Abstract
Mimivirus, a parasite of Acanthamoeba polyphaga, is the largest DNA virus known; it encodes dozens of proteins with imputed functions in nucleic acid transactions. Here we produced, purified, and characterized mimivirus DNA topoisomerase IB (TopIB), which we find to be a structural and functional homolog of poxvirus TopIB and the poxvirus-like topoisomerases discovered recently in bacteria. Arginine, histidine, and tyrosine side chains responsible for TopIB transesterification are conserved and essential in mimivirus TopIB. Moreover, mimivirus TopIB is capable of incising duplex DNA at the 5′-CCCTT cleavage site recognized by all poxvirus topoisomerases. Based on the available data, mimivirus TopIB appears functionally more akin to poxvirus TopIB than bacterial TopIB, despite its greater primary structure similarity to the bacterial TopIB group. We speculate that the ancestral bacterial/viral TopIB was disseminated by horizontal gene transfer within amoebae, which are permissive hosts for either intracellular growth or persistence of many present-day bacterial species that have a type IB topoisomerase.
DNA topoisomerases are found in all free-living organisms; they modify DNA topology by introducing reversible enzyme-linked breaks in the DNA phosphodiester backbone (reviewed in reference 6). Topoisomerases accomplish this either by cleaving one strand of the DNA duplex and passing the intact complementary strand through the nick (type I topoisomerase) or by cleaving both strands and passing an intact duplex segment through the double-strand break (type II topoisomerase). Type II topoisomerases are oligomeric proteins that form covalent tyrosyl-5′-phosphodiester linkages at the two sites of strand scission. Type I enzymes are monomeric and are classified as having either type IA or type IB mechanisms. Type IA enzymes form a tyrosyl-5′-phosphodiester linkage at the break site, whereas type IB topoisomerases generate a tyrosyl-3′-phosphodiester (6).
DNA viruses encounter the same topological problems during replication and transcription as their hosts. Whereas small DNA viruses rely primarily on host cell enzymes for their replication and transcription (e.g., phiX174 and simian virus 40), the largest DNA viruses encode most, if not all, of the enzymatic machinery for viral nucleic acid transactions, including a DNA topoisomerase. Vaccinia virus topoisomerase, a type I enzyme, was discovered in 1977 (1) and, over the past 20 years, has been exploited as a model system for mechanistic studies of the topoisomerase IB enzyme family (5, 35-38). The type II topoisomerase of bacteriophage T4 was discovered in 1979 (22, 34). Type II topoisomerases have since been identified in the proteomes of eukaryotic viruses, including African swine fever virus (2, 10), Paramecium bursaria Chlorella virus (8, 9, 21), and Emiliana huxleyi Coccolithovirus (39). Unlike most free-living organisms, which have multiple topoisomerases from at least two of the major families, it appeared that viruses acquired no more than one topoisomerase in aid of their replication. This notion has been called into question by the discovery and genome analysis of mimivirus, a eukaryotic DNA virus with an extraordinarily large genome (1.2 Mb) and protein-coding capacity (19, 29).
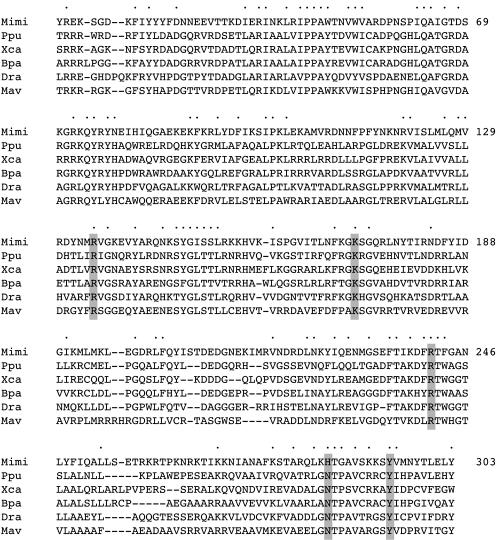
Remarkably, mimivirus simultaneously encodes type IA, type IB, and type II topoisomerases (29). It is not obvious why this virus would need three different types of topoisomerases. Mimivirus is a parasite of the protozoan Acanthamoeba polyphaga, and virtually nothing has been published about the viral replication cycle. The putative mimivirus type IB topoisomerase (MimiTopIB) displays primary structure similarity to poxvirus topoisomerases and, to an even greater extent, to the recently discovered family of bacterial poxvirus-like TopIB enzymes (15) (Fig. 1). Mimivirus TopIB (336 amino acids [aa]), poxvirus TopIB (314 aa), and bacterial TopIB proteins are of similar size, and they share the Arg-Lys-Arg-(His/Asn)-Tyr “catalytic pentad” responsible for the type IB DNA transesterification reaction (Fig. 1).
FIG. 1.
Mimivirus TopIB. The amino acid sequence of the mimivirus (Mimi) TopIB-like protein is aligned with the poxvirus-like bacterial TopIB polypeptides encoded by genes of Pseudomonas putida (Ppu), Xanthomonas campestris (Xca), Bordetella parapertussis (Bpa), Deinococcus radiodurans (Dra), and Mycobacterium avium (Mav). Gaps in the alignment are indicated by dashes. Positions of side chain identity and/or similarity in all six proteins are indicated by dots. The conserved equivalents of the active site of vaccinia virus TopIB are highlighted in shaded boxes.
Vaccinia exemplifies the Poxviridae, which replicate in the cytoplasm of the host cell. To accommodate the cytoplasmic lifestyle, poxviruses encode and encapsidate the enzymatic machinery for transcription of the viral genome and processing of viral mRNAs (24). This machinery includes a multisubunit DNA-dependent RNA polymerase, mRNA capping enzymes, and a poly(A) polymerase. The poxvirus TopIB is also encapsidated within the virus particle (1, 30, 31), where it plays an important role in the synthesis of viral early mRNAs (7). The fact that mimivirus encodes its own putative multisubunit RNA polymerase and RNA capping enzymes (29), in addition to a putative TopIB, highlights the likelihood that the mimivirus gene expression strategy will have a poxvirus-like flavor. We therefore thought it worthwhile to examine the biochemical properties of the mimivirus TopIB and the extent to which it resembles the vaccinia homolog.
Here, we report that MimiTopIB relaxes DNA supercoils in the absence of divalent cation or nucleotide cofactor. A notable finding was that MimiTopIB is able to cleave DNA at the same 5′-CCCTT↓ site recognized by all poxvirus DNA topoisomerases (13, 14, 18, 26, 28, 33). We determined the kinetic and equilibrium parameters for transesterification by MimiTopIB at this site. The supercoil relaxation and transesterification activities of MimiTopIB required the tyrosine nucleophile and at least three other constituents of the catalytic pentad. Our results attest to the evolutionary and functional relatedness of the mimivirus, poxvirus, and bacterial topoisomerases and suggest a model for dissemination of TopIB by horizontal transfer in an ancestral amoebal host.
MATERIALS AND METHODS
T7-based vector for expression of mimivirus TopIB.
Oligonucleotide primers complementary to the 5′ and 3′ ends of the mimivirus gene R194 encoding a 336-aa TopIB-like polypeptide (GenBank accession no. NC_006450) were used to amplify the complete open reading frame. A pUC19 clone containing the genomic locus was used as the template for PCR. The sense-strand primer 5′-d(GGAGATCTAGTCTTATGACGAAAAACATCTCATGGAA) was designed to introduce a BglII restriction site upstream of the translation start codon of the TopIB gene. The antisense primer 5′-d(CCTAGATCTATTCTCCAAGTACATCTTTTCTATATGA) introduced a BglII site 3′ of the stop codon. The 1-kbp PCR product was digested with BglII and then cloned into the BamHI site of the T7-based expression plasmid pET28b-His10-Smt3. Single alanine mutations R135A, R241A, H285A, and Y294A were introduced into the MimiTopoIB gene via two-stage PCR overlap extension (12). The entire viral DNA inserts in the wild-type and mutant pET28b-His10-Smt3-MimiTopIB plasmids were sequenced in order to exclude the introduction of unwanted coding changes during amplification and cloning.
Recombinant MimiTopIB.
The pET28b-His10-Smt3-MimiTopIB plasmids were transformed into Escherichia coli BL21(DE3)-RIL (Stratagene). Cultures (500 ml) derived from single transformants were grown at 37°C in Luria-Bertani medium containing 50 μg/ml kanamycin and 12.5 μg/ml chloramphenicol until the A600 reached 0.6. The cultures were adjusted to 0.5 mM isopropyl-1-thio-β-d-galactopyranoside and 2% ethanol and placed on ice for 2 h, after which incubation was continued for 20 h at 17°C with constant shaking. The cells were harvested by centrifugation, and the pellets were stored at −80°C. All of the subsequent steps were performed at 4°C. Thawed bacteria were resuspended in 25 ml of buffer A (50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 10% glycerol). Phenylmethylsulfonyl fluoride and lysozyme were added to final concentrations of 250 μM and 100 μg/ml, respectively. After 30 min, Triton X-100 was added to a final concentration of 0.1% and the lysate was sonicated to reduce viscosity. Insoluble material was removed by centrifugation for 45 min at 16,000 rpm in a Sorval SS34 rotor. The soluble extract was mixed for 1 h with 4 ml of Ni2+-nitrilotriacetic acid-agarose resin (QIAGEN) that had been preequilibrated with buffer A. The resin was recovered by centrifugation, resuspended in buffer A, and then poured into a column and washed with 45 ml of buffer A containing 5 mM imidazole. Bound material was eluted with 500 mM imidazole in buffer A. The eluate was diluted with buffer A to achieve a protein concentration of 1 mg/ml and then dialyzed against buffer A containing 1 mM dithiothreitol (DTT) and 0.01% Triton X-100. The His10-Smt3-tag was removed by treatment of the recombinant protein with Ulp1 (a Smt3-specific protease) (25) for 15 min on ice at a Ulp1:MimiTopIB ratio of 1:2,000. The digests were applied to 1-ml columns of Ni2+-nitrilotriacetic acid-agarose equilibrated with buffer A. The tag-free MimiTopIB proteins were recovered in the flowthrough fractions. The MimiTopIB proteins were stored at −80°C. Protein concentrations were determined by using the Bio-Rad dye reagent with bovine serum albumin (BSA) as the standard.
Glycerol gradient sedimentation.
An aliquot (50 μg) of the tag-free wild-type MimiTopIB was mixed with catalase (40 μg), BSA (40 μg), and cytochrome c (40 μg). The mixture was applied to a 5-ml 15 to 30% glycerol gradient containing 50 mM Tris-HCl, pH 7.5, 1 M NaCl, 2 mM DTT, 1 mM EDTA, and 0.05% Triton X-100. The gradient was centrifuged at 50,000 rpm for 12 h at 4°C. Fractions (∼0.19 ml) were collected from the bottom of the tube.
DNA relaxation.
Reaction mixtures containing (per 20 μl) 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.3 μg pUC19 plasmid DNA, and TopIB as specified were incubated at 37°C. The reactions were quenched by addition of a solution containing glycerol, xylene cyanol, bromophenol blue, and SDS (0.2% final concentration). The products were analyzed by electrophoresis through horizontal 1.0% agarose gels in TBE buffer (90 mM Tris borate, 2.5 mM EDTA). The gel was stained with 0.5 μg/ml ethidium bromide and then photographed under short-wave UV illumination.
Suicide DNA cleavage.
An 18-mer CCCTT-containing DNA oligonucleotide was 5′ end-labeled by enzymatic phosphorylation in the presence of [γ-32P]ATP and T4 polynucleotide kinase and then gel purified and hybridized to a complementary 30-mer strand (present at sevenfold molar excess). Cleavage reaction mixtures containing (per 20 μl) 50 mM Tris-HCl, pH 7.5, 0.3 pmol 18-mer/30-mer DNA, and TopIB as specified were incubated at 37°C. The reactions were quenched by adding sodium dodecyl sulfate (SDS) to a 1% final concentration. The products were analyzed by electrophoresis through a 10% polyacrylamide gel containing 0.1% SDS. Covalent complex formation was revealed by transfer of radiolabeled DNA to the TopIB polypeptide. The extent of covalent complex formation was quantified by scanning the gel using a Fujifilm BAS2500 imager. A plot of the percentage of input DNA cleaved versus time established the endpoint values for cleavage. The data were then normalized to the endpoint values (defined as 100%), and the cleavage rate constants (kcl) were calculated by fitting the normalized data to the equation 100 − % cleavage(norm) = 100e−kt.
Single-turnover religation by the suicide intermediate.
Cleavage reaction mixtures (90 μl) containing 50 mM Tris-HCl, pH 7.5, 2.7 pmol 32P-labeled 18-mer/30-mer DNA, and 4.1 μg MimiTopIB (wild type or H285A) were incubated at 37°C for 30 min to form the suicide intermediate. An aliquot (10 μl; religation time zero) was withdrawn and quenched with SDS. Religation was initiated by adding 80 μl of a prewarmed (37°C) solution containing 0.5 M NaCl and 240 pmol of the 5′-OH 18-mer acceptor strand 5′-d(ATTCCGATAGTGACTACA) (i.e., a 100-fold molar excess over the input DNA substrate). Aliquots (20 μl) were withdrawn at various times and quenched immediately with 5 μl of 2% SDS. The samples were digested for 60 min at 37°C with 10 μg of proteinase K and then mixed with an equal volume of 95% formamide, 20 mM EDTA. The samples were heat denatured, and aliquots (10 μl) were analyzed by electrophoresis through a 40-cm 17% polyacrylamide gel containing 7 M urea in TBE. Religation of the covalently bound 12-mer strand to the 18-mer acceptor DNA yielded a 5′ 32P-labeled 30-mer strand transfer product. The products were quantified by scanning the gel. The extent of religation (expressed as the percent of the covalent intermediate converted into 30-mer) was plotted as a function of reaction time. The data were normalized to the endpoint values, and krel was determined by fitting the data to the equation 100 − % religated(norm) = 100e−kt.
Equilibrium cleavage.
A 34-mer CCCTT-containing oligonucleotide was 5′ 32P-labeled and then gel purified and annealed to an unlabeled complementary 60-mer strand (present in sevenfold molar excess). Equilibrium cleavage reaction mixtures (20 μl) containing 50 mM Tris-HCl, pH 7.5, 0.3 pmol 34-mer/60-mer DNA, and TopIB as specified were incubated for 10 min at 37°C. The reactions were quenched by adding SDS to a 0.4% final concentration. The samples were digested for 60 min at 37°C with 10 μg of proteinase K. The digests were then adjusted to 47% formamide and heat denatured, and aliquots (10 μl) were analyzed by electrophoresis through a 40-cm 17% polyacrylamide gel containing 7 M urea in TBE. The cleavage product, comprising a cluster of 32P-labeled 12-mer oligonucleotide bound to short peptides, was resolved from the labeled input 34-mer scissile strand. The extent of strand cleavage was quantified by scanning the gel. The cleavage equilibrium constant (Kcl) is defined as the ratio of covalently bound DNA to noncovalently bound DNA at the reaction endpoint under conditions of saturating enzyme and was calculated according to the equation Kcl = % cleaved/(100 − % cleaved).
RESULTS
Recombinant MimiTopIB relaxes supercoiled DNA.
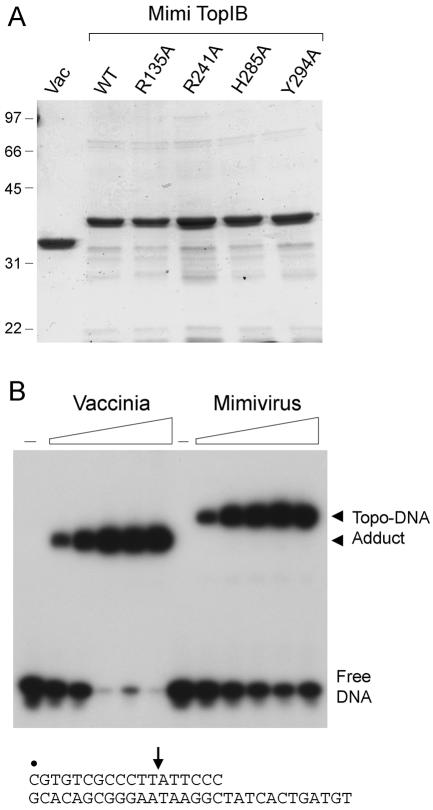
MimiTopIB was produced in E. coli as a His10-Smt3 fusion and purified from a soluble bacterial lysate by Ni2+-agarose chromatography. The His10-Smt3 tag was removed by the Smt3-specific protease Ulp1 (25), and the tag-free MimiTopIB was separated from the tag by a second step of Ni2+-agarose chromatography. SDS-polyacrylamide gel electrophoresis (PAGE) showed that the preparation consisted of a major 37-kDa polypeptide corresponding to MimiTopIB (Fig. 2A). The recombinant protein was reacted with supercoiled pUC19 plasmid DNA in mixtures containing no divalent cation or nucleoside triphosphate; the products were analyzed by agarose gel electrophoresis and visualized by staining the gel with ethidium bromide (Fig. 3A). Topoisomerase activity was evinced by near quantitative conversion of the supercoiled (SC) plasmid substrate to slower migrating relaxed (Rel) products. The extent of relaxation during a 30-min reaction increased with the amount of input MimiTopIB (Fig. 3A, left panel). Relaxed DNA accumulated steadily with time (Fig. 3A, right panel), and a subpopulation of partially relaxed topoisomers was evident among the products. Analysis of the reaction products by agarose gel electrophoresis in the presence of ethidium bromide verified that the products were covalently closed relaxed circles, not nicked circles (data not shown). MimiTopIB relaxed DNA over a broad pH range, from pH 5.5 to pH 9.0 in 50 mM Tris-acetate or Tris-HCl buffers. No activity was detected in Tris-acetate buffer below pH 5.0 (data not shown). MgCl2 was not required for supercoil relaxation, and the addition of MgCl2 up to 5 mM did not increase the activity (data not shown).
FIG. 2.
Purification and DNA cleavage activity of MimiTopIB. (A) Aliquots (5 μg) of the tag-free wild-type (WT) MimiTopIB and the R135A, R241A, H285A, and Y294A mutants were analyzed by SDS-PAGE in parallel with recombinant vaccinia virus TopIB (Vac). The Coomassie blue-stained gel is shown. The positions and sizes (in kilodaltons) of marker polypeptides are indicated on the left. (B) Reaction mixtures (20 μl) containing 50 mM Tris-HCl, pH 7.5, 0.3 pmol 5′-labeled 18-mer/30-mer suicide DNA substrate (depicted at bottom, with the 5′ label indicated by the dot and the presumptive cleavage site denoted by the arrow) and increasing amounts of vaccinia or mimivirus TopIB (75, 150, 300, 450, or 600 ng, proceeding from left to right) were incubated at 37°C for 10 min. TopIB was omitted from control reaction mixtures in lanes labeled with dashes. The reactions products were resolved by SDS-PAGE. An autoradiograph of the gel is shown. The positions of the Topo-DNA adducts and the free DNA are indicated on the right.
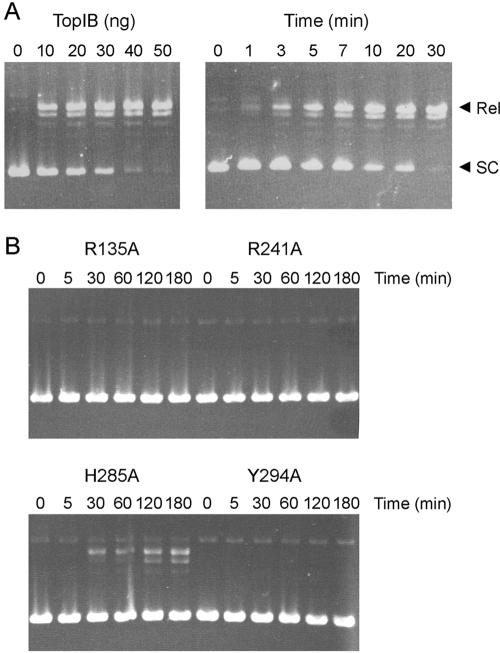
FIG. 3.
Supercoil relaxation activity of MimiTopIB. (A) The left panel shows reaction mixtures (20 μl) containing 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.3 μg pUC19 DNA, and 0, 10, 20, 30, 40, or 50 ng of wild-type MimiTopIB incubated for 30 min at 37°C. The right panel shows a reaction mixture (160 μl) containing 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 2.4 μg pUC19 DNA, and 200 ng of wild-type protein incubated at 37°C. Aliquots (20 μl) were withdrawn at the times specified. The DNA products were resolved by agarose gel electrophoresis and stained with ethidium bromide. The positions of supercoiled (SC) and relaxed (Rel) circular DNAs are indicated on the right. (B) Reaction mixtures (160 μl) containing 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 2.4 μg pUC19 DNA, and 400 ng of the indicated MimiTopIB-Ala proteins were incubated at 37°C. Aliquots (20 μl) were withdrawn at the times specified.
Velocity sedimentation.
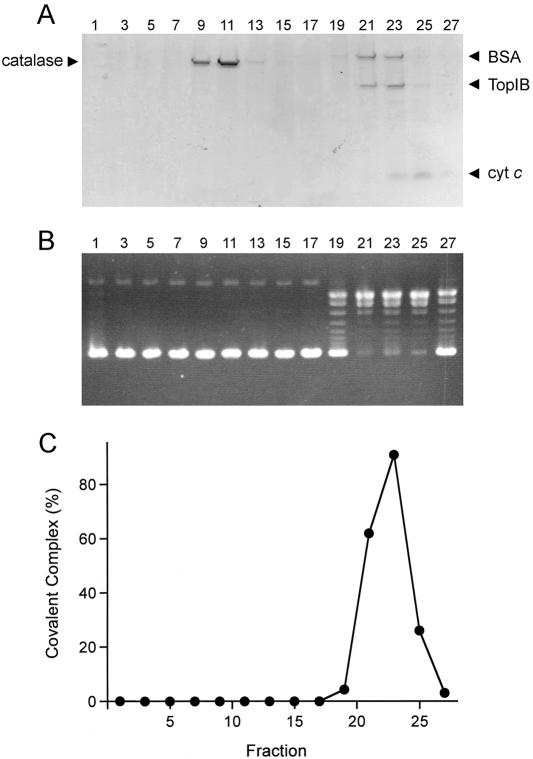
The native size of MimiTopIB was gauged by zonal velocity sedimentation through a 15 to 30% glycerol gradient containing 1 M NaCl. Marker proteins catalase (248 kDa), BSA (66 kDa), and cytochrome c (12 kDa) were included as internal standards. The odd-numbered gradient fractions were analyzed by SDS-PAGE and assayed for DNA relaxation. The 37-kDa MimiTopIB polypeptide sedimented as a single component with a peak at fraction 23 (Fig. 4A), coincident with a single symmetrical component of topoisomerase activity peaking in fractions 21 to 25 (Fig. 4B). No relaxation activity was detected in the gradient fractions on the heavy side of the BSA marker. The activity peak at fraction 23 overlapped the “light” side of the BSA peak. The sedimentation results are consistent with MimiTopIB being a monomer in solution, although we cannot exclude the prospect that MimiTopIB is an asymmetrically shaped homodimer. Note that the sedimentation behavior of MimiTopIB is acutely dependent on the salt concentration of the gradient buffer; when the protein was sedimented in a gradient containing 200 mM NaCl, the MimiTopIB polypeptide (and the relaxation activity) were evenly distributed in fractions extending from the bottom of the gradient to the light side of the BSA peak (not shown). We surmise that MimiTopIB is prone to oligomerize at lower ionic strength.
FIG. 4.
Glycerol gradient sedimentation. MimiTopIB was sedimented through a 15 to 30% glycerol gradient as described in Materials and Methods. (A) Aliquots (18 μl) of the odd-numbered gradient fractions were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown. The polypeptides corresponding to MimiTopIB and marker proteins catalase, BSA, and cytochrome c (cyt c) are indicated by arrowheads flanking the gel. (B) DNA relaxation activity. Reaction mixtures (20 μl) containing 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.3 μg pUC19 DNA, and 2.5 μl of the odd-numbered gradient fractions were incubated at 37°C for 10 min. The DNA products were resolved by agarose gel electrophoresis and stained with ethidium bromide. (C) DNA cleavage activity. Reaction mixtures (20 μl) containing 50 mM Tris-HCl, pH 7.5, 0.3 pmol 32P-labeled 18-mer/30-mer suicide substrate, and 2.5 μl of the odd-numbered gradient fractions were incubated at 37°C for 15 min. Activity was quantified as the percentage of the input labeled material that was converted to a covalent TopIB-DNA complex.
Conservation of the active site in mimivirus and poxvirus TopIB.
Previous studies of vaccinia TopIB showed that conserved amino acid side chains Arg-130, Lys-167, Arg-223, and His-265 catalyze the attack of Tyr-274 on the scissile phosphodiester (4, 27, 40). These five residues are conserved in MimiTopIB as Arg-135, Lys-172, Arg-241, His-285, and Tyr294 (Fig. 1). To determine whether the catalytic mechanism of MimiTopIB is similar to that of vaccinia TopIB, we introduced alanine substitutions for MimiTopIB residues Arg-135, Arg-241, His-285, and Tyr-294 and tested their effects on activity. The recombinant proteins R135A, R241A, H285A, and Y294A were produced in E. coli as His10-Smt3 fusions, purified by Ni2+-agarose chromatography, treated with Ulp1 to remove the tag, and then freed of the tag by a second Ni2+-agarose step. SDS-PAGE showed that the purity of the mutant proteins was comparable to that of wild-type MimiTopIB (Fig. 2A).
A kinetic analysis of supercoil relaxation by the mutant enzymes is shown in Fig. 3B. Whereas 25 ng of the wild-type MimiTopIB relaxed 0.3 μg of the plasmid DNA to completion in 30 min, 50 ng of the R135A, R241A, and Y294A mutants generated no relaxed product after 3 h. The H285A mutant displayed a low level of supercoil relaxation, which agrees with the mutational data for His265 of vaccinia TopIB (27). We conclude that the active sites of MimiTopIB and vaccinia TopIB are composed of similar functional groups.
MimiTopIB transesterifies at the 5′-CCCTT target site of poxvirus TopIB.
Type IB topoisomerases alter DNA topology by cleaving and rejoining one strand of the DNA duplex. Cleavage occurs via a transesterification reaction in which the scissile phosphodiester is attacked by a tyrosine of the enzyme, resulting in the formation of a DNA-(3′-phosphotyrosyl) enzyme intermediate and the expulsion of a 5′-OH DNA strand. In the rejoining step, the DNA 5′-OH group attacks the covalent intermediate to expel the active site tyrosine and restore the DNA phosphodiester backbone. Vaccinia TopIB is distinguished by its specificity for DNA transesterification at a pentapyrimidine target sequence 5′-(T/C)CCTT↓ (33). The Tp↓ nucleotide is linked to Tyr274 of the enzyme. Topoisomerases encoded by other genera of vertebrate and invertebrate poxviruses recognize the same DNA target sequence (13, 14, 18, 26, 28), despite the large variations in overall G+C contents of poxvirus genomes. However, mammalian TopIB is incapable of transesterifying at the DNA target sites exploited by vaccinia TopIB (23).
The cleavage reaction of vaccinia TopIB is amenable to study under single-turnover conditions using a “suicide substrate” composed of a 5′ 32P-labeled 18-mer strand containing a CCCTTp↓A cleavage site annealed to an unlabeled 30-mer complementary strand (Fig. 2B). Transesterification at the TpA linkage results in covalent attachment of a 5′ 32P-labeled 12-mer (5′-pCGTGTCGCCCTTp) to the enzyme to form a radiolabeled Topo-DNA adduct that is resolved from free DNA during SDS-PAGE (Fig. 2B). The instructive finding was that incubation of MimiTopIB with this suicide substrate resulted in label transfer to the MimiTopIB polypeptide. The MimiTopIB-DNA adduct migrated more slowly than the vaccinia TopIB-DNA complex during SDS-PAGE (Fig. 2B), consistent with the relative migration of the free vaccinia and mimivirus TopIB polypeptides (Fig. 2A). The transesterification activity profile coincided with the MimiTopIB polypeptide and the DNA relaxation activity during glycerol gradient sedimentation (Fig. 4C), verifying that the reaction was intrinsic to MimiTopIB.
Transesterification kinetics.
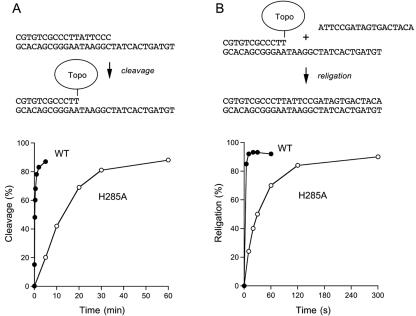
In the suicide cleavage reaction, the unlabeled 6-mer 5′-OH leaving strand (5′ATTCCC) dissociates spontaneously from the protein-DNA complex and thereby drives the reaction toward the covalent state, so that the reaction can be treated kinetically as a unidirectional process (35, 40). The reaction of a molar excess of wild-type MimiTopIB with the suicide substrate resulted in a time-dependent accumulation of the TopIB-DNA complex; the reaction attained an endpoint of 87% covalent adduct after 5 to 10 min (Fig. 5A). The data fit well to a single exponential with an apparent cleavage rate constant of 0.06 s−1. The cleavage rate constant of MimiTopIB is about one-sixth the kcl value determined for vaccinia TopIB on the same substrate.
FIG. 5.
Kinetic analysis of single-turnover cleavage and religation. (A) Single-turnover cleavage. The 5′ 32P-labeled suicide substrate and the covalent topoisomerase-DNA adduct are depicted at the top of the figure. Reaction mixtures (160 μl) containing 50 mM Tris-HCl, pH 7.5, 2.4 pmol 18-mer/30-mer DNA, and 3.6 μg of wild-type or H285A MimiTopIB were incubated at 37°C. Aliquots were withdrawn at 0, 5, 10, 20, 30, 60, 120, and 300 s for the wild-type (WT) MimiTopIB and at 0, 5, 10, 20, 30, and 60 min for the H285A mutant and quenched immediately by adding SDS. The percentage of the total radiolabel in the covalent TopIB-DNA complex is plotted as a function of time. (B) Single-turnover religation. The suicide covalent intermediate, the 18-mer acceptor strand, and the religation reaction product are depicted at the top of the figure. The suicide cleavage complex was formed during a 30-min preincubation at 37°C as described in Material and Methods. Religation was initiated by the simultaneous addition of NaCl to 0.25 M and a 100-fold molar excess of the 18-mer acceptor strand. Aliquots were withdrawn at the times specified. The extent of religation is plotted as a function of time postaddition of the acceptor strand.
The R135A, R241A, and Y294A mutants of MimiTopIB formed no detectable protein-DNA adduct during a 24-h incubation with the suicide substrate (data not shown). Thus, the failure of these three proteins to relax supercoiled DNA (Fig. 3B) is probably attributable to their inability to perform transesterification chemistry. The H285A mutant cleaved the suicide substrate, albeit slowly. Covalent adduct accumulated steadily over 30 min; 88% of the input substrate was cleaved after 1 h (Fig. 5A). The H285A data fit well to a single exponential with an apparent cleavage rate constant of 0.001 s−1. The finding that the H285A mutation slowed the rate of cleavage by a factor of 60 compared to wild-type MimiTopIB likely accounts for the slowed DNA relaxation activity of the H285A enzyme (Fig. 3B).
The religation step of the TopIB catalytic cycle can be studied under single-turnover conditions by assaying the ability of a preformed TopIB-DNA complex to transfer the covalently held 5′ 32P-labeled strand to a heterologous acceptor strand, as depicted in Fig. 5B. Wild-type MimiTopIB and the H285A mutant were incubated with the suicide cleavage substrate for 30 min in order to attain near-equivalent levels of the covalent intermediate. We then added a 100-fold molar excess of an 18-mer acceptor strand complementary to the 5′ tail of the covalent donor complex (Fig. 5B) while simultaneously increasing the ionic strength to 0.25 M NaCl. (Addition of NaCl during the religation phase promotes dissociation of the topoisomerase after strand closure and prevents recleavage of the strand transfer product.) Religation to the 18-mer yielded the 32P-labeled 30-mer depicted in Fig. 5B. The strand transfer product was resolved from the input 32P-labeled 18-mer strand by denaturing gel electrophoresis. The extent of religation by wild-type MimiTopIB at the earliest time point analyzed (5 s) was 91% of the endpoint value. We used this datum to estimate a religation rate constant of at least 0.5 s−1. Strand transfer by H285A was much slower. The religated 30-mer accumulated steadily over 2 min and attained an endpoint at 5 min. The apparent religation rate constant was 0.03 s−1. We surmise that His285 promotes both transesterification steps of the TopIB reaction.
Transesterification equilibrium.
A 5′ 32P-labeled CCCTT-containing duplex containing 12 bp of DNA upstream of the cleavage site and 22 bp of DNA downstream of the cleavage site was employed to assay transesterification under equilibrium conditions (Fig. 6). This DNA is an equilibrium substrate, because the 5′-OH leaving strand generated upon cleavage at CCCTT remains stably associated with the topoisomerase-DNA complex via base-pairing to the nonscissile strand. We determined by enzyme titration that MimiTopIB cleaved 5% of the unmodified substrate at saturation. We thereby derived a value of 0.05 for the cleavage-religation equilibrium constant (Kcl), which is defined as the ratio of covalent complex/noncovalent complex when all potential cleavage sites are occupied by TopIB. (The assumption is that all labeled strands are annealed to form a reactive duplex and that all sites are occupied at saturating enzyme, so that the fraction of noncovalent complex can be taken as 100% minus the percent of covalent adduct.) In a parallel experiment, vaccinia topoisomerase cleaved 25% of the input substrate at saturation (Kcl = 0.33) (data not shown).
FIG. 6.
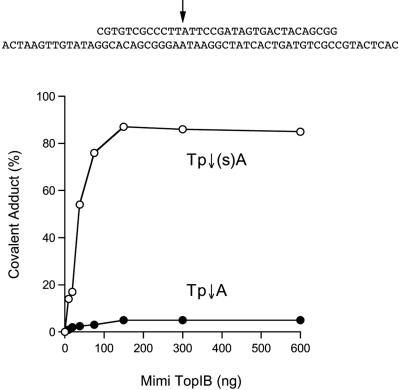
Equilibrium cleavage. A 5′ 32P-labeled 34-mer scissile strand, containing either a standard Tp↓A phosphodiester or a 5′-bridging phosphorothiolate Tp↓(S)A at the presumptive cleavage site, was hybridized to a sevenfold molar excess of the complementary 60-mer strand to form the equilibrium cleavage substrate shown at the top of the figure. The cleavage site is indicated by the arrow. Cleavage reactions were performed as described in Materials and Methods. The extent of covalent adduct formation is plotted as a function of input MimiTopIB protein.
The cleavage-religation equilibrium constant Kcl is also defined as the ratio of the cleavage and religation rate constants (kcl/krel). Although we regard the observed kcl value of 0.06 s−1 as accurate, the apparent religation rate of ≥0.5 s−1 must be regarded as a lower bound estimate, insofar as the reaction was virtually complete after 5 s, and we are unable to manually measure product formation at shorter time intervals. However, we can estimate krel using the ratio of experimental values kcl/Kcl (0.06/0.05), which yields a religation rate of ∼1.2 s−1. The latter value is similar to the religation rate derived for vaccinia TopIB at the same cleavage site (40). We surmise that the ∼6-fold lower equilibrium constant of mimivirus versus vaccinia TopIB reflects the ∼6-fold slower rate of the forward cleavage transesterification reaction of MimiTopIB.
Effect of a 5′-bridging phosphorothiolate on the cleavage-religation equilibrium.
Introduction of a single 5′-bridging phosphorothiolate linkage (C3′-O-[PO2]-S-C5′) at the scissile phosphodiester of an equilibrium duplex substrate results in the trapping of mammalian and poxvirus TopIB in the covalently bound state (3, 16, 18). An explanation for the altered equilibrium is that the 5′-SH leaving strand in the cleavage reaction is a relatively poor nucleophile in the religation step. Here we compared the reactivity of MimiTopIB with equilibrium substrates containing a 34-mer scissile strand with either a standard phosphodiester or a 5′-bridging phosphorothiolate at the predicted cleavage site (Fig. 6). The enzyme dependence of covalent complex formation on the thio substrate was similar to that of the standard DNA (i.e., both substrates were saturated at 150 ng of input TopIB), implying that the thio substitution had no major impact on site affinity, but the yield of covalent adduct at saturating levels of MimiTopIB was increased to 87% on the thio substrate, compared to 5% on the phosphodiester substrate (Fig. 6). Thus, Kcl was increased more than 100-fold, from 0.05 to 6.7. This result verifies that MimiTopIB incises the CCCTT-containing strand at the same internucleotide linkage as the poxvirus topoisomerases.
DISCUSSION
Mimivirus encodes dozens of proteins with imputed functions in nucleic acid transactions (29). Here we produced, purified, and characterized mimivirus TopIB, which we find to be a structural and functional homolog of vaccinia TopIB, insofar as (i) amino acid side chains responsible for transesterification chemistry by vaccinia TopIB are conserved and essential in MimiTopIB and (ii) MimiTopIB is capable of recognizing and incising duplex DNA at the 5′-CCCTT cleavage site shared by all known poxvirus DNA topoisomerases. This is the first case of a viral TopIB from a nonpoxvirus source and the first instance in which a nonpoxviral TopIB has been shown to cleave at the 5′-CCCTT target site.
An initial mutational analysis of MimiTopIB shows that loss of the Arg135, Arg241, and Tyr294 side chains abolishes DNA relaxation and cleavage activity. The elimination of the tyrosine nucleophile is expected to elicit such an effect. The degree of reliance on Arg135 and Arg241 is consistent with studies of vaccinia TopIB, showing that each of these two arginines (Arg130 and Arg233 in vaccinia TopIB) contributes a 105-fold enhancement of the rate of transesterification (4, 40). Arg130 (Arg135 in MimiTopIB) is part of a proton relay that catalyzes the expulsion of the 5′-O of the leaving DNA strand (16, 17), whereas Arg223 promotes transesterification by neutralizing the extra negative charge developed in the proposed pentacoordinate phosphorane transition state (38). The H285A mutation in MimiTopIB suppresses DNA relaxation and slows the DNA cleavage rate by a factor of 60. The equivalent histidine of vaccinia TopIB (His265) accelerates transesterification by a factor of 102 by donating a hydrogen bond to a nonbridging oxygen of the scissile phosphodiester to stabilize the transition state (36, 37). We infer that the active site and catalytic mechanism of MimiTopIB adhere closely to that proposed for the poxvirus enzyme.
MimiTopIB joins a growing TopIB enzyme family that includes eukaryotic nuclear and mitochondrial TopIB (41), poxvirus topoisomerases, and poxvirus-like topoisomerases encoded by the genomes of many bacteria (15). Mimivirus, poxvirus, and bacterial TopIB proteins are of similar size (314 to 417 aa). Human TopIB (a 765-aa polypeptide) and other nuclear TopIB enzymes are much larger and contain multiple structural modules added on to a protein scaffold that is otherwise quite similar to that of the vaccinia TopIB (5, 6, 15). It has been proposed that nuclear TopIB enzymes evolved from a bacterial or poxvirus-like precursor (15). MimiTopIB is more closely related in overall primary structure to the bacterial group of poxvirus-like topoisomerases (30 to 33% identity in pairwise alignments) than it is to the poxvirus topoisomerases themselves (23% identity). Thus far, only one member of the bacterial TopIB group has been characterized biochemically, the TopIB from Deinococcus radiodurans (DraTopIB) (15). DraTopIB relaxes supercoiled DNA and relies on the same set of conserved catalytic residues as the poxvirus enzyme. However, DraTopIB is conspicuously incapable of cleaving CCCTT-containing suicide substrates (even phosphorothiolate-modified substrates) that are efficiently incised by vaccinia TopIB and, as shown here, by MimiTopIB. Based on the available data, we surmise that MimiTopIB is functionally more akin to its poxvirus cousins than it is to an exemplary bacterial TopIB, despite greater structural conservation with the latter.
Although MimiTopIB can cleave the poxvirus TopIB consensus target site in model substrates, we do not yet know if its cleavage specificity is confined to the poxvirus recognition site or whether MimiTopIB can also transesterify at additional sites that are not recognized by vaccinia TopIB. Preliminary efforts to map specific sites of MimiTopIB cleavage in linear plasmid DNA have not been fruitful, perhaps because the cleavage-religation equilibrium favors the noncovalent state, as we observed for the model CCCTT-containing equilibrium cleavage substrate. It is presumed that the cleavage specificity of viral TopIB proteins is somehow related to their biological functions. Vaccinia TopIB promotes early mRNA synthesis within the virion particle (7). Mimivirus encodes a poxvirus-like RNA polymerase and RNA capping enzyme, but it is not yet known if the mRNA synthetic machinery, or MimiTopIB, are packaged within the mimivirus particle.
Given the likelihood that the poxvirus, mimivirus, and bacterial TopIB proteins comprise a discrete subgroup of structurally conserved low-molecular-weight topoisomerases with a shared ancestry, it is of interest to consider how they came to be distributed among present-day bacterial species and eukaryal viruses. A reasonable assumption is that this flavor of TopIB was disseminated by horizontal gene transfer (15). The open questions are whether the spread was mediated via a bacterium or a virus (or both) and in which ecological niche(s) the transfer was likely to have occurred. The closer primary structure similarity between mimivirus and bacterial TopIB leads us to propose that amoebae are plausible incubators for transfer of TopIB between bacteria and eukaryotic viruses. Free-living amoebae are ubiquitous in marine and soil environments, where they tend to gravitate to biofilms or water-plant interfaces and feed on assorted bacteria, fungi, and algae. Many microorganisms have been identified that have the capacity to survive or grow within amoebae and then exit after internalization (reviewed in reference 11). Amoeba-resistant bacteria include Pseudomonas aeruginosa, Burkholderia cepacia, Bradyrhizobium japonicum, Parachlamydia, Mycobacterium avium, and Mycobacterium smegmatis. Each of these bacteria has been isolated by (or shown to be resistant to) coculture with Acanthamoeba, the waterborne amoeba from which mimivirus was isolated (11). Each of these amoeba-resistant bacteria has a poxvirus/mimivirus-like TopIB. Acanthamoeba is a natural host for Ralstonia picketii; the related species Ralstonia eutropha has a poxvirus/mimivirus-like TopIB. Cytophaga species have been identified as endosymbionts of amoebae; Cytophaga hutchinsonii has a poxvirus/mimivirus-like TopIB.
The potential for transient coexistence within amoebae of diverse bacterial species and a eukaryal DNA virus sets the stage for the transfer of TopIB between microbial genomes. One plausible scenario is that a DNA virus was the ancestral source and that diverse bacterial genera acquired the TopIB gene when they and the virus found themselves in the same amoebal niche. Alternatively, a bacterium was the original source for dissemination to other bacteria that enter amoebae as well as transfer to an ancestral DNA virus. Given the cleavage specificity data presented here, we envision that an ancestral poxvirus either acquired its TopIB gene from a mimi-like virus or that the ancestral poxvirus and mimivirus derive from a common progenitor virus that replicated in a unicellular eukaryal host. The impediment to connecting the dots between mimivirus and poxviruses is that no poxviruses have yet been identified in primitive metazoa (below the level of arthropoda), protozoa, or fungi.
The mechanistic similarities and overlapping cleavage site specificities of poxvirus and mimivirus TopIB are potentially relevant to the discovery of novel TopIB poisons as antiviral agents. Topoisomerase poisons, which trap the covalent protein-DNA intermediates of either type II topoisomerases or nuclear TopIB, are mainstays of antibacterial and anticancer therapy. Camptothecin, the prototypal poison of human TopIB, is ineffective against the poxvirus TopIB (23, 32). Clinically significant natural poxvirus infections of humans include smallpox, monkeypox, and molluscum contagiosum, plus the various complications of intentional infection with vaccinia virus for smallpox prophylaxis. Mimivirus was recently detected in the bronchoalveolar lavage specimen of a patient with hospital-acquired pneumonia (20) and in a laboratory technician working with mimivirus who developed acute pneumonia (29a), highlighting its potential for causing human disease. We expect that further comparative studies, structural and functional, of viral TopIB enzymes will provide useful insights to their site specificity and the evolution of the TopIB enzyme family.
Acknowledgments
This work was supported by NIH grant GM46330. S.S. is an American Cancer Society Research Professor.
REFERENCES
- 1.Bauer, W. R., E. C. Ressner, J. Kates, and J. V. Patzke. 1977. A DNA nicking-closing enzyme encapsidated in vaccinia virus: partial purification and properties. Proc. Natl. Acad. Sci. USA 74:1841-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baylis, S. A., L. K. Dixon, S. Vydelingum, and G. L. Smith. 1992. African swine fever virus encodes a gene with extensive homology to type II DNA topoisomerases. J. Mol. Biol. 228:1003-1010. [DOI] [PubMed] [Google Scholar]
- 3.Burgin, A. B., B. H. Huizenga, and H. A. Nash. 1995. A novel suicide substrate for DNA topoisomerases and site-specific recombinases. Nucleic Acids Res. 15:2973-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng, C., L. K. Wang, J. Sekiguchi, and S. Shuman. 1997. Mutational analysis of 39 residues of vaccinia DNA topoisomerase identifies Lys-220, Arg-223, and Asn-228 as important for covalent catalysis. J. Biol. Chem. 272:8263-8269. [DOI] [PubMed] [Google Scholar]
- 5.Cheng, C., P. Kussie, N. Pavletich, and S. Shuman. 1998. Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases. Cell 92:841-850. [DOI] [PubMed] [Google Scholar]
- 6.Corbett, K. D., and J. M. Berger. 2004. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 33:95-118. [DOI] [PubMed] [Google Scholar]
- 7.Da Fonseca, F., and B. Moss. 2003. Poxvirus DNA topoisomerase knockout mutant exhibits decreased infectivity associated with reduced early transcription. Proc. Natl. Acad. Sci. USA 100:11291-11296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fortune, J. M., O. V. Lavrukhin, J. R. Gurnon, J. L. Van Etten, R. S. Lloyd, and N. Osheroff. 2001. Topoisomerase II from Chlorella virus PBCV-1 has an exceptionally high DNA cleavage activity. J. Biol. Chem. 276:24401-24408. [DOI] [PubMed] [Google Scholar]
- 9.Fortune, J. M., J. S. Dickey, O. V. Lavrukhin, J. L. Van Etten, R. S. Lloyd, and N. Osheroff. 2002. Site-specific DNA cleavage by Chlorella virus topoisomerase II. Biochemistry 41:11761-11769. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Beato, R., J. M. Freije, C. Lopez-Otin, R. Blasco, E. Vinuela, and M. L. Salas. 1992. A gene homologous to topoisomerase II in African swine fever virus. Virology 188:938-947. [DOI] [PubMed] [Google Scholar]
- 11.Greub, G., and D. Raoult. 2004. Microorganisms resistant to free-living amoebae. Clin. Microbiol. Rev. 17:413-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and R. L. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 13.Hwang, Y., B. Wang, and F. D. Bushman. 1998. Molluscum contagiosum virus topoisomerase: purification, activities, and response to inhibitors. J. Virol. 72:3401-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klemperer, N., D. J. Lyttle, D. Tauzin, P. Traktman, and A. J. Robinson. 1995. Identification and characterization of the orf virus type I topoisomerase. Virology 206:203-215. [DOI] [PubMed] [Google Scholar]
- 15.Krogh, B. O., and S. Shuman. 2002. A poxvirus-like type IB topoisomerase family in bacteria. Proc. Natl. Acad. Sci. USA 99:1853-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krogh, B. O., and S. Shuman. 2000. Catalytic mechanism of DNA topoisomerase IB. Mol. Cell 5:1035-1041. [DOI] [PubMed] [Google Scholar]
- 17.Krogh, B. O., and S. Shuman. 2002. Proton relay mechanism of general acid catalysis by DNA topoisomerase IB. J. Biol. Chem. 277:5711-5714. [DOI] [PubMed] [Google Scholar]
- 18.Krogh, B. O., C. Cheng, A. Burgin, and S. Shuman. 1999. Melanoplus sanguinipes entomopoxvirus DNA topoisomerase: site-specific DNA transesterification and effects of 5′-bridging phosphorothiolates. Virology 264:441-451. [DOI] [PubMed] [Google Scholar]
- 19.La Scola, B., S. Audic, C. Robert, L. Jungang, X. de Lamballerie, M. Drancourt, R. Birtles, J. M. Claverie, and D. Roualt. 2003. A giant virus in amoebae. Science 299:2033. [DOI] [PubMed] [Google Scholar]
- 20.La Scola, B., T. J. Marrie, J. P. Auffray, and D. Raoult. 2005. Mimivirus in pneumonia patients. Emerg. Infect. Dis. 11:449-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavrukhin, O. V., J. M. Fortune, T. G. Wood, D. E. Burbank, J. L. Van Etten, N. Osheroff, and R. S. Lloyd. 2000. Topoisomerase II from Chlorella virus PBCV-1: characterization of the smallest known type II topoisomerase. J. Biol. Chem. 275:6915-6921. [DOI] [PubMed] [Google Scholar]
- 22.Liu, L. F., C. C. Liu, and B. M. Alberts. 1979. T4 DNA topoisomerase: a new ATP-dependent enzyme essential for initiation of T4 bacteriophage DNA replication. Nature 281:456-461. [DOI] [PubMed] [Google Scholar]
- 23.Morham, S. G., and S. Shuman. 1992. Covalent and noncovalent DNA binding by mutants of vaccinia DNA topoisomerase I. J. Biol. Chem. 267:15984-15992. [PubMed] [Google Scholar]
- 24.Moss, B. 2001. Poxviridae: the viruses and their replication, p. 2849-2883. In D. M. Knipe, D. M., P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 25.Mossessova, E., and C. D. Lima. 2000. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5:865-876. [DOI] [PubMed] [Google Scholar]
- 26.Palaniyar, N., C. Fisher, R. Parks, and D. H. Evans. 1996. SFV topoisomerase: sequence specificity in a genetically marked interval. Virology 221:351-354. [DOI] [PubMed] [Google Scholar]
- 27.Petersen, B. Ø., and S. Shuman. 1997. Histidine-265 is important for covalent catalysis by vaccinia topoisomerase and is conserved in all eukaryotic type I enzymes. J. Biol. Chem. 272:3891-3896. [DOI] [PubMed] [Google Scholar]
- 28.Petersen, B. Ø., R. L. Hall, R. W. Moyer, and S. Shuman. 1997. Characterization of a DNA topoisomerase encoded by Amsacta moorei entomopoxvirus. Virology 230:197-206. [DOI] [PubMed] [Google Scholar]
- 29.Raoult, D., S. Audic, C. Robert, C. Abergel, P. Renesto, H. Ogata, B. La Scola, M. Suzan, and J. M. Claverie. 2004. The 1.2-megabase genome sequence of mimivirus. Science 306:1344-1350. [DOI] [PubMed] [Google Scholar]
- 29a.Raoult, D., P. Renesto, and P. Brouqui. Laboratory infection of a technician by a giant mimivirus. Ann. Intern. Med., in press. [DOI] [PubMed]
- 30.Shaffer, R., and P. Traktman. 1987. Vaccinia virus encapsidates a novel topoisomerase with the properties of a eukaryotic type I enzyme. J. Biol. Chem. 262:9309-9315. [PubMed] [Google Scholar]
- 31.Shuman, S., and B. Moss. 1987. Identification of a vaccinia virus gene encoding a type I DNA topoisomerase. Proc. Natl. Acad. Sci. USA 84:7478-7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shuman, S., M. Golder, and B. Moss. 1988. Characterization of vaccinia virus DNA topoisomerase I expressed in Escherichia coli. J. Biol. Chem. 263:16401-16407. [PubMed] [Google Scholar]
- 33.Shuman, S., and J. Prescott. 1990. Specific DNA cleavage and binding by vaccinia virus DNA topoisomerase I. J. Biol. Chem. 265:17826-17836. [PubMed] [Google Scholar]
- 34.Stetler, G. L., G. J. King, and W. M. Huang. 1979. T4 DNA-delay proteins, required for specific DNA replication, form a complex that has ATP-dependent DNA topoisomerase activity. Proc. Natl. Acad. Sci. USA 76:3737-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stivers, J. T., S. Shuman, and A. S. Mildvan. 1994. Vaccinia DNA topoisomerase I: single-turnover and steady-state kinetic analysis of the DNA strand cleavage and ligation reactions. Biochemistry 33:327-339. [DOI] [PubMed] [Google Scholar]
- 36.Stivers, J. T., G. J. Jagadeesh, B. Nawrot, J. W. Stec, and S. Shuman. 2000. Stereochemical outcome and kinetic effects of Rp and Sp phosphorothioate substitutions at the cleavage site of vaccinia type I DNA topoisomerase. Biochemistry 39:5561-5572. [DOI] [PubMed] [Google Scholar]
- 37.Tian, L., C. D. Claeboe, S. M. Hecht, and S. Shuman. 2003. Guarding the genome: electrostatic repulsion of water by DNA suppresses a potent nuclease activity of topoisomerase IB. Mol. Cell 12:199-208. [DOI] [PubMed] [Google Scholar]
- 38.Tian, L., C. D. Claeboe, S. M. Hecht, and S. Shuman. 2005. Mechanistic plasticity of DNA topoisomerase IB: phosphate electrostatics dictate the need for a catalytic arginine. Structure 13:513-520. [DOI] [PubMed] [Google Scholar]
- 39.Wilson, W. H., D. C. Schroeder, M. J. Allen, M. T. Holden, J. Parkhill, B. G. Barrell, C. Churcher, N. Hamlin, K. Mungall, H. Norbertczak, M. A. Quail, C. Price, E. Rabbinowitsch, D. Walker, M. Craigon, D. Roy, and P. Ghazal. 2005. Complete genome sequence and lytic phase transcription profile of a Coccolithovirus. Science 309:1090-1092. [DOI] [PubMed] [Google Scholar]
- 40.Wittschieben, J., and S. Shuman. 1997. Mechanism of DNA transesterification by vaccinia topoisomerase: catalytic contributions of essential residues Arg-130, Gly-132, Tyr-136, and Lys-167. Nucleic Acids Res. 25:3001-3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, H., J. M. Barcelo, B. Lee, G. Kohlhagen, D. B. Zimonjic, N. C. Popescu, and Y. Pommier. 2001. Human mitochondrial topoisomerase I. Proc. Natl. Acad. Sci. USA 98:10608-10613. [DOI] [PMC free article] [PubMed] [Google Scholar]