Abstract
The first step in the infection process of tailed phages is recognition and binding to the host receptor. This interaction is mediated by the phage antireceptor located in the distal tail structure. The temperate Lactococcus lactis phage TP901-1 belongs to the P335 species of the Siphoviridae family, which also includes the related phage Tuc2009. The distal tail structure of TP901-1 is well characterized and contains a double-disk baseplate and a central tail fiber. The structural tail proteins of TP901-1 and Tuc2009 are highly similar, but the phages have different host ranges and must therefore encode different antireceptors. In order to identify the antireceptors of TP901-1 and Tuc2009, a chimeric phage was generated in which the gene encoding the TP901-1 lower baseplate protein (bppLTP901-1) was exchanged with the analogous gene (orf532009) of phage Tuc2009. The chimeric phage (TP901-1C) infected the Tuc2009 host strain efficiently and thus displayed an altered host range compared to TP901-1. Genomic analysis and sequencing verified that TP901-1C is a TP901-1 derivative containing the orf532009 gene in exchange for bppLTP901-1; however, a new sequence in the late promoter region was also discovered. Protein analysis confirmed that TP901-1C contains ORF532009 and not the lower baseplate protein BppLTP901-1, and it was concluded that BppLTP901-1 and ORF532009 constitute antireceptor proteins of TP901-1 and Tuc2009, respectively. Electron micrographs revealed altered baseplate morphology of TP901-1C compared to that of the parental phage.
The first physical contact between a phage and its bacterial host is a crucial step in the life cycle of the phage. For tailed phages, this initial host interaction is mediated by a so-called antireceptor located in the distal part of the tail. This antireceptor recognizes and interacts with a specific structure or receptor on the surface of the host cell as a prerequisite to the injection of the phage genome. In several Escherichia coli phages, e.g., λ, P2, T4, and the T-even-type phages, the phage antireceptors have been identified in tail fibers (20, 26, 48). In contrast to this, the antireceptors of the E. coli phages T5 and BF23 have been shown to be located in the tail shaft just above a straight tail fiber (22, 24). Compared to these extensively studied E. coli phages, the knowledge of the antireceptors of phages infecting lactic acid bacteria (LAB), and gram-positive bacteria in general, is limited. Most speculations regarding genes encoding host-interacting proteins of LAB phages originate from sequence comparisons among the rapidly increasing number of completely sequenced LAB phages (10, 11, 31, 33, 39). Phage antireceptor proteins of a few Streptococcus thermophilus and Lactococcus lactis phages have, however, recently been identified. Duplessis and Moineau (12) identified ORF18 as the determinant of host specificity for the S. thermophilus phages DT1 and MD4 by the generation of chimeric phages with an altered host range. These chimeric phages were readily propagated on the new host strain, and a C-terminal variable region of orf18 was found to be responsible for host recognition. In similar studies, Stuer-Lauridsen et al. (44) and Dupont et al. (14) identified receptor binding proteins of the virulent L. lactis phages bIL67 and CHL92 of the c2 species and sk1 and bIL170 of the 936 species, respectively. The chimeric phages in those studies displayed an altered host range in plaque assays, but the phages were either partially or completely deficient in their ability to propagate lytically on the new host strain, thus indicating that additional factors may contribute to the host range determination of these phages. Although the receptor binding protein from sk1 was shown to be located at the tip of the phage tail (14), these antireceptors of LAB phages have not yet been assigned to more specific tail structures.
The temperate L. lactis pac type phage TP901-1 belonging to the P335 species of the Siphoviridae family is among the best-studied LAB phages. The 37.7-kbp genome has been fully sequenced (GenBank accession no. NC_002747) (7), and many aspects of the TP901-1 life cycle have been investigated experimentally (8, 28, 34, 37). We have recently completed a detailed structural characterization of the distal tail of TP901-1, which consists of a double-disk baseplate and a central tail fiber (47). The baseplate was found to assemble onto a conical structure situated below the tail tube, and the proteins BppU and BppL were shown to form the upper and lower baseplate disks, respectively. The baseplate was furthermore demonstrated to be necessary for host infection, and the TP901-1 antireceptor was therefore expected to be located in the baseplate (47). The protein TalTP901-1 was found to share many characteristics with the tail-associated lysin protein Tal2009 of phage Tuc2009 (29), and it was shown that TalTP901-1 constitutes the TP901-1 tail fiber, which protrudes below the baseplate (47). Moreover, TalTP901-1, the distal tail protein (Dit), and the tape measure protein were proposed to form a tail assembly initiator complex constituting the conical structure and the tail fiber (38, 47). The individual tail proteins of TP901-1 are thus related to specific tail structures, and TP901-1 is therefore suitable as a model phage for the study of antireceptors of LAB phages. The genomes of TP901-1 and the related L. lactis phage Tuc2009 are organized in a similar manner, with particular congruity to gene order and amino acid homology between the encoded tail proteins (7, 31, 42). The phages, however, have different host ranges and are therefore expected to have dissimilar antireceptor proteins.
In this study, we identify antireceptor proteins of TP901-1 and Tuc2009 by the generation of a chimeric TP901-1 phage with an altered host range. This new phage has the TP901-1 lower baseplate gene exchanged with the analogous Tuc2009 gene, and it is shown to efficiently infect the Tuc2009 host strain L. lactis UC509.9. Genomic and structural analysis of the chimeric phage confirmed the genetic exchange as well as an alteration of the protein content of the distal tail.
MATERIALS AND METHODS
Bacterial strains, culture conditions, plasmids, and phage preparations.
The bacterial strains, plasmids, and phages used in this study are listed in Table 1. L. lactis strains were grown in M17 medium (Oxoid Ltd., Basington, Hampshire, England) supplemented with 0.5% (wt/vol) glucose (GM17) (45), or GSB medium (10 g glucose, 10 g beef extract, 5 g yeast extract, 5 g tryptone, 5 g tryptose, 7.2 g sodium-β-glycerophosphate, and 2 g glycine per liter), which is a glucose/glycine modification of LSB (2); 5 μg/ml chloramphenicol, 2 μg/ml tetracycline, or 1 μg/ml erythromycin was added where appropriate. For phage propagations, calcium chloride was added to a final concentration of 5 mM. Preparation of competent cells and electroporation were performed as previously described (47). Strains containing plasmid pGhost8 or derivatives thereof were grown at 28°C, while all other strains were grown at 30°C, unless otherwise stated. The phages TP901-1 wild type (wt) and TP901-1erm were induced from their respective lysogenic L. lactis 901-1 strains by 3 μg/ml mitomycin C (Sigma-Aldrich, St. Louis, Mo.), while phage Tuc2009 and the chimeric phage TP901-1C were propagated lytically on L. lactis UC509.9. Purification of phages from cell lysates by isopycnic centrifugation through cesium chloride density gradients was performed as previously described (47).
TABLE 1.
Bacterial strains, plasmids, and phages
| Bacterial strain, plasmid, or phage | Relevant feature(s)a | Reference or source |
|---|---|---|
| L. lactis subsp. cremoris strains | ||
| 901-1 | Lysogenic for phage TP901-1 | 5 |
| 3107 | Host strain for phage TP901-1 | 9 |
| UC509.9 | Host strain for phage Tuc2009 | 2 |
| MG1363 | Transformational strain | 17 |
| CSV52-6 | 901-1, lysogenic for phage TP901-1erm | This study |
| CSV72-1 | 3107 + pCSV71-8 | This study |
| CSV75 | UC509.9, lysogenic for phage TP901-1C | This study |
| Plasmids | ||
| pCI372 | 5.7 kbp; cloning vector; Cmr | 21 |
| pGhost8 | 5.2 kbp; TS replicon; Tcr | 35 |
| pBC197 | 13.2 kbp; pCI372::TP901-1 (nt 33,954-2,704) orf55erm (nt 35,637); Ermr Cmr | 30 |
| pCSV27-31 | 8.4 kbp; pGhost8::TP901-1 (nt 33,954-36,842) orf55erm (nt 35,637); Ermr Tcr | This study |
| pCSV71-8 | 7.9 kbp; pCI372::TP901-1 (nt 29,561-30,390) Tuc2009 (nt 33,461-34,000) TP901-1 (nt 30,897-31,748); Cmr | This study |
| Phages | ||
| TP901-1 | Temperate phage; isolated following induction of L. lactis 901-1 | 5 |
| Tuc2009 | Temperate phage; isolated following induction of L. lactis UC509 | 2 |
| TP901-1erm | TP901-1erm in orf55TP901-1 | This study |
| TP901-1C | Chimeric TP901-1erm with orf532009 | This study |
Abbreviations: Cmr, chloramphenicol resistance; erm, adenine methylase gene from pAMβ1 (6); Ermr, erythromycin resistance; Tcr, tetracycline resistance; TS, temperature sensitive. nt, nucleotide position in genome.
DNA technology and sequencing.
Phage DNA was isolated from purified phage preparations by phenol-chloroform extraction as described previously for phage λ by Sambrook and Russell (41), while plasmid DNA was isolated with the QIAprep Spin Miniprep or Plasmid Midi kit (QIAGEN GmbH, Hilden, Germany), according to the manufacturer's instructions, following a 20-min incubation at 37°C with 20 mg/ml lysozyme (Sigma-Aldrich). Phage restriction endonuclease profiles were performed on 1.5 to 2 μg phage DNA, and the digests were carried out according to the enzyme manufacturer's recommendations (New England Biolabs, Beverly, Mass.). Restriction fragments were separated by electrophoresis on 0.7% (wt/vol) agarose (Invitrogen, Carlsbad, Calif.) gels in Tris-amino-EDTA buffer (0.04 M Tris-acetate, 0.02 M EDTA, pH 8.0). PCR amplifications of phage DNA used for cloning were performed with the Pwo DNA polymerase (Roche, Mannheim, Germany), while a Taq DNA polymerase (Invitrogen) was used for all other amplifications. Water suspensions of L. lactis colonies were either boiled or treated with 10 units of mutanolysin (Sigma-Aldrich) prior to colony PCR amplifications. Phage DNA sequences were determined by MWG (Ebersberg, Germany) or with the CEQ 2000 Dye Terminator cycle sequencing kit on a Beckman Coulter CEQ 2000 DNA analysis system (Beckman Coulter Inc., Fullerton, Calif.).
Construction of a TP901-1 prophage mutant encoding an erythromycin resistance marker.
A TP901-1 derivative carrying an adenine methylase gene transcribed from a constitutive promoter was constructed as a prophage mutant of a lysogenic L. lactis 901-1 strain. The adenine methylase gene, originating from Enterococcus faecalis plasmid pAMβ1 (GenBank accession no Y00116) (6), causes bacterial resistance to erythromycin (Ermr) and is hereafter referred to as erm. A TP901-1 EcoRI-EcoRV (genomic position 33,954 to 36,842) DNA fragment containing the 1.2-kbp erm gene inserted into orf55TP901-1 (genomic position 35,637) was subcloned from plasmid pBC197 (30) into the pGhost8 vector, thus forming the construct pCSV27-31. Following transformation of L. lactis MG1363 and subsequent plasmid purification, pCSV27-31 was introduced into the TP901-1 lysogenic strain L. lactis 901-1 by transformation. The erm gene was finally transferred from pCSV27-31 directly into orf55TP901-1 of the lysogenic TP901-1 genome by homologous recombination, which was obtained by succeeding incubation steps at alternating temperatures as previously described (3, 38). The lysogenic strain containing TP901-1erm was verified by colony PCR for the presence of the erm gene in orf55TP901-1 and the loss of the pGhost8-derived vector (results not shown). The phage TP901-1erm was induced from the lysogenic strain and purified by isopycnic centrifugation. According to previous observations (30), TP901-1erm was found to infect L. lactis 3107 with the same efficiency as TP901-1 wt (results not shown).
Generation and isolation of chimeric TP901-1/Tuc2009 phage.
By use of the splicing-by-overlap-extension PCR technique (27), a fragment of recombinant phage DNA was amplified from purified TP901-1 and Tuc2009 DNA with the following oligonucleotides: F-CV007 (5′-CGCGGATCCATGCGGATGTCAATAGTCAAGCCATTGTTG), R-CV035 (5′-CCTATTTCTATTAAGCTACAAAAACATAGC), F-CV036 (5′-TGTAGCTTAATAGAAATAGGAGAATAAAATG), R-CV037 (5′-CCCCTACTTTCTAATTCCGATAAAGTTTTAC), F-CV038 (5′-TCGGAATTAGAAAGTAGGG-GTTATGGAGG), and R-CV039 (5′-ACGCGTCGACAAATTTTCAGGACTAATACC) (the incorporated flanking BamHI and SalI sites are underlined). This PCR fragment contained the Tuc2009 gene orf532009 with the Shine-Dalgarno sequence inserted between bppUTP901-1 and orf50TP901-1, hence creating a 2.3-kbp recombinant phage sequence in which orf532009 was inserted in exchange for bppLTP901-1 (Fig. 1 and Table 1). The recombinant phage fragment was inserted as a BamHI-SalI fragment into the vector pCI372, thereby producing pCSV71-8. Following the transformation of L. lactis MG1363 and subsequent plasmid purification, pCSV71-8 was finally introduced into the TP901-1 host strain L. lactis 3107 by transformation.
FIG. 1.
Tail proteins of the phages TP901-1 and Tuc2009. Percentages of amino acid identity of homologous proteins are indicated between proteins. The values were obtained from a BLASTp search (1) and are stated with respect to the longest protein sequence. Unique proteins, with no or minor sequence similarity to proteins from the opposite phage, are shown in black. Abbreviations: BppL, lower baseplate protein of TP901-1; BppU, upper baseplate protein of TP901-1; Dit, distal tail protein of TP901-1; G, putative equivalent to gpG of λ; MTP, major tail protein; NPS; neck passage structure protein; T, putative equivalent to “gpT” of λ; Tal2009, tail-associated lysin from Tuc2009; TalTP901-1, tail fiber protein of TP901-1. Unnamed proteins are referred to by the numbers of their encoding open reading frames. TP901-1 and 2009 subscripts are omitted where space is limited.
In order to obtain chimeric phages with BppLTP901-1 exchanged for ORF532009, the TP901-1erm phage was propagated in the presence of the recombinant TP901-1/Tuc2009 fragment. L. lactis 3107 cells harboring pCSV71-8 were grown in GSB medium supplemented with 5 μg/ml chloramphenicol until early in the exponential phase (optical density at 600 nm [OD600] of 0.1), at which stage TP901-1erm phages were added at a multiplicity of infection (MOI) of less than 0.02. The infected culture was wrapped in foil and incubated at room temperature (about 23°C) until complete lysis was obtained. Cellular debris was removed by centrifugation for 10 min at 7,500 × g, and the lysate was filtered by gravity at 4°C through a 0.45-μm-pore-size filter (Frisenette, Ebeltoft, Denmark). The titers of phages in the lysate were determined by plaque assay on TP901-1 host strain L. lactis 3107.
The lysate of TP901-1erm propagated in the presence of the recombinant phage fragment was analyzed for chimeric phages with an altered host range by testing for phages which could lysogenize and hence confer an Ermr lysogenic conversion phenotype to the Tuc2009 host strain. Cells of L. lactis UC509.9 were grown to the exponential phase (OD600 of about 0.3), at which stage 3.5 ml cells was mixed with 1 ml phage lysate and calcium chloride to a final concentration of 5 mM. The mixture was incubated for 1 to 1.5 h at 30°C in order to allow for phage adsorption, DNA injection, and erm expression. The cells were subsequently collected by centrifugation for 10 min at 3,800 × g, resuspended in 0.5 ml GM17, added to 3 ml GM17 soft medium plus calcium chloride, and poured onto GM17 solid medium plus calcium chloride supplemented with 1 μg/ml erythromycin. Following an overnight incubation at 30°C, the plates were incubated for 3 days at room temperature. Colonies were inoculated in GM17 medium plus 1 μg/ml erythromycin and incubated at 30°C overnight. The supernatants were assayed for spontaneously released phages that could infect L. lactis UC509.9 in plaque assays.
Numerous unsuccessful experiments using the method described above as well as direct testing for TP901-1 wt-derived chimeric phages by plaque assays on L. lactis UC509.9 were carried out prior to the isolation of a single chimeric phage. The primary problem was to obtain a sufficiently high titer in the phage lysate containing the chimeric phages. Lysate titers were generally at the level of 107 to 109 PFU/ml, as is regularly observed for lytic propagation of TP901-1 on L. lactis 3107 (L. Brøndsted, unpublished data), and only once was a lysate with a titer of 6 × 1011 PFU/ml obtained.
Plaque, adsorption, and lysogenic conversion assays.
Phage titers were determined by plaque assay on TP901-1 host strain L. lactis 3107 or Tuc2009 host strain L. lactis UC509.9 in GM17 medium supplemented with 5 mM calcium chloride and agarose (Invitrogen), as described previously by Lillehaug (32). Phage adsorption assays for L. lactis 3107 and L. lactis UC509.9 were performed as described previously by Garvey et al. (16). In brief, late-exponential-phase cells or GM17 medium was mixed with phages (approximately 105 PFU/ml) and calcium chloride. Following a 15-min incubation at room temperature, the mixture was cleared of cells by centrifugation, and the supernatant was assayed for phages by plaque assay. The level of adsorption was calculated as 1 minus the ratio between phages (PFU/milliliter) in the cell supernatants and phages (PFU/milliliter) in the cell-free supernatant.
The frequency of lysogenic conversion (Ermr) of L. lactis 3107 and L. lactis UC509.9 was determined by infecting 0.1 ml of early-exponential-phase cells (OD600 of 0.2 to 0.3) with 0.1 ml of phages (diluted in 10 mM calcium chloride-0.9% [wt/vol] sodium chloride) at an MOI of 0.5. Following a 20-min preincubation at room temperature, the mixture was added to GM17 soft medium plus calcium chloride and poured onto GM17 solid medium plus calcium chloride supplemented with 1 μg/ml erythromycin. After 3 days of incubation, the number of Ermr CFU was determined, and the frequency of lysogenic conversion was calculated as the number of Ermr CFU divided by the total number of added phages (PFU).
SDS-PAGE and Western blotting.
Protein profiles of the phages were determined with approximately 1010 PFU of purified and denatured phage particles. The proteins were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) under reducing conditions with the NuPAGE system in 10% Novex Bis-Tris polyacrylamide precast gels and MES (morpholineethanesulfonic acid) SDS running buffer (Invitrogen, Carlsbad, Calif.) as previously described (47). Following electrophoresis, the protein bands were either silver stained with the SilverQuest Silver Staining kit (Invitrogen) or electroblotted onto a polyvinylidene difluoride membrane. A Western blot with polyclonal antibodies raised in rabbits against ORF532009 and a secondary labeled anti-rabbit antibody was carried out as previously described (47).
Transmission electron microscopy.
Purified phage preparations were dialyzed against SM buffer (100 mM sodium chloride, 10 mM magnesium sulfate, 50 mM Tris [pH 7.5], 0.01% [wt/vol] gelatin), and a carbon film was floated from a mica sheet into the dialyzed phage suspension and incubated for 10 min. The film was subsequently rinsed in demineralized water and stained for 30 s with 2% (wt/vol) uranyl acetate (Agar Scientific, Stansted, United Kingdom). A 400-mesh copper grid (Agar Scientific) was used to pick up the carbon film, and the phages were examined with a transmission electron microscope (Tecnai 10; FEI Eindhoven, The Netherlands) at an acceleration voltage of 80 kV. Micrographs were taken with a MegaView II charge-coupled-device camera (SIS, Münster, Germany).
Nucleotide sequence accession number.
A sequence of the TP901-1C late promoter region has been deposited in the GenBank database under accession no. DQ093874.
RESULTS
Comparison of TP901-1 and Tuc2009 tail proteins.
In order to identify potential candidates for the antireceptors of TP901-1 and Tuc2009, the tail proteins of the two phages were compared. TP901-1 and Tuc2009 have different host ranges, and their antireceptor proteins were therefore expected to show limited sequence similarity. Tail proteins encoded from the major tail protein (MTP) gene to the neck passage structure (NPS) gene were compared by their amino acid sequences, and it was observed that the majority of these proteins displayed a high degree of identity (74 to 100% over the entire sequence) (Fig. 1). Tail fiber proteins have often been found to constitute antireceptors of E. coli phages (for a review, see reference 23), but the TP901-1 tail fiber protein TalTP901-1 was found to display 93% sequence identity with Tal2009 of Tuc2009, and it was therefore considered unlikely that the tail fiber proteins were determinants for the host range difference between these two phages. While no homologous protein of Tuc2009 ORF522009 was found among the TP901-1 tail proteins, TP901-1 BppLTP901-1 and Tuc2009 ORF532009 showed a weak identity (20%) over the entire sequence and 50% identity over the first 61 N-terminal amino acids. Recently, we identified BppLTP901-1 as the protein of the lower baseplate disk of TP901-1, and our results indicated that BppLTP901-1 may be the TP901-1 antireceptor (47). BppLTP901-1 and ORF532009 are almost equal in size (163 and 174 amino acids) and share some degree of similarity; moreover, the two proteins have approximately the same genomic position. It was therefore hypothesized that these proteins were the antireceptor proteins of TP901-1 and Tuc2009, respectively.
Generation and isolation of a chimeric TP901-1 phage with an altered host range.
In order to determine whether BppLTP901-1 and ORF532009 constitute the respective antireceptors of TP901-1 and Tuc2009, attempts were made to isolate a TP901-1 phage with the host range of Tuc2009 by exchanging BppLTP901-1 with ORF532009. First, a recombinant phage fragment was created in which orf532009 and the associated Shine-Dalgarno sequence were inserted between the up- and downstream flanking sequences of bppLTP901-1, thereby replacing bppLTP901-1 with orf532009 (Fig. 1). The fragment was generated using splicing-by-overlap-extension PCR in order to preserve the original sequence composition, i.e., to avoid the insertion of additional restriction sites in the surrounding sequences of orf532009. The recombinant fragment was cloned into the vector pCI372, forming pCSV71-8, and introduced into the TP901-1 host strain L. lactis 3107. The relatively low-copy cloning vector pCI372 was used for this experiment because of previous difficulties with the transformation of L. lactis 3107 with several high-copy lactococcal vectors (37). It was reasoned that bppLTP901-1 of the TP901-1 genome could be exchanged for orf532009 by homologous recombination upon TP901-1 infection of L. lactis 3107 harboring pCSV71-8. When infected at a low MOI, the resulting lysate would contain chimeric TP901-1 phages, both genotypic and phenotypic orf532009, and according to the proposed hypothesis, these phages were expected to have an altered host range. A TP901-1 derivative (TP901-1erm) containing an Ermr marker was used as the parental phage for the generation of these chimeric phages in order to overcome potential problems with the lytic propagation of the chimeric phage, as previously noted for other such lactococcal phages (14, 44). Lysates generated in these experiments were consequently analyzed for phages that could confer the Ermr lysogenic conversion phenotype to the Tuc2009 host strain L. lactis UC509.9. Infection of L. lactis UC509.9 with a total of 6 × 1012 PFU (determined on L. lactis 3107) from a lysate of TP901-1erm propagated on L. lactis 3107 harboring pCSV71-8 resulted in a single Ermr colony, which was found to spontaneously release L lactis UC509.9 infectious phages when cultured in liquid media. The phage of this lysogenic strain was named TP901-1C.
Genomic analysis of the chimeric phage TP901-1C.
The genome of the chimeric phage was isolated from purified TP901-1C phages, and the region of the TP901-1C lower baseplate gene was amplified and sequenced to determine if the isolated phage had obtained the orf532009 gene in exchange for bppLTP901-1. The sequencing results confirmed that orf532009 was correctly inserted between bppUTP901-1 and orf50TP901-1 of TP901-1C. A single point mutation was discovered (T→C at position 33,664 of the Tuc2009 sequence) (GenBank accession no. NC_002703), but this mutation did not change the amino acid sequence, and it was therefore considered to be insignificant (results not shown).
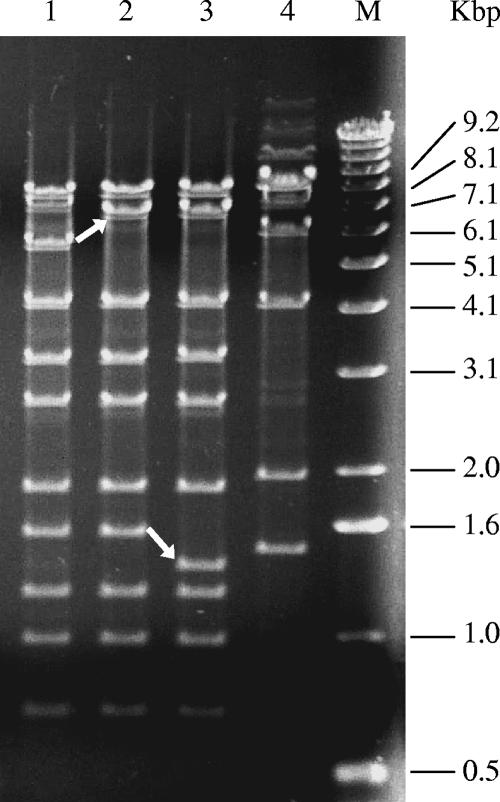
The genome of TP901-1C was analyzed with restriction endonuclease digestion in order to verify that TP901-1C was a TP901-1erm derivative with an altered host range. Phage genomic DNA of TP901-1 wt, TP901-1erm, TP901-1C, and Tuc2009 was isolated from purified phage preparations, digested with EcoRV, and analyzed by agarose gel electrophoresis (Fig. 2). Consistent with prior observations (30), the inserted 1.2-kbp erm gene in orf55TP901-1 resulted in a changed mobility of a 5.7-kbp fragment, which was found to migrate together with a larger fragment in the TP901-1erm and TP901-1C profiles (Fig. 2, upper arrow). The restriction profiles clearly demonstrate that TP901-1C is a TP901-1erm derivative, as the TP901-1C profile was found to be very similar to the TP901-1erm profile and significantly different from the Tuc2009 profile. Equivalent EcoRI and PstI restriction profiles further confirmed these observations (results not shown). A single fragment of the TP901-1C EcoRV profile was, however, found to show a changed mobility compared to the TP901-1 wt and TP901-1erm profiles (Fig. 2, lower arrow). To investigate the reason for this altered single fragment mobility, the corresponding region of TP901-1C was amplified and sequenced. It was thus discovered that TP901-1C contained a 2.5-kbp region (GenBank accession no. DQ093874) that differed from the TP901-1 wt sequence. The impacts of this new sequence were the losses of orf27TP901-1 and orf28TP901-1, two small genes (97 and 47 codons, respectively) of unknown function in the middle transcript region of TP901-1 (7, 34). Furthermore, TP901-1C had acquired a new alt gene (activator of late transcription), which showed 99% nucleotide identity and 100% amino acid identity with the corresponding alt gene from phage Tuc2009 (8, 42), and a changed −35 sequence of the late promoter (ACATCA) with only a single nucleotide mismatch (underlined) with the corresponding region (ACCTCA) of phage Tuc2009. TP901-1C thus contained a late promoter region and regulator, which was highly similar to the corresponding region of phage Tuc2009 (Fig. 3). In order to investigate the origin of this unexpected new sequence, colony PCRs were performed with TP901-1C discriminative primers on L. lactis 3107 and L. lactis UC509.9. These PCRs revealed that the new sequence originated from L. lactis 3107, a result that was confirmed by sequencing of the fragment (1.2 kbp) amplified from this strain (results not shown). These results indicate that TP901-1C had obtained the new sequence by homologous recombination with the L. lactis 3107 genome.
FIG. 2.
EcoRV endonuclease restriction profiles of phage DNA genomes separated by agarose gel electrophoresis. Lane 1, TP901-1 wt; lane 2, TP901-1erm; lane 3, TP901-1C; lane 4, Tuc2009; M, 1-kb DNA ladder (Invitrogen). Sizes of double-stranded DNA fragments are indicated to the right of the gel. The upper arrow indicates the changed mobility of the fragment covering the inserted erm gene in TP901-1erm and TP901-1C. The lower arrow indicates the changed mobility of a TP901-1C fragment due to a secondary recombination.
FIG. 3.
The sequenced late promoter region of TP901-1C compared to the corresponding regions of TP901-1 wt and Tuc2009. The late promoters (PLate) and relevant encoded proteins are indicated with protein names or the numbers of their encoding open reading frames. Identical amino acid sequences are shown with identical shades. ORF27TP901-1 and ORF28TP901-1 are absent from the TP901-1C sequence. The activators of late transcription (Alt) of TP901-1C and Tuc2009 display 100% amino acid identity. The late promoter sequence of TP901-1C is more closely related to that of Tuc2009 than to that of TP901-1 wt. The large-subunit terminase terL genes of TP901-1C and TP901-1 wt display a 10-nucleotide difference in the 5′ sequence.
Examination of TP901-1C adsorption and infection.
The phage TP901-1C was found to form clearly visible plaques on the Tuc2009 host strain L. lactis UC509.9 and was readily propagated to a titer of 5 × 109 PFU/ml. In contrast, TP901-1C could not infect the TP901-1 wt host strain, as no plaques were observed in plaque assays with TP901-1C and L. lactis 3107 (Table 2). It was therefore concluded that TP901-1C has an altered host range compared to that of the parental TP901-1 phage.
TABLE 2.
Infection, adsorption, and lysogenic conversion frequencies of TP901-1, TP901-1erm, Tuc2009, and TP901-1C
| Phage | Infectiona
|
Adsorption (%)b
|
Ermr lysogenic conversion frequency (%)
|
|||
|---|---|---|---|---|---|---|
| 3107c | UC509.9d | 3107c | UC509.9d | 3107c | UC509.9d | |
| TP901-1 wt | + | − | 93 ± 3 | 3 ± 5 | NDg | ND |
| TP901-1erm | + | − | 93 ± 2 | 8 ± 11e | 0.002 | <10−7f |
| TP901-1C | − | + | 16 ± 16 | 88 ± 3 | <10−7f | 0.01 |
| Tuc2009 | − | + | 8 ± 12 | 81 ± 10 | ND | <10−7f |
Infection ability determined by plaque assays. +, positive; −, negative.
Means of three independent experiments and standard deviations.
L. lactis 3107.
L. lactis UC509.9.
Means of two experiments.
Outside the limits of the assay.
ND, not determined.
In order to test whether the inability of TP901-1C to infect L. lactis 3107 was due to reduced phage adsorption or the blocking of later steps in the infection process, an adsorption assay was carried out. The results of this assay (Table 2) showed that TP901-1C displayed approximately the same adsorption characteristics as Tuc2009, i.e., a high level of adsorption to L. lactis UC509.9 but very limited adsorption to L. lactis 3107. These data strongly indicate that the failure of TP901-1C to infect L. lactis 3107 was due to its inability to adsorb to the TP901-1 host strain.
A lysogenic conversion assay was carried out to analyze the frequency of Ermr lysogenic conversion and hence the infection efficiency of TP901-1C in comparison to that of the parental phage, TP901-1erm (Table 2). The obtained results revealed that the TP901-1C-mediated lysogenic conversion of L. lactis UC509.9 was as efficient as the TP901-1erm-mediated lysogenic conversion of L. lactis 3107, thus indicating that TP901-1C can infect L. lactis UC509.9 as efficiently as TP901-1erm can infect L. lactis 3107. The frequency of Ermr lysogenic conversion by TP901-1erm obtained in this assay was approximately 103 times lower than that previously observed (30). Given that the result was confirmed by independent experiments (results not shown), we presume that the large difference in frequency is caused by different experimental conditions, e.g., the preincubation time and MOI value.
TP901-1C baseplate proteins.
In order to verify that TP901-1C was a chimeric TP901-1 derivative, the structural proteins of TP901-1 wt, TP901-1erm, TP901-1C, and Tuc2009 were separated and evaluated by SDS-PAGE. A comparison of the protein profiles (Fig. 4A) revealed that the profiles of TP901-1 wt and TP901-1erm were identical, while the profile of TP901-1C diverged from these phages by the size of the smallest protein band (Fig. 4A, lower arrow). The smallest protein bands of TP901-1 wt and TP901-1erm were estimated to be approximately 17 kDa, while the corresponding band of TP901-1C was estimated to be approximately 19 kDa and was found to show the same size as a protein band in the Tuc2009 profile. This correlates well with the predicted 17.1- and 18.9-kDa molecular masses of BppLTP901-1 and ORF532009, respectively (7, 42). Because the remaining protein bands of the TP901-1C profile, including the 35-kDa upper baseplate protein BppUTP901-1 (Fig. 4A, upper arrow), were found to be indistinguishable from the TP901-1 wt and TP901-1erm profiles, the SDS-PAGE analysis revealed that TP901-1C is a TP901-1 derivative with a different lower baseplate protein.
FIG. 4.
(A) Silver-stained 10% SDS-PAGE gel with structural phage protein profiles. Lane 1, TP901-1 wt; lane 2, TP901-1erm; lane 3, TP901-1C; lane 4, Tuc2009. Molecular masses are indicated to the left of the gel. The upper arrow indicates the 35-kDa upper baseplate protein BppUTP901-1 of TP901-1 wt, TP901-1erm, and TP901-1C. The lower arrow indicates the 19-kDa ORF532009 protein of TP901-1C and Tuc2009. (B) Western blot with polyclonal antibodies raised against ORF532009. Lane 1, TP901-1 wt; lane 2, TP901-1erm; lane 3, TP901-1C; lane 4, Tuc2009. Molecular masses are indicated to the left of the gel.
To determine if the distinctive 19-kDa protein band of TP901-1C was in fact ORF532009, the protein profiles of TP901-1 wt, TP901-1erm, TP901-1C, and Tuc2009 were analyzed by Western blot using polyclonal antibodies raised against ORF532009. As illustrated in Fig. 4B, the antibodies reacted strongly with the 19-kDa ORF532009 band of Tuc2009 and less intensively with the 23-kDa major tail protein (MTPTP901-1) and the 17-kDa BppLTP901-1 of both TP901-1 wt and TP901-1erm. A strong antibody reaction to the distinctive 19-kDa band of TP901-1C was furthermore observed, and since no secondary reaction was observed with a band of 17 kDa from TP901-1C, the chimeric phage was concluded to contain the protein ORF532009 in exchange for BppLTP901-1.
TP901-1C morphology.
We have previously shown that BppLTP901-1 constitutes the TP901-1 lower baseplate disk (47). To investigate if the exchange of BppLTP901-1 for ORF532009 had caused any morphological changes in the TP901-1C baseplate, the purified phages of TP901-1 wt, TP901-1erm, and TP901-1C were examined and compared by transmission electron microscopy (Fig. 5). The baseplate of TP901-1 wt consists of two well-defined disks (Fig. 5A) (47), and the electron micrographs of TP901-1erm revealed that the inserted erm gene altered neither the baseplate nor the overall virion morphology, as TP901-1erm was found to be indistinguishable from TP901-1 wt (Fig. 5B). In contrast to this, the baseplate of TP901-1C was found to be clearly different (Fig. 5C), as the lower part of the chimeric baseplate looked like small hanging droplets rather than a defined disk. Moreover, the diameter of this lower part was found to be more narrow (23 ± 3 nm [n = 38]) than the lower baseplate disk from TP901-1 wt (28 ± 3 nm) (47). The overall appearance of the TP901-1C baseplate was estimated as an intermediate between the TP901-1 wt and Tuc2009 baseplate morphology (47; S. Mc Grath et al., unpublished data). The remaining structures of TP901-1C were found to be identical to those of TP901-1 wt, supporting the previous observations of the lower baseplate being the only structural change of TP901-1C.
FIG. 5.
Transmission electron micrographs of phages negatively stained with uranyl acetate. (A) TP901-1 wt; (B) TP901-1erm; (C) TP901-1C. White arrows point to the double-disk baseplate of TP901-1 and TP901-1erm. Black arrows point to the “hanging droplets” of the TP901-1C baseplate.
DISCUSSION
The infection processes of LAB phages and phages infecting gram-positive bacteria in general are poorly understood, and one of the primary reasons for this lack of insight is the limited knowledge of the host-interacting proteins and structures of these phages. This study describes the isolation and analysis of a chimeric L. lactis phage TP901-1 derivative with an altered host range.
The chimeric phage TP901-1C was constructed by homologous recombination between TP901-1erm (a TP901-1 phage carrying an erm gene) and a recombinant phage DNA fragment, wherein the bppLTP901-1 gene was exchanged for orf532009 from the related phage Tuc2009. DNA sequencing, SDS-PAGE, and Western blot analyses of TP901-1C verified that the chimeric phage carried ORF532009 and not the TP901-1 lower baseplate protein BppLTP901-1, while the remaining structural protein profile of TP901-1C was identical to the profiles of TP901-1 wt and TP901-1erm. A transmission electron microscopy examination showed that the chimeric phage has a baseplate structure different from the characteristic double disks of TP901-1. The morphology of the lower baseplate of TP901-1C was determined to be a more narrow structure with the appearance of small droplets hanging from the upper disk, while the remaining TP901-1C virion was found to be morphologically indistinguishable from those of TP901-1 wt and TP901-1erm. On the basis of these analyses, we conclude that TP901-1C is a chimeric TP901-1 phage in which BppLTP901-1 is exchanged for ORF532009, and this exchange causes a morphological alteration of the TP901-1C baseplate.
TP901-1C was isolated as a prophage integrant of the lysogenic L. lactis UC509.9 strain. The chimeric phage had lost the ability to infect the TP901-1 host strain L. lactis 3107 while it acquired the ability to infect the Tuc2009 host strain L. lactis UC509.9. Furthermore, TP901-1C was readily propagated to sufficient titers (>109 PFU/ml) for phage purification by isopycnic centrifugation, and it is therefore estimated that TP901-1C and Tuc2009 infect L. lactis UC509.9 with approximately the same efficiency. Based on these results, we conclude that TP901-1C has an altered host range compared to that of the parental TP901-1 phage.
Restriction endonuclease digestion of the TP901-1C genome verified that the chimeric phage was a TP901-1erm derivative. However, the EcoRV profile and subsequent sequencing revealed that TP901-1C contains an unexpected non-TP901-1 sequence region located approximately 16 kbp upstream of the exchanged baseplate gene. By sequencing of an L. lactis 3107 fragment, it was revealed that TP901-1C most likely had acquired this sequence by homologous recombination while propagated on L. lactis 3107 harboring pCSV71-8. This observation is in agreement with several previous reports of lactococcal phages of the P335 species frequently recombining with putative prophage sequences of their L. lactis host strains (4, 15, 36). The outcome of this secondary recombination was a changed late promoter region highly similar to the corresponding region of phage Tuc2009 and the loss of orf27TP901-1 and orf28TP901-1, two very small genes of unknown function.
The chimeric phage TP901-1C was isolated as a single phage with an altered host range from 6 × 1012 PFU; i.e., the phage was obtained with a frequency of approximately 10−13. This frequency is several orders of magnitude lower than those reported from similar experiments with LAB phages (12, 14, 44). We believe this low frequency may be caused by the following factors. (i) The method used to isolate the chimeric phage in this study requires both a double-crossover recombination event and Ermr lysogenic conversion, i.e., infection, lysogenization, and erm expression, an aspect supported by the finding of a 10−4 lysogenic conversion frequency of TP901-1C. (ii) The parental pCI372 vector used for the construction of the recombinant phage plasmid is not a high-copy vector like, e.g., the vector used in a related study by Duplessis and Moineau (12). (iii) L. lactis UC509.9 has been found to carry a restriction/modification system which decreases the efficiency of plating by 104-fold for unmodified phages (43).
It has previously been argued that the host specificity of phages involves more host-compatible features than the phage antireceptor (14, 44), and it would seem natural to suggest that the altered late promoter region of TP901-1C is a prerequisite for L. lactis UC509.9 infection. The chimeric phage was, however, isolated as a prophage integrant of a lysogenic L. lactis UC509.9 strain, and transcription of the lytic genes from the late promoter was therefore not required for TP901-1C infection, i.e., lysogenization of L. lactis UC509.9. The erm gene of TP901-1C is expressed constitutively from its own promoter (6), and the Ermr phenotype of the TP901-1C lysogenic strain is therefore also independent of transcription from a phage promoter. This is in agreement with the results of the lysogenic conversion assay, which verified that TP901-1C efficiently lysogenizes L. lactis UC509.9. Recently, it was reported by Rakonjac et al. (40) that the acquirement of a new sequence adjacent to the cosR site in L. lactis phage c2 could overcome a host infection inhibition mechanism acting on the level of DNA injection or circularization by cos-end ligation. The corresponding pac site of TP901-1 is expected to be located in the vicinity of the late promoter (7), i.e., within the secondary recombination of TP901-1C. It is therefore possible that the secondary recombination of TP901-1C reflects a requirement for an altered pac site in order to overcome further inhibition mechanisms of L. lactis UC509.9. Nevertheless, neither the late promoter region nor the pac site is expected to have any effect on the initial host receptor interaction, and we therefore conclude that BppLTP901-1 and ORF532009 are antireceptor proteins of TP901-1 and Tuc2009, respectively, and these proteins can promote adsorption to the host cells. Certainly, it cannot be excluded that other proteins participate in the adsorption and host receptor binding, but any such proteins are expected to play a minor host-specific role in this initial infection process, as the substitution of BppLTP901-1 and ORF532009 was sufficient to completely alter the host adsorption of TP901-1C. The genomic comparison of TP901-1 and Tuc2009 showed that Tuc2009 carries an additional tail gene (orf522009) encoded between the upper baseplate protein homologue (ORF512009) and the antireceptor ORF532009 (Fig. 1). The genomic position of this gene indicates that ORF522009 participates in host infection, as previously suggested by Seegers et al. (42), but the present study shows that ORF522009 is in fact not required for TP901-1C infection of the Tuc2009 host strain L. lactis UC509.9. It is, however, possible that ORF522009 has a function in the host-interacting process of Tuc2009, a function that could possibly be structural stabilization/flexibilization of the Tuc2009 baseplate or host range extension/enhancement of the adsorption efficiency, as suggested for the dispensable long tail fibers of E. coli phage λ (25). Investigations have been initiated in order to explore these possible functions of ORF522009 in the infection process of Tuc2009. Moreover, it has been suggested that the TP901-1 tail fiber protein TalTP901-1 and the lytic active Tal2009 protein of Tuc2009 participate in the initial host infection processes by degradation of peptidoglycan to promote access to the host receptor or by assisting the DNA injection process analogous to the straight tail fiber of E. coli phage T5 (29, 47). The functions of TalTP901-1 and Tal2009 are, however, expected to be host unspecific, as these proteins share 93% amino acid identity (Fig. 1).
The results of the present study have identified antireceptor proteins of the temperate L. lactis phages TP901-1 and Tuc2009. This is the first identification of baseplate-located antireceptors from lactococcal phages and the first identification of antireceptor proteins of phages from the P335 species. To our knowledge, this is also the first time an exchange of such proteins has been shown to result in altered phage morphology. The results of the present study have prompted further investigations of the host-interacting mechanisms of TP901-1 and Tuc2009. BppLTp901-1 and ORF532009 will be employed to identify the respective host receptors, which are largely unknown for lactococcal phages of the P335 species. Of particular interest is whether these phages use a membrane protein receptor equivalent to those of phages of the c2 species (18) or bind to cell wall components as previously suggested for phages of the 936 species (13, 19, 46). The distal tail structures and proteins of TP901-1 and Tuc2009 are possibly the best characterized among LAB phages (29, 38, 47; Mc Grath et al., unpublished). It will therefore be possible to consider the functions of individual tail proteins in future host interaction studies of these phages, which will ultimately lead to the elucidation of infection mechanisms and hence a better understanding of the host interaction mechanisms of LAB phages and phages infecting gram-positive bacteria in general.
Acknowledgments
This work was supported by The Royal Veterinary and Agricultural University of Denmark and the Centre for Advanced Food studies. S.M. is the recipient of an EMBARK postdoctoral fellowship from the Irish Research Council for Science and Technology. D.V.S. is the recipient of an Investigator grant from Science Foundation Ireland.
Bashir Aideh (KVL, Copenhagen, Denmark) and Bernd Fahrenholz (FRCNF, Kiel, Germany) are acknowledged for their technical assistance.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arendt, E. K., C. Daly, G. F. Fitzgerald, and M. van de Guchte. 1994. Molecular characterization of lactococcal bacteriophage Tuc2009 and identification and analysis of genes encoding lysin, a putative holin, and two structural proteins. Appl. Environ. Microbiol. 60:1875-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biswas, I., A. Gruss, S. D. Ehrlich, and E. Maguin. 1993. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 175:3628-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouchard, J. D., and S. Moineau. 2000. Homologous recombination between a lactococcal bacteriophage and the chromosome of its host strain. Virology 270:65-75. [DOI] [PubMed] [Google Scholar]
- 5.Braun, V., Jr., S. Hertwig, H. Neve, A. Geis, and M. Teuber. 1989. Taxonomic differentiation of bacteriophages of Lactococcus lactis by electron microscopy, DNA-DNA hybridization, and protein profiles. J. Gen. Microbiol. 135:2551-2560. [Google Scholar]
- 6.Brehm, J., G. Salmond, and N. Minton. 1987. Sequence of the adenine methylase gene of the Streptococcus faecalis plasmid pAM beta 1. Nucleic Acids Res. 15:3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brøndsted, L., S. Østergaard, M. Pedersen, K. Hammer, and F. K. Vogensen. 2001. Analysis of the complete DNA sequence of the temperate bacteriophage TP901-1: evolution, structure, and genome organization of lactococcal bacteriophages. Virology 283:93-109. [DOI] [PubMed] [Google Scholar]
- 8.Brøndsted, L., M. Pedersen, and K. Hammer. 2001. An activator of transcription regulates phage TP901-1 late gene expression. Appl. Environ. Microbiol. 67:5626-5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christiansen, B., M. G. Johnsen, E. Stenby, F. K. Vogensen, and K. Hammer. 1994. Characterization of the lactococcal temperate phage TP901-1 and its site-specific integration. J. Bacteriol. 176:1069-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crutz-Le Coq, A. M., B. Cesselin, J. Commissaire, and J. Anba. 2002. Sequence analysis of the lactococcal bacteriophage bIL170: insights into structural proteins and HNH endonucleases in dairy phages. Microbiology 148:985-1001. [DOI] [PubMed] [Google Scholar]
- 11.Desiere, F., W. M. McShan, D. van Sinderen, J. J. Ferretti, and H. Brüssow. 2001. Comparative genomics reveals close genetic relationships between phages from dairy bacteria and pathogenic streptococci: evolutionary implications for prophage-host interactions. Virology 288:325-341. [DOI] [PubMed] [Google Scholar]
- 12.Duplessis, M., and S. Moineau. 2001. Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol. Microbiol. 41:325-336. [DOI] [PubMed] [Google Scholar]
- 13.Dupont, K., T. Janzen, F. K. Vogensen, J. Josephsen, and B. Stuer-Lauridsen. 2004. Identification of Lactococcus lactis genes required for bacteriophage adsorption. Appl. Environ. Microbiol. 70:5825-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dupont, K., F. K. Vogensen, H. Neve, J. Bresciani, and J. Josephsen. 2004. Identification of the receptor-binding protein in 936-species lactococcal bacteriophages. Appl. Environ. Microbiol. 70:5818-5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durmaz, E., and T. R. Klaenhammer. 2000. Genetic analysis of chromosomal regions of Lactococcus lactis acquired by recombinant lytic phages. Appl. Environ. Microbiol. 66:895-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garvey, P., C. Hill, and G. F. Fitzgerald. 1996. The lactococcal plasmid pNP40 encodes a third bacteriophage resistance mechanism, one which affects phage DNA penetration. Appl. Environ. Microbiol. 62:676-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geller, B. L., R. G. Ivey, J. E. Trempy, and B. Hettinger-Smith. 1993. Cloning of a chromosomal gene required for phage infection of Lactococcus lactis subsp. lactis C2. J. Bacteriol. 175:5510-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geller, B. L., H. T. Ngo, D. T. Mooney, P. Su, and N. Dunn. 2005. Lactococcal 936-species phage attachment to surface of Lactococcus lactis. J. Dairy Sci. 88:900-907. [DOI] [PubMed] [Google Scholar]
- 20.Haggård-Ljungquist, E., C. Halling, and R. Calendar. 1992. DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J. Bacteriol. 174:1462-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayes, F., C. Daly, and G. F. Fitzgerald. 1990. Identification of the minimal replicon of Lactococcus lactis subsp. lactis UC317 plasmid pCI305. Appl. Environ. Microbiol. 56:202-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heller, K. J. 1984. Identification of the phage gene for host receptor specificity by analyzing hybrid phages of T5 and BF23. Virology 139:11-21. [DOI] [PubMed] [Google Scholar]
- 23.Heller, K. J. 1992. Molecular interaction between bacteriophage and the Gram-negative cell envelope. Arch. Microbiol. 158:235-248. [DOI] [PubMed] [Google Scholar]
- 24.Heller, K. J., and H. Schwarz. 1985. Irreversible binding to the receptor of bacteriophages T5 and BF23 does not occur with the tip of the tail. J. Bacteriol. 162:621-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hendrix, R. W., and R. L. Duda. 1992. Bacteriophage lambdaPaPa: not the mother of all lambda phages. Science 258:1145-1148. [DOI] [PubMed] [Google Scholar]
- 26.Henning, U., and S. Hashemolhosseini. 1994. Receptor recognition by T-even-type coliphages, p. 291-298. In J. D. Karam (ed.), Molecular biology of bacteriophage T4. American Society for Microbiology, Washington, D.C.
- 27.Horton, R. M., Z. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8:528-535. [PubMed] [Google Scholar]
- 28.Johansen, A. H., L. Brøndsted, and K. Hammer. 2003. Identification of operator sites of the CI repressor of phage TP901-1: evolutionary link to other phages. Virology 311:144-156. [DOI] [PubMed] [Google Scholar]
- 29.Kenny, J. G., S. McGrath, G. F. Fitzgerald, and D. van Sinderen. 2004. Bacteriophage Tuc2009 encodes a tail-associated cell wall-degrading activity. J. Bacteriol. 186:3480-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koch, B., B. Christiansen, T. Evison, F. K. Vogensen, and K. Hammer. 1997. Construction of specific erythromycin resistance mutations in the temperate lactococcal bacteriophage TP901-1 and their use in studies of phage biology. Appl. Environ. Microbiol. 63:2439-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Labrie, S., and S. Moineau. 2002. Complete genomic sequence of bacteriophage ul36: demonstration of phage heterogeneity within the P335 quasi-species of lactococcal phages. Virology 296:308-320. [DOI] [PubMed] [Google Scholar]
- 32.Lillehaug, D. 1997. An improved plaque assay for poor plaque-producing temperate lactococcal bacteriophages. J. Appl. Microbiol. 83:85-90. [DOI] [PubMed] [Google Scholar]
- 33.Lucchini, S., F. Desiere, and H. Brüssow. 1999. Comparative genomics of Streptococcus thermophilus phage species supports a modular evolution theory. J. Virol. 73:8647-8656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madsen, P. L., and K. Hammer. 1998. Temporal transcription of the lactococcal temperate phage TP901-1 and DNA sequence of the early promoter region. Microbiology 144:2203-2215. [DOI] [PubMed] [Google Scholar]
- 35.Maguin, E., H. Prévost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moineau, S., S. Pandian, and T. R. Klaenhammer. 1994. Evolution of a lytic bacteriophage via DNA acquisition from the Lactococcus lactis chromosome. Appl. Environ. Microbiol. 60:1832-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Østergaard, S., L. Brøndsted, and F. K. Vogensen. 2001. Identification of a replication protein and repeats essential for DNA replication of the temperate lactococcal bacteriophage TP901-1. Appl. Environ. Microbiol. 67:774-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pedersen, M., S. Østergaard, J. Bresciani, and F. K. Vogensen. 2000. Mutational analysis of two structural genes of the temperate lactococcal bacteriophage TP901-1 involved in tail length determination and baseplate assembly. Virology 276:315-328. [DOI] [PubMed] [Google Scholar]
- 39.Proux, C., D. van Sinderen, J. Suarez, P. Garcia, V. Ladero, G. F. Fitzgerald, F. Desiere, and H. Brüssow. 2002. The dilemma of phage taxonomy illustrated by comparative genomics of Sfi21-like Siphoviridae in lactic acid bacteria. J. Bacteriol. 184:6026-6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rakonjac, J., P. W. O'Toole, and M. Lubbers. 2005. Isolation of lactococcal prolate phage-phage recombinants by an enrichment strategy reveals two novel host range determinants. J. Bacteriol. 187:3110-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Seegers, J. F., S. Mc Grath, M. O'Connell-Motherway, E. K. Arendt, M. van de Guchte, M. Creaven, G. F. Fitzgerald, and D. van Sinderen. 2004. Molecular and transcriptional analysis of the temperate lactococcal bacteriophage Tuc2009. Virology 329:40-52. [DOI] [PubMed] [Google Scholar]
- 43.Seegers, J. F., D. van Sinderen, and G. F. Fitzgerald. 2000. Molecular characterization of the lactococcal plasmid pCIS3: natural stacking of specificity subunits of a type I restriction/modification system in a single lactococcal strain. Microbiology 146:435-443. [DOI] [PubMed] [Google Scholar]
- 44.Stuer-Lauridsen, B., T. Janzen, J. Schnabl, and E. Johansen. 2003. Identification of the host determinant of two prolate-headed phages infecting Lactococcus lactis. Virology 309:10-17. [DOI] [PubMed] [Google Scholar]
- 45.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valyasevi, R., W. E. Sandine, and B. L. Geller. 1990. The bacteriophage kh receptor of Lactococcus lactis subsp. cremoris KH is the rhamnose of the extracellular wall polysaccharide. Appl. Environ. Microbiol. 56:1882-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vegge, C. S., L. Brøndsted, H. Neve, S. Mc Grath, D. van Sinderen, and F. K. Vogensen. 2005. Structural characterization and assembly of the distal tail structure of the temperate lactococcal bacteriophage TP901-1. J. Bacteriol. 187:4187-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, J., M. Hofnung, and A. Charbit. 2000. The C-terminal portion of the tail fiber protein of bacteriophage lambda is responsible for binding to LamB, its receptor at the surface of Escherichia coli K-12. J. Bacteriol. 182:508-512. [DOI] [PMC free article] [PubMed] [Google Scholar]