded with progressively increasing amounts in the supernatants (Fig. 4B). When CCHF virus-infected supernatants were immunoprecipitated with antibody GP38/379-392, a similar pattern was observed for GP38, GP85, and GP160 (Fig. 4C).
FIG. 4.
Pulse-chase analysis of GP38-associated proteins in cell and supernatant. SW-13 cells were infected with CCHF virus, proteins were pulse-labeled with [35S]cysteine for 20 min and chased for various times indicated at the top of the gels. Brefeldin A (BFA) and 32 μM dec-RVKR-CMK were added during starvation, labeling, and chase periods. The samples were run on a 7% NuPAGE gel system. (A) Proteins in cell lysates were immunoprecipitated with MAb 6C11. (B) Proteins in supernatants were immunoprecipitated with MAb 6C11. (C) Supernatant proteins were precipitated with antibody GP38/379-392.
In order to investigate whether exit from the endoplasmic reticulum (ER) and transit to the late Golgi compartment is required for the processing of the GP38 protein, we treated the CCHF virus-infected cells with brefeldin A (a fungal metabolite that blocks the transport of proteins from the ER) during starvation, pulse, and chase periods and immunoprecipitated the proteins with MAb 6C11. The appearance of GP38 was completely blocked in CCHF virus-infected cells treated with brefeldin A compared to in untreated cells (Fig. 4A, compare brefeldin A-treated time points—30 min, 2 h, and 6.5 h of chase—to the same time points of the untreated cells). Previous data showed that the N terminus of Gn (and thus the C terminus of GP38) is cleaved by SKI-1 in the ER or early Golgi compartment and is brefeldin A treatment independent (34). The present data indicate that a subsequent processing event at the N terminus of GP38 occurs in a post-ER compartment, and blockage of transport by brefeldin A effectively abrogates the processing of GP38. In addition, our data also imply that furin or furin-like PCs residing in the late Golgi might be responsible for facilitating this processing. Under brefeldin A treatment conditions, we observed a protein slightly larger than GP160 at the 30-min chase time point but it had almost disappeared by the 6.5-h chase point. The identity of this larger protein is unclear. Because brefeldin A impairs the transport of proteins from the ER to subsequent cellular compartments, no virus-specific glycoproteins were recovered from the medium (Fig. 4B).
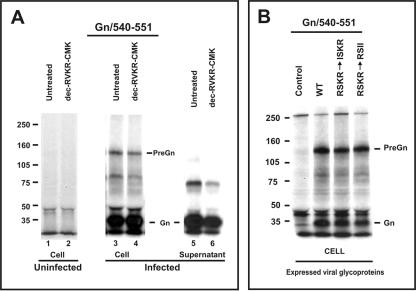
Since our data indicated that the generation of mature GP38 is a late event in the biosynthetic pathway and since the presence of RSKR at the N terminus of GP38 would suggest a potential cleavage by furin or furin-like PCs, we further investigated the role of PCs in the processing of GP38. CCHF virus-infected SW-13 cells were treated during starvation, pulse, and chase periods with 32 μM dec-RVKR-CMK, a compound that inhibits the function of furin and other PCs. Next the proteins were immunoprecipitated with MAb 6C11. In the cells treated with dec-RVKR-CMK, PreGn, GP85, and GP160 appear similar to the untreated samples (Fig. 4A, compare dec-RVKR-CMK lanes with the untreated lanes). However, GP38 was completely absent in the dec-RVKR-CMK-treated cells. Furthermore, when proteins in the infected supernatants were analyzed in the presence of dec-RVKR-CMK, GP160 and GP85 were found to persist, while GP38 was completely inhibited (Fig. 4B). For additional confirmation, supernatants from infected cells treated with or without dec-RVKR-CMK were immunoprecipitated with the antibody GP38/379-392 at the indicated chase periods. Again, GP160, GP85, and GP38 were seen in the untreated supernatants but only GP160 and GP85 were observed under treatment conditions, consistent with the reactivity of MAb 6C11 (Fig. 4C). The appearance of GP160 and GP85 in the medium under dec-RVKR-CMK treatment conditions further confirms that the lack of GP38 in the medium is due to lack of processing and not due to a global transport defect. Thus, the generation of GP38 is mediated by furin or furin-like PCs whose proteolytic functions are abrogated by dec-RVKR-CMK, demonstrating that this additional posttranslational processing event occurs within the CCHF virus glycoprotein precursor.
Mutational analysis of the potential furin cleavage site (RSKR247).
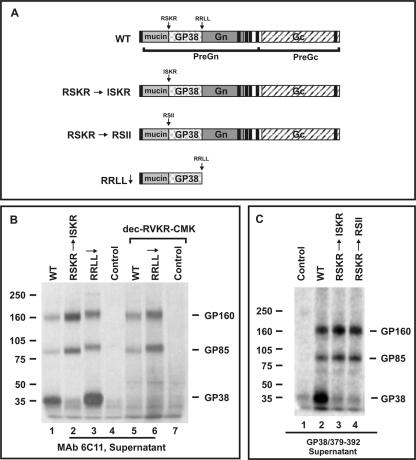
After establishing that GP38 is selectively inhibited by dec-RVKR-CMK, we sought to confirm the potential cleavage site involved in the processing. The RSKR motif (aa 244 to 247) was of interest because it has been identified in other furin substrates, including many cellular and viral glycoproteins (20). Since the P4 arginine (R) is important for furin/PC recognition of their substrates, we altered R-244 of the RSKR sequence within the full-length glycoprotein construct to isoleucine (I), resulting in the mutant ISKR (Fig. 5A). The processing profile of GP160, GP85, and GP38 in the ISKR mutant was evaluated by pulse-chase analysis and compared with the results for the WT and RRLL↓ constructs. Supernatant proteins were analyzed with MAb 6C11 after 5 h of chase (Fig. 5B). In the case of WT- or RRLL↓-expressing cells, GP160, GP85, and GP38 were detected in the medium as expected (Fig. 5B, lanes 1 and 3). However, when the ISKR mutant was analyzed, only GP160 and GP85 were visible in the supernatant, while GP38 was eliminated (Fig. 5B, lane 2). The lack of GP38 in the medium was due to the abrogation of intracellular processing of the mutant protein, not to a broader transport defect, as GP160 and GP85 were readily detected in the medium. In addition, as with the infection studies presented above, GP160 and GP85, but not GP38, were detected in the supernatants from the furin inhibitor dec-RVKR-CMK-treated cells that were transfected with the WT and RRLL↓ constructs (Fig. 5B, compare lanes 1 and 3 with lanes 5 and 6). These findings provide convincing evidence that GP38 is processed at the RSKR247 site and that the arginine residue at the P4 position is required to enable the processing. These data are entirely consistent with furin or furin-like PC cleavage at this site, resulting in the generation of GP38.
FIG. 5.
Effect of mutations on the generation of GP38. (A) Schematic of the mutations generated in the CCHF virus glycoprotein ORF. (B) SW-13 cells were transfected with the indicated constructs, and proteins were pulse-labeled with [35S]cysteine for 30 min and chased for 5 h. The supernatant proteins were immunoprecipitated with MAb 6C11. WT, RRLL↓, and untransfected control were also treated with 50 μM dec-RVKR-CMK. Proteins were run on a 3 to 8% NuPAGE gel. (C) SW-13 cells were transfected with the indicated constructs, and proteins were pulse-labeled with [35S]cysteine for 30 min and chased for 3 h. Proteins were immunoprecipitated with the antibody GP38/379-392 and then run on a 3 to 8% NuPAGE gel.
The results obtained with the ISKR mutant construct using MAb 6C11 were consistently reproduced when the peptide antibody GP38/379-392 was used in the analysis, confirming the importance of the P4 arginine for processing at this site (Fig. 5C, lane 3). In addition, we examined an additional mutant (RSII) in which the K and R in the −2 and −1 positions, respectively, in the context of the RSKR motif, were replaced by isoleucines (Fig. 5A). Consistent with furin requiring the presence of two basic amino acids in addition to the P4 arginine for recognition, GP38 was not processed from this mutant (Fig. 5C, lane 4). Similar results were obtained using antibody GP38/294-309 (data not shown). Taken together, the data confirm the role of furin or furin-like proteases in generating GP38 and the requirement of critical amino acids at the RSKR247 cleavage site to facilitate the processing.
Inhibition of furin-like PC processing of GP38 by dec-RVKR-CMK treatment or cleavage site mutagenesis does not interfere with the SKI-1 processing of mature Gn.
In order to verify that dec-RVKR-CMK is specific to inhibiting GP38 processing and not that of Gn, SW-13 cells were infected with CCHF virus and labeled with [35S]cysteine. Cells were either untreated or treated with 30 μM dec-RVKR-CMK. Cell lysates and supernatants were immunoprecipitated with the Gn-specific peptide antibody Gn/540-551 (24). Mature Gn was observed in the cells and media with or without inhibitor treatment (Fig. 6A, lanes 3 to 6). As a control, inhibition of GP38 processing by dec-RVKR-CMK was observed under similar experimental conditions using antibody GP38/379-392 (data not shown). In addition, since the RSKR247 site is topologically located N terminally to the SKI-1 cleavage site RRLL519 (see Fig. 1B), we also tested the mutants RSKR→ISKR and RSKR→RSII with the Gn-specific antibody to ensure that these mutations did not result in any structural or functional alterations of PreGn. In those mutants, mature Gn was as efficiently processed as with the WT construct (Fig. 6B). These experiments indicate that dec-RVKR-CMK inhibition is selective to furin/PCs and does not affect SKI-1-mediated processing, and the mutations of the furin/PC recognition motif also do not affect Gn processing.
FIG. 6.
Inhibition of GP38 processing does not prevent the processing of mature Gn. (A) SW-13 cells were infected with CCHF virus, pulse-labeled with [35S]cysteine for 30 min, chased for 4 h and the cell lysates/supernatants were immunoprecipitated with antibody Gn/540-551, and run on a 3 to 8% NuPAGE gel. Samples were either untreated or treated with 30 μM dec-RVKR-CMK. (B) SW-13 cells were transfected with the indicated constructs, and proteins were pulse-labeled with [35S]cysteine for 30 min, chased for 3 h, immunoprecipitated with antibody Gn/540-551, and run on a 3 to 8% NuPAGE gel.
DISCUSSION
Many cellular and viral glycoproteins are synthesized as inactive precursors which are later cleaved by host cell proteases, resulting in active proteins (18). In the case of enveloped viruses, proteolytic cleavage can modulate biological characteristics of the virus (2, 12, 16, 19, 30, 31, 35, 38, 39). Usually viral glycoprotein precursors undergo only one proteolytic processing step (besides signal peptide removal), resulting in two protein subunits. Most virus glycoproteins are processed by cellular furin or furin-like PCs with similar substrate specificity. The presence of the R-X-(K/R)-R motif in the extracellular domain of a protein would make it a likely substrate for furin or PC activation (1, 20). Among the hemorrhagic fever viruses, the glycoproteins of Ebola and Marburg viruses are cleaved by furin or furin-like PCs (2, 21, 25, 35, 36, 41). Ebola virus utilizes furin and other PCs to process its envelope and secreted glycoproteins, GP and sGP, respectively (35, 37). In addition, it was recently shown that shedding of the Ebola virus glycoprotein occurs via TACE enzyme, although this is an extracellular event (9). In contrast to this common pattern of viral glycoprotein proteolytic processing by furin or furin-like PCs, the glycoproteins of CCHF virus and arenaviruses such as Lassa and lymphocytic choriomeningitis viruses were shown to be cleaved by SKI-1 (4, 17, 34). SKI-1 prefers hydrophobic residues for cleavage recognition in contrast to furin/PCs which require basic residues (28). Details of this novel viral glycoprotein processing have emerged in a series of recent studies with CCHF virus (3, 14, 24, 34). The CCHF virus mature 37-kDa Gn and 75-kDa Gc proteins have been shown to be processed from the 140-kDa PreGn and 85-kDa PreGc precursors by SKI-1 and SKI-1-like proteases, respectively (24, 34).
In the present study, three additional CCHF virus-specific proteins, GP38, GP85, and GP160, were shown to be reactive to antibodies raised against selective epitopes present within aa 248 to 519 which lie within PreGn upstream of Gn. We demonstrate that for CCHF virus strain IbAr10200, the RSKR motif located at the junction of the mucin domain and the remainder of the polyprotein (aa position 244 to 247) is cleaved by furin/PCs. Comparison of the sequences of this PreGn region of several different CCHF virus strains indicates complete conservation of this site, which would be a suitable cleavage recognition site for furin or other mammalian PCs including PC1, PC2, PC4, PACE4, PC5, and PC7 (2, 28, 38) in a variety of cell or tissue types. Such conservation would suggest that PC cleavage at this site is of biological importance. The dec-RVKR-CMK inhibitor and RSKR mutational analysis data clearly indicate that GP38 is generated by furin/PC cleavage at the RSKR247 motif and SKI-1 cleavage at RRLL519 (see Fig. 1B). This cleavage event occurs late in the secretory pathway, as GP38 generation was abolished when transport was blocked by brefeldin A, consistent with the need for the substrate to reach the trans Golgi network (TGN). That finding agrees with the localization and functional activity of furin and most of the PCs in host cells (28). Our studies indicate that GP38 is generated by a combination of an early cleavage event in the ER/cis Golgi at RRLL519, followed by a late cleavage event in the TGN at RSKR247.
Our data demonstrate that despite similar gel mobilities, GP85 is distinct from the 85-kDa PreGc (24), as GP85 is detected even in the absence of PreGc/Gc sequences. GP85 and GP160 are not homo-oligomers of the mucin domain or GP38 proteins alone, as antibody reactivity patterns indicate that they contain both mucin and GP38 domains. In addition, the appearance of GP85 and GP160 does not involve furin/PC cleavage at the RSKR247 motif, suggesting that these proteins may not be hetero-oligomers consisting of mature mucin protein and GP38.
In addition to the predicted extensive O-glycosylation of the mucin domain and modest O-glycosylation of GP38 (24), N-linked glycosylation was also predicted at five potential sites in the mucin domain and two in GP38. Glycosylation differences alone can result in large protein mobility shifts, as evidenced by respiratory syncytial virus G protein with a core molecular mass of 32 kDa resolving as a 90-kDa fully glycosylated form (7, 40). Our enzyme digestion data demonstrate for the first time that substantial amounts of both O- and N-glycans are added to the mucin and GP38 domains. Deglycosylated GP85 appears to resolve as a 50-kDa protein, which could represent the core molecular mass of the protein comprising the mucin and GP38 domains. While PNGase F is expected to digest all N-glycans, the O-glycosidases used do not digest all possible O-linked structures, for instance O-linked N-acetylgalactosamine (such as that found in Ebola and Marburg virus glycoproteins) (33). The fact that GP160 protein treated with all the available O- and N-glycosidases did not comigrate on gels with similarly treated GP85 could be indicative of the presence of resistant complex carbohydrates on GP160. Nevertheless, we cannot rule out the possibility that GP160 is a dimer of GP85, although the molecular sizes of the glycosidase-treated GP160 and GP85 do not add up to exhibit a monomer-dimer relationship. The analysis of proteins in urea gels also might suggest that GP160 is not a dimer of GP85. Additional approaches such as direct amino acid sequencing or cross-linking studies will be needed to more precisely determine the identity of CCHF virus GP85 and GP160 proteins.
GP38, GP85, and GP160 are likely to be soluble proteins given that the confirmed SKI-1 processing at RRLL519 would leave these proteins without any predicted transmembrane domain. We cannot rule out that they could associate with cell membranes and become incorporated into virions via disulfide bonds or ionic interactions between them and mature Gn or Gc. For instance, with arenaviruses, the virion association of GP1 depends on such interactions with the membrane-anchored GP2 (5, 6). However, analysis of the partially purified CCHF viral pellet failed to detect the presence of GP160, GP85, and GP38, with only the mature forms of Gn, Gc, and NP being seen as the major virion components. Although it is possible that GP38 was masked by the similarly sized mature Gn (37 kDa), no secondary amino acid sequence was detected when the excised Gn band was analyzed by direct N-terminal protein sequencing. To date, no biological function has been attributed to the soluble GP38, GP85, or GP160 protein. Previous studies have shown that the mucin domain of Ebola virus is a major determinant for pathogenicity in organ culture (43) and in cell culture (29). It remains to be determined whether the variable mucin-like domain within CCHF virus glycoproteins has similar cytopathic potential or function in host response modulation as suggested for the sGP of Ebola virus (42). Interestingly, the heavily O-glycosylated mucin domain of Ebola and Marburg virus GP appears to play an important role in binding to target cells, such as macrophages, via interaction with host cell C-type lectins (32, 33). The CCHF virus mucin-containing proteins are not expected to play a similar role given the likely soluble nature of these proteins.
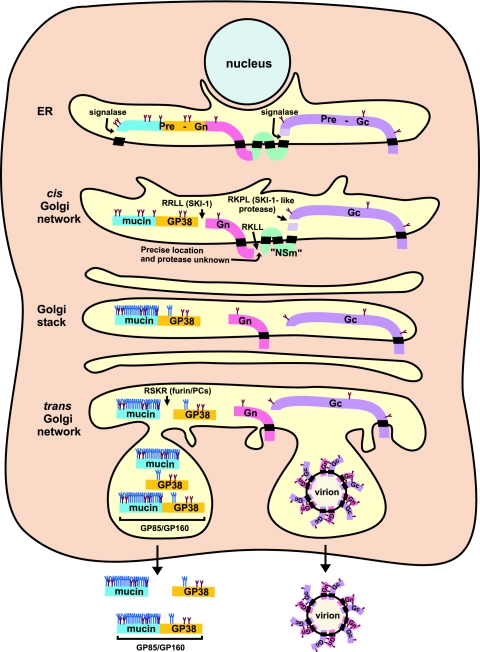
Figure 7 shows our current working model of the unique processing events of the CCHF virus glycoprotein precursor (references 3, 14, 24, and 34 and this study). In the ER, cotranslational cleavage of the precursor polyprotein by signalase and N-glycosylation are expected to occur similar to that of other virus glycoproteins. As a result of signalase cleavage of the full-length polyprotein, PreGn (confirmed to contain mucin, GP38, and Gn) and PreGc are generated. Mature Gc is processed from PreGc by cleavage at an RKPL1040 motif in the ER. Mature Gc is predominantly localized in the ER and requires Gn to enable its transport to the Golgi analogous to other members of the family Bunyaviridae. The C terminus of PreGn is predicted to span the membrane four times based on the presence of hydrophobic amino acid stretches. In the ER/cis Golgi, mature Gn is processed from PreGn at the motif RRLL519 by SKI-1. The C terminus of mature Gn has not been precisely determined, and mutations in the SKI-1-like site RKLL807 did not abrogate the generation of Gn (34). Prediction models indicate that the RKLL807 motif is on the cytoplasmic side of the membrane where SKI-1 is not known to be localized (28). Nevertheless, it is interesting to note that this C-terminal motif is conserved in all CCHF virus strains as R(R/K)LL, which is the same conserved motif at the N-terminal end of Gn (V. Deyde and S. Nichol, unpublished data). Once the mature Gn is generated, it may directly or indirectly interact with Gc and transfer to the cellular compartment where virus assembly occurs. The multiple membrane-spanning region located at the C terminus of PreGn, which would presumably be generated after the C-terminal cleavage of Gn, has been tentatively hypothesized as “NSm” since this potential protein has some similarities to the NSm protein of other Bunyaviridae family members (13). After SKI-1-mediated cleavage, the N-terminal part of PreGn consisting of the mucin domain and GP38 is released. Substantial O-glycans are added to the mucin portion of the glycoprotein during its transport through the secretory pathway. In the TGN, the RSKR247 motif is recognized by furin or furin-like PCs resulting in the cleavage of the mucin domain and GP38. The processed Gn and Gc, in conjunction with NP, L, and the RNA genome, can initiate virus assembly and be transported to the plasma membrane to be released into the medium (10, 23). GP38 and presumably the mucin protein as well as the uncleaved forms GP85 and GP160 are secreted to the medium in significant quantities.
FIG. 7.
Current model of CCHF virus glycoprotein processing.
In conclusion, in comparison to members of other genera of the family Bunyaviridae, the CCHF virus (genus Nairovirus) genome M segment encodes an unusually large polyprotein (1,684 amino acids in length) which undergoes complex proteolytic processing. CCHF virus glycoprotein processing is rather unique among RNA viruses in that in addition to cotranslational cleavage by signalase, it involves posttranslational cleavage by at least two additional classes of proteases, namely furin/PCs and SKI-1. Comparison of the deduced glycoprotein amino acid sequence of CCHF and Dugbe viruses suggests that many of the unusual features of CCHF virus glycoprotein processing may be shared with other tick-borne members of the genus Nairovirus. The likely role of the soluble glycoproteins and those with mucin-like properties (in addition to the the mature virion glycoproteins) in the pathogenesis of CCHF virus and other nairoviruses make it important to continue to more precisely define their synthesis and biologic functions.
Acknowledgments
We thank Jonathan Smith for kindly providing MAbs for CCHF virus proteins, Kathleen Murray for excellent editing of the manuscript, Varough Deyde for sharing unpublished CCHF virus sequence information, and Anthony Sanchez for helpful suggestions with the figures.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the funding agency.
REFERENCES
- 1.Barr, P. J. 1991. Mammalian subtilisins: the long-sought dibasic processing endoproteases. Cell 66:1-3. [DOI] [PubMed] [Google Scholar]
- 2.Basak, A., M. Zhong, J. S. Munzer, M. Chrétien, and N. G. Seidah. 2001. Implication of the proprotein convertases furin, PC5 and PC7 in the cleavage of surface glycoproteins of Hong Kong, Ebola and respiratory syncytial viruses: a comparative analysis with fluorogenic peptides. Biochem. J. 353:537-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertolotti-Ciarlet, A., J. Smith, K. Strecker, J. Paragas, L. A. Altamura, J. M. McFalls, N. Frias-Stäheli, A. García-Sastre, C. S. Schmaljohn, and R. W. Doms. 2005. Cellular localization and antigenic characterization of Crimean-Congo hemorrhagic fever virus glycoproteins. J. Virol. 79:6152-6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beyer, W. R., D. Pöpplau, W. Garten, D. von Laer, and O. Lenz. 2003. Endoproteolytic processing of the lymphocytic choriomeningitis virus glycoprotein by the subtilase SKI-1/S1P. J. Virol. 77:2866-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchmeier, M. J., M. D. Bowen, and C. J. Peters. 2001. Arenaviridae: the viruses and their replication, p. 1635-1668. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 2nd ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 6.Burns, J. W., and M. J. Buchmeier. 1993. Glycoproteins of the arenaviruses, p. 17-35. In M. S. Salvato (ed.), The arenaviridae. Plenum Press, New York, N.Y.
- 7.Collins, P. L., R. M. Chanock, and B. R. Murphy. 2001. Respiratory syncytial virus, p. 1443-1485. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 2nd ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 8.Comer, F. I., and G. W. Hart. 2000. O-Glycosylation of nuclear and cytosolic proteins. Dynamic interplay between O-GlcNAc and O-phosphate. J. Biol. Chem. 275:29179-29182. [DOI] [PubMed] [Google Scholar]
- 9.Dolnik, O., V. Volchkova, W. Garten, C. Carbonnelle, S. Becker, J. Kahnt, U. Ströher, H.-D. Klenk, and V. Volchkov. 2004. Ectodomain shedding of the glycoprotein GP of Ebola virus. EMBO J. 23:2175-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donets, M. A., M. P. Chumakov, M. B., Korolev, and S. G. Rubin. 1977. Physicochemical characteristics, morphology and morphogenesis of virions of the causative agent of Crimean hemorrhagic fever. Intervirology 8:294-308. [DOI] [PubMed] [Google Scholar]
- 11.Feldmann, H., S. T. Nichol, H.-D. Klenk, C. J. Peters, and A. Sanchez. 1994. Characterization of filoviruses based on differences in structure and antigenicity of the virion glycoprotein. Virology 199:469-473. [DOI] [PubMed] [Google Scholar]
- 12.Freed, E. O., and R. Risser. 1987. The role of envelope glycoprotein processing in murine leukemia virus infection. J. Virol. 9:2852-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garry, C. E., and R. F. Garry. 2004. Proteomics computational analyses suggest that the carboxyl terminal glycoproteins of bunyaviruses are class II viral fusion protein (beta-penetrenes). Theor. Biol. Med. Model. 1:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haferkamp, S., L. Fernando, T. F. Schwarz, H. Feldmann, and R. Flick. 2005. Intracellular localization of Crimean-Congo hemorrhagic fever (CCHF) virus glycoproteins. Virol. J. 2:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang, Y., L. Xu, Y. Sun, and G. J. Nabel. 2002. The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and posttranslational modification of nucleoprotein. Mol. Cell 10:307-316. [DOI] [PubMed] [Google Scholar]
- 16.Klenk, H.-D., and W. Garten. 1994. Host cell proteases controlling virus pathogenicity. Trends Microbiol. 2:39-43. [DOI] [PubMed] [Google Scholar]
- 17.Lenz, O., J. ter Meulen, H.-D. Klenk, N. G. Seidah, and W. Garten. 2001. The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc. Natl. Acad. Sci. USA 98:12701-12705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marriott, A. C., A. A. El-Ghorr, and P. A. Nuttall. 1992. Dugbe Nairovirus M RNA: nucleotide sequence and coding strategy. Virology 190:606-615. [DOI] [PubMed] [Google Scholar]
- 19.McCune, J. M., L. B. Rabin, M. B. Feinburg, M. Lieberman, J. C. Kosek, G. R. Reyes, and I. L. Weissman. 1988. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell 53:55-67. [DOI] [PubMed] [Google Scholar]
- 20.Nakayama, K. 1997. Furin: a mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem. J. 327:625-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neumann, G., H. Feldmann, S. Watanabe, I. Lulashevich, and Y. Kawaoka. 2002. Reverse genetics demonstrates that proteolytic processing of the Ebola virus glycoprotein is not essential for replication in cell culture. J. Virol. 76:406-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nichol, S. T. 2001. Bunyaviruses, p. 1603-1633. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 2nd ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 23.Pettersson, R. F., and L. Melin. 1996. Synthesis, assembly, and intracellular transport of Bunyaviridae membrane proteins, p 159-188. In R. M. Elliot (ed.), The Bunyaviridae. Plenum Press, New York, N.Y.
- 24.Sanchez, A. J., M. J. Vincent, and S. T. Nichol. 2002. Characterization of the glycoproteins of Crimean-Congo hemorrhagic fever virus. J. Virol. 76:7263-7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sänger, C., E. Mühlberger, B. Lötfering, H. D. Klenk, and S. Becker. 2002. The Marburg virus surface protein GP is phosphorylated at its ectodomain. Virology 295:20-29. [DOI] [PubMed] [Google Scholar]
- 26.Schmaljohn, C. S., and J. W. Hooper. 2001. Bunyaviridae: the viruses and their replication, p. 1581-1602. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 2nd ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 27.Seidah, N. G., S. J. Mowla, J. Hamelin, A. M. Mamarbachi, S. Benjannet, B. B Touré, A. Basak, J. S. Munzer, J. Marcinkiewicz, M. Zhong, J.-C. Barale, C. Lazure, R. A. Murphy, M. Chrétien, and M. Marcinkiewicz. 1999. Mammalian subtilisin/kexin isozyme SKI-1: a widely expressed proprotein convertase with a unique cleavage specificity and cellular localization. Proc. Natl. Acad. Sci. USA 96:1321-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seidah, N. G., and A. Prat. 2002. Precursor convertases in the secretory pathway, cytosol and extracellular milieu. Essays Biochem. 38:79-94. [DOI] [PubMed] [Google Scholar]
- 29.Simmons, G., R. J. Wool-Lewis, F. Baribaud, R. C. Netter, and P. Bates. 2002. Ebola virus glycoproteins induce global surface protein down-modulation and loss of cell adherence. J. Virol. 76:2518-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stadler, K., S. L. Allison, J. Schalich, and F. X. Heinz. 1997. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 71:8475-8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stieneke-Grober, A., M. Vey, H. Angliker, E. Shaw, G. Thomas, C. Roberts, H.-D. Klenk, and W. Garten. 1992. Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO J. 11:2407-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki, N., K. Yamamoto, S. Toyoshima, T. Osawa, and T. Irimura. 1996. Molecular cloning and expression of cDNA encoding human macrophage C-type lectin. J. Immunol. 156:128-135. [PubMed] [Google Scholar]
- 33.Takada, A., K. Fujioka, M. Tsuiji, A. Morikawa, N. Higashi, H. Ebihara, D. Kobasa, H. Feldmann, T. Irimura, and Y. Kawaoka. 2004. Human macrophage C-type lectin specific for galactose and N-acetylgalactosamine promotes filovirus entry. J. Virol. 78:2943-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vincent, M. J., A. J. Sanchez, B. R. Erickson, A. Basak, M. Chretien, N. G. Seidah, and S. T. Nichol. 2003. Crimean-Congo hemorrhagic fever virus glycoprotein proteolytic processing by subtilase SKI-1. J. Virol. 77:8640-8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Volchkov, V. E., H. Feldmann, V. A. Volchkova, and H. D. Klenk. 1998. Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc. Natl. Acad. Sci. USA 95:5762-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Volchkov, V. E., V. A. Volchkova., U. Stroher., S. Becker., O. Dolnik., Cieplik., W. Garten., H. D. Klenk, and H. Feldmann. 2000. Proteolytic processing of Marburg virus glycoprotein. Virology 268:1-6. [DOI] [PubMed] [Google Scholar]
- 37.Volchkova, V. A., H.-D. Klenk, and V. E. Volchkov. 1999. Delta-peptide is the carboxy-terminal cleavage fragment of the nonstructural small glycoprotein sGP of Ebola virus. Virology 265:164-171. [DOI] [PubMed] [Google Scholar]
- 38.Vollenweider, F., S. Benjannet, E. Decroly, D. Savaria, C. Lazure, G. Thomas, M. Chrétien, and N. G. Seidah. 1996. Comparative cellular processing of the human immunodeficiency virus (HIV-1) envelope glycoprotein gp160 by the mammalian subtilisin/kexin-like convertases. Biochem. J. 314:521-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe, M., A. Hirano, S. Stenglein, J. Nelson, G. Thomas, and T. C. Wong. 1995. Engineered serine protease inhibitor prevents furin-catalyzed activation of the fusion glycoprotein and production of infectious measles virus. J. Virol. 69:3206-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wertz, G. W., M. Krieger, and L. A. Ball. 1989. Structure and cell surface maturation of the attachment glycoprotein of human respiratory syncytial virus in a cell line deficient in O glycosylation. J. Virol. 63:4767-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wool-Lewis, R. J., and P. Bates. 1999. Endoproteolytic processing of the Ebola virus envelope glycoprotein: cleavage is not required for function. J. Virol. 73:1419-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang, Z.-Y., R. Delgado, L. Xu, R. F. Todd, E. G. Nabel, A. Sanchez, and G. J. Nabel. 1998. Distinct cellular interactions of secreted and transmembrane Ebola virus glycoproteins. Science 279:1034-1037. [DOI] [PubMed] [Google Scholar]
- 43.Yang, Z.-Y., H. J. Duckers, N. J. Sullivan, A. Sanchez, E. G. Nabel, and G. J. Nabel. 2000. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nature Med. 6:886-889. [DOI] [PubMed] [Google Scholar]