When observed under the microscope, cell division is just another dark event in the secret life of a bacterium: the cell grows up to a certain size, and then a constriction appears in its center that finally separates the cell into two daughters without any other perceptible changes. Upon a more careful examination, division stands out for its regularity, an indication that it is a strictly controlled event. It took years for bacterial geneticists to identify a deceptively small number of genes that seemed to indicate that the division process is rather simple at the molecular level too. This apparent simplicity has been brightened during the last decade by the use of fluorescence microscopy, which has illuminated the process of bacterial division with many colorful details, some of which are summarized in this review. We will focus on results obtained with Escherichia coli, which has been the most intensively studied organism in this field, although research is not by any means limited to this organism.
In rod-shaped bacteria such as E. coli, division involves the invagination of the cell membrane, closely followed by septation, which is the synthesis of the cell wall that will separate the daughter cells. Septation involves a change in the direction of synthesis of peptidoglycan, which takes place without disrupting the osmotic barrier of the cell envelope. Separation of the daughter cells by cleavage of the central part of the septal cell wall can occur together with septation and constriction, as in E. coli, or at a later stage, as in Bacillus subtilis. In E. coli, these processes involve the localization of at least 15 proteins at the division site (13, 56, 92). They are cytoplasmic, membrane, and periplasmic proteins. Each one localizes into a ring across the cell width at midcell (Fig. 1). It is assumed that all these rings are coincident at the same place and collectively are called the division ring. The ring constricts during division and disappears when the cells separate, although some remnants may eventually be detected at the poles of the newborn cells. Several studies combining genetics and fluorescence microscopy suggest that the division proteins form multiprotein complexes at the division site, a view supported by two-hybrid analyses and other assays (14, 25, 32, 46). These complexes have not been physically isolated and subjected to biochemical and structural analyses due to several drawbacks: some of the division proteins are low-abundance membrane proteins; others (e.g., FtsI and FtsN) are in close contact with the murein sacculus, and their complexes might be broken or distorted upon cell fractionation; in addition, the division ring is a dynamic and energy-dependent structure that quickly disassembles when the nucleotide triphosphate pools are depleted (78), as happens when the cells are broken and their contents diluted.
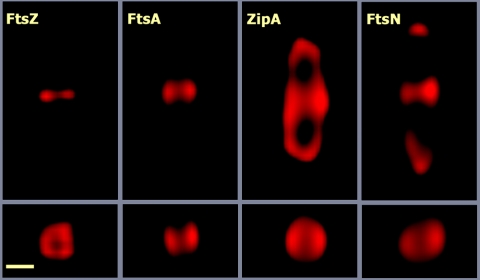
FIG. 1.
Imaging the division ring by three-dimensional deconvolution microscopy. The images correspond to E. coli MC1061 cells from an exponentially growing culture. The cells were stained with polyclonal antisera against FtsZ, FtsA, FtsN, or ZipA obtained in the authors' laboratory and an Alexa 594-conjugated anti-rabbit secondary antibody. Images from optical sections spaced 73 nm apart were taken with an Olympus BX-61 wide-field motorized microscope equipped with a DP70 charge-coupled-device camera. The image stacks were deconvoluted using the Huygens Professional software package. The top row shows images of the central, in-focus, section of each stack, whereas the bottom row shows the cross section of the same cells taken at the points of maximal fluorescence intensity. The elongation along the z axis introduced by the optical system was corrected in the deconvoluted stacks by multiplying the vertical scale by 0.42 (the correction factor calculated from the measurement of control spherical particles). The scale bar corresponds to 0.5 μm.
In E. coli, the division proteins are recruited into the ring following a sequential and almost linear pathway, as deduced from the localization of green fluorescent protein fusions or the immunostaining of the proteins in cell division mutants (13, 19). The flow is as follows: FtsZ > [FtsA, ZapA, ZipA] > (FtsE, FtsX) > FtsK > FtsQ > (FtsB, FtsL) > FtsW > FtsI > FtsN > AmiC > EnvC, where the proteins within brackets are independent of each other but dependent on FtsZ and those within parentheses assemble simultaneously. Although it is not known if the incorporation of a particular protein into the ring requires the continuous presence of all those that precede it in the sequence, a failure in the localization of a protein causes a catastrophic effect on the assembly of those that incorporate later, preventing their localization into the ring. The exception is ZapA, which is nonessential for division and therefore nonessential for the recruitment of the downstream proteins. This hierarchical localization might reflect a pathway of enzymatic reactions taking place at the division site, in which each reaction modifies some component of the septum, allowing the entry of the next protein (temporal dimension), or it might reflect a sequence of protein-protein interactions that leads to the assembly of a multiprotein complex (spatial dimension), the divisome (68), or the septosome (94). Both possibilities could concur in the E. coli assembly pathway. In B. subtilis, the localization does not follow a linear sequence but rather a concerted or cooperative mode, as most division proteins are interdependent for septal localization (27), a fact that argues in favor of the physical protein interaction sequence.
THE GENES
Most of the genes that code for the cell division proteins have been identified in E. coli and B. subtilis in screenings for conditional mutants unable to divide under nonpermissive conditions, usually high temperatures. These mutants are normal at the permissive temperature but grow as multinucleated filaments at the restrictive temperature, and so they were named fts for filamentous temperature sensitive (though the name has been extended to other cell division genes even if they were identified by other methods and there are no known temperature-sensitive alleles, e.g., ftsB and ftsN). In E. coli at least 15 genes are known to be involved in septation: ftsA, -B, -E, -I, -K, -L, -N, -Q, -W, -X, -Z, zipA, zapA, amiC, and envC (27, 56, 89, 92). Other fts genes, like ftsH, ftsJ, and ftsY, turned out to be involved in general processes that have pleiotropic effects on cell division (17, 21, 85). Additional genes are required for division but are not specific for this process, like groEL and hscA, which code for the chaperones GroE and HscA, respectively (70, 86), and mrcA and -B, which code for PBP 1a and 1b, two bifunctional enzymes (penicillin-binding proteins [PBPs]) that are essential for both lateral and septal peptidoglycan synthesis (42).
Comparison of different bacterial genomes has shown that the division genes are well conserved among bacteria, indicating that most bacterial groups share common division machinery and mechanisms. Nevertheless, none of the known division genes is present in all the groups, probably because the machinery is flexible to some extent, and has evolved to fit the diverse nature of the bacterial cell envelopes, cell shapes, and life cycles. Even the ftsZ gene, which is found in the archaea and some eukaryotes (where the FtsZ protein is needed for organelle division), is missing in some groups of wall-less bacteria, like the planctomycetes and the chlamydiae, implying that different division mechanisms have evolved in some bacteria. The only candidate that might be a universal bacterial division gene is mraW, which in most bacteria is associated with other division genes, forming a group called the dcw cluster (4, 84). The mraW gene is found in all the bacterial genomes sequenced to date (our unpublished observations) and codes for an S-adenosyl-methionine-dependent methyltransferase of unknown function (18).
A recent survey of 70 completely sequenced bacterial genomes identified six highly conserved genes whose products might constitute the core of the bacterial division machinery (59): ftsA, ftsI, ftsK, ftsQ, ftsW, and ftsZ. Except for ftsK, all the others belong to the dcw gene cluster. Another gene that might be part of the core group is ftsL. This gene is quite short and poorly conserved at the sequence level, and so it is difficult to identify unequivocally. For example, the ftsL genes from the gram-positive bacteria do not have significant homology to the ones from the proteobacteria, although they share a common secondary structure and are found in equivalent positions within the dcw cluster (59). Interestingly, there are several genomes that contain short open reading frames at the same position in the cluster and have similar predicted secondary structures (our unpublished observations), suggesting that ftsL is indeed conserved and that there are several unidentified (or even unannotated) homologs.
The cluster contains genes involved in cell division and genes involved in the synthesis of the cell wall precursors (4). The gene order and gene contents of the cluster are conserved among bacteria from very distant groups, with a strong trend toward conservation among rod-shaped bacteria and to dispersion in other morphological groups (62, 63, 84). To explain the amazing degree of conservation of the cluster among very distantly related groups of bacteria, as well as the correlation between cluster structure and cell shape, a model that states that in rod-shaped bacteria the clustering of these genes favors the cotranslational assembly and localization of their products at the division site (the genomic channeling hypothesis) (Fig. 2) has been postulated (62, 63). The broad phylogenetic distribution of the cluster, together with the association of genes involved in two different functions, suggests that the cell division machinery is an ancient structure that evolved in parallel with the cell wall and its biosynthetic pathways and became fixed in a rod-shaped cell, presumably the ancestor of extant bacteria (48, 62).
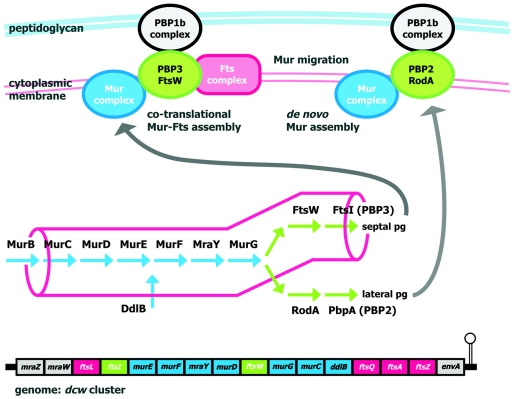
FIG. 2.
Schematic view of the genomic channeling hypothesis. Localized assembly of the enzymes that form the pathway of peptidoglycan (pg) precursor synthesis (indicated in the central part of the figure) into a multienzymatic complex (Mur complex) might drive the precursors directly to the sites of peptidoglycan synthesis by substrate channeling. This would be important in rod-shaped cells, where the highly localized machinery responsible for the synthesis of septal peptidoglycan should compete for precursors with the lateral peptidoglycan synthesis machinery, which is distributed over a much larger surface (both machineries are sketched at the top). The genomic channeling hypothesis (62, 63) states that the clustering of genes in rod-shaped cells (the E. coli dcw cluster is represented at the bottom) favors the cotranslational assembly and localization of the division protein complexes (Fts complex) and the precursor synthesis complex (Mur complex), driving the flux of precursors toward the septum synthesis machinery (PBP3-FtsW/PBP1b complexes) during cell division. After division the Mur complex might migrate to the sites of lateral peptidoglycan synthesis (PBP2-RodA/PBP1b complexes), or it might disassemble and assemble de novo at these sites. Note that in E. coli the murB gene is not found within the dcw cluster.
THE DIVISION RING
A build-up of some proteins forming a ring at the cell center before constriction begins can be detected (1, 22). The ring is visualized by fluorescence microscopy using fusions to fluorescent proteins or antibodies against the different cell division proteins (Fig. 1). It usually appears either as a sharp band that crosses the cell from side to side and is called a closed ring (22) or as two bright dots at both sides of some cells, what is called an open ring (22). A slight delay between the appearance of the open and closed rings of FtsZ, FtsA, and ZipA has been reported (22, 78), suggesting that they might correspond to different, successive stages in ring assembly. The two-dot image of the open ring may be generated by the two-dimensional projection of a three-dimensional ring where the horizontal parts are blurred due to the elongation of the microscope point spread function along the z axis. The closed ring can be resolved in some cases, though not always, into two-dot images by three-dimensional deconvolution (our unpublished observations). This suggests that the two forms of the ring might indeed be very similar, if not identical, structures.
It is not known whether the ring is a single continuous circular structure or several short, unconnected segments, because fluorescent images of the ring lack molecular detail and the use of electron microscopy has not revealed any salient feature. Recently, it has been shown that in Streptococcus pneumoniae there are two different rings, the FtsZ ring and another one formed by the high-molecular-weight PBPs. These two rings seem to be independent, because in mutants in the streptococcal PBP3 protein (homolog of E. coli PBP5), they do not always colocalize (64). It is not known whether the membrane proteins (PBPs) of the streptococcal division ring have the ability to localize and form a ring by themselves, whether they localize in an FtsZ-dependent manner, and whether, once formed, their ring can detach from the FtsZ ring.
The division ring is assembled outwards, starting from the cytoplasmic components and then progressing toward the outer membrane by recruiting the membrane and periplasmic division proteins (Fig. 3 and 4). The assembly of the cytoplasmic side is relatively simple, with only five components: FtsZ, FtsA, ZapA, and the cytoplasmic domains of ZipA and FtsK. The main component is FtsZ, which is the most abundant cell division protein, with 4,000 to 15,000 molecules per cell (Table 1) (54, 76, 78). So, the bulk of the ring lies in the cytoplasmic side, while the membrane and periplasmic parts constitute a smaller fraction, at least in mass terms, because some of its components, PBP3, FtsQ, and FtsL, are found in very small amounts in the cell (5, 93) or are very small proteins, such as FtsL and FtsB (Table 1) (15, 35).
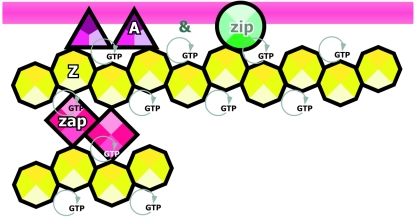
FIG. 3.
Proteins of the early assembly step of the division ring. Assembly of the FtsZ ring depends on either FtsA or ZipA or both, while the localization of these two depends on FtsZ. The localization of ZapA is dependent on FtsZ. FtsA and ZipA are bound to the inner cell membrane (magenta stripe), while FtsZ interacts with these two proteins. These three proteins assemble into the ring at the same time in a first step that might span a significant part of the cell cycle before the assembly of the late proteins shown in Fig. 4. Maintenance of the FtsZ ring is energy dependent (60), probably because FtsZ polymerization requires the presence of GTP at the nucleotide-binding site. Protein names are abbreviated as follows: A, FtsA; Z, FtsZ; Zip, ZipA; and Zap, ZapA. Icons are ordered from left to right following the linear assembly sequence of the proteins. An ampersand indicates that both proteins assemble simultaneously (see the text).
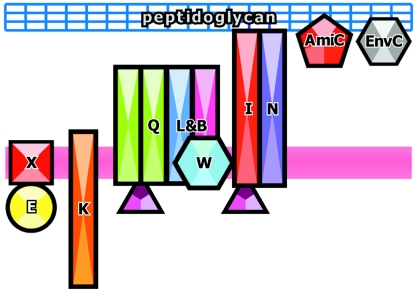
FIG. 4.
Proteins of the late assembly step of the division ring. Schematic view of the assembly of the late cell division proteins showing their relation to the cell membrane (magenta stripe) and peptidoglycan (blue grid). The protein icons are ordered from left to right according to the commonly accepted assembly sequence. An ampersand indicates that both proteins assemble simultaneously (see the text). Proteins that have been shown to interact physically are drawn in contact, while those whose assembly has been determined only by genetic methods are drawn separated. Protein names have been abbreviated by excluding “Fts” from them. FtsA is drawn as in Fig. 3.
TABLE 1.
E. coli division ring ensemble
| Protein | Molecular mass (kDa)a | No. of molecules/cellb | Reference(s) |
|---|---|---|---|
| FtsAd | 45.3 | 50-700 | 78, 91 |
| FtsB | 11.6 | ND | |
| FtsE | 24.4 | ND | |
| FtsIe | 63.9 | 25-100 | 5, 93 |
| FtsK | 146.6 | ND | |
| FtsL | 13.6 | 20-40 | 35 |
| FtsN | 35.8 | 1,000-6,000 | 1, 87 |
| FtsQ | 31.4 | 25 | 5 |
| FtsW | 46.0 | ND | |
| FtsX | 38.5 | ND | |
| FtsZ | 40.3 | 3,200-15,000 | 54, 76, 78 |
| ZipA | 36.5 | 100-1,500 | 36, 78 |
| ZapA | 12.6 | 250c | 34 |
| AmiC | 49.0 | ND | |
| EnvC | 47.5 | ND |
The molecular masses of the monomeric proteins calculated from the gene sequences are indicated.
ND, not determined.
The indicated abundance is for ZapA in B. subtilis cells.
FtsA might be phosphorylated, but the fraction of phosphorylated protein is not known (79).
The division ring is a dynamic structure that requires active metabolism for its maintenance. Some of its components (FtsZ and ZipA) have been shown to be replaced from the ring in just a few seconds (3), and the entire ring quickly disappears upon depletion of the cellular ATP pools (78). During sporulation in B. subtilis the ring can be reprogrammed; the medial ring migrates to the poles in a process that involves a dynamic spiral FtsZ structure (6).
INITIATION OF THE Z-RING
The first known event in the differentiation of the division site is the localization of FtsZ at the future division site to form the Z-ring. FtsZ is a soluble, cytoplasmic protein. It is a structural homolog of eukaryotic tubulin (52), has GTPase activity (66), and in vitro forms a variety of polymers in a GTP-dependent manner (26, 33, 61, 66, 72). It is then logical to think that assembly of the Z-ring involves the localized polymerization of FtsZ. However, such polymers have not been visualized in thin sections of dividing cells even when fast-freezing treatments are used (our unpublished observations), perhaps because they do not form a prominent pattern whose cross section could be differentiated against the cytoplasmic background.
The correct localization of the Z-ring at midcell depends on two inhibitory mechanisms, namely, the MinCDE system and nucleoid occlusion (Noc), mediated in E. coli by SlmA (9, 27, 39, 56). The first mechanism prevents the assembly at places other than the midcell position, and its absence gives rise to abnormal polar divisions that yield minicells. The second mechanism prevents assembly at regions in which a nucleoid is present; when it is inactivated, the antiguillotine checkpoint is abolished and septa can form in the region containing the nucleoid. In vivo, both mechanisms prevent the assembly of Z-rings, but while MinC and -D seem to act by blocking FtsZ polymerization, the action of SlmA in vitro is exactly the opposite; i.e., it promotes the bundling of FtsZ filaments into higher-order structures (9, 43). Their combined actions result, nevertheless, in the ring assembly at midcell only when nucleoids are segregated. When these systems are absent, multiple rings are formed, suggesting that FtsZ has the potential to assemble at any point along the cell (97). Interestingly, under these conditions FtsZ does not spread over the whole nucleoid-free cell surface but condenses into discrete and regularly spaced rings. This means that the sharpness of the rings is not the result of the narrowness of the permitted region in which the assembly is not inhibited, because even when a broad region is available, thin, discrete rings are formed. Multiple sharp rings may arise as a result of a pattern-forming reaction with a self-enhancing (autocatalytic) process, which might be FtsZ polymerization, combined with a long-range inhibition that might be the local depletion of free FtsZ in the vicinity of the ring (58).
FtsZ rings have been found in all the mutants that lack some of the downstream proteins, but not in the ftsA zipA double mutant, indicating that at least one of the two proteins, FtsA or ZipA, must be present, together with FtsZ, to assemble the Z-ring (Fig. 3) (75) and that FtsZ cannot do it alone. Interestingly, in the double ftsA zipA mutant, FtsZ accumulates into regularly spaced clumps between chromosomes, indicating that although it cannot bind to the membrane to form rings, the site selection systems are still functional and the protein is driven to the correct places (75).
In addition to these proteins, the widely conserved, though not essential, protein ZapA (Fig. 3), which binds FtsZ polymers and promotes their bundling in vitro, might positively modulate Z-ring assembly in vivo (34, 51), while in B. subtilis, the EzrA protein is a negative assembly regulator (50). Other proteins downstream from FtsA and ZipA might also contribute to regulate or stabilize the ring, because the frequency of Z-rings formed in mutants, in which they are defective, is often lower than in the wild-type strain (12, 19).
ASSEMBLY OF FtsA AND ZipA
Assembly of FtsA and ZipA into the ring is dependent solely on FtsZ and not on each other or on the rest of the downstream proteins (37). Both proteins are bound to the membrane, ZipA through its N-terminal transmembrane domain (36) and FtsA through a conserved C-terminal amphipathic helix (74), and both have been found to stabilize the Z-ring, probably by a direct interaction with the carboxy-terminal tail of FtsZ (38, 55, 65). This suggests that one role of these proteins might be to anchor FtsZ to the membrane (Fig. 3) (71, 75), though this should be a rather dynamic anchor, because the turnover of FtsZ and ZipA in the ring is very fast (83).
FtsA belongs to the actin/Hsp70/sugar kinase superfamily (11, 79). It has been shown to bind ATP (79, 96) and should be expected to have some ATPase activity. We have not found such activity in purified FtsA proteins from different sources (E. coli, S. pneumoniae, or Thermotoga maritima) expressed in E. coli (49), although two other groups have described ATPase activity in B. subtilis FtsA (28) or phosphatase activity in the protein from Pseudomonas aeruginosa expressed in E. coli (73). Nucleoside triphosphatase activities are abundant in E. coli extracts (81), and caution should be taken when interpreting the results of nucleoside triphosphatase assays. In this case, essential controls should include the analysis of the activities of inactive or altered mutants. The difficulties in finding an enzymatic activity for the FtsA proteins from some sources suggest that they may require an unknown cofactor to be active. Purified S. pneumoniae FtsA is able to bind ATP and polymerizes in an ATP-dependent manner (49). This and the fact that streptococcal FtsA is relatively abundant compared to FtsZ suggest that it might have a structural/mechanical role in ring function. Conversely, based on the phenotypes of the different division mutants and on its relatively low abundance in E. coli (Table 1), it has been proposed that FtsA might be a switch, acting like a checkpoint able to lock the Z-ring and prevent its contraction until the downstream proteins are correctly assembled (56).
The three-dimensional structure of FtsA from T. maritima has been solved (88). The protein has the classical two-domain structure of actin family members, although one of the subdomains is in a different orientation, so the protein has two core subdomains (1A and 2A), forming the nucleotide-binding cleft, and two subdomains (1C and 2B) at opposite sides of the core. Deletion of all of domain 1C of E. coli FtsA does not affect the ability of the remaining fragment to localize to the ring, but then the downstream proteins are not recruited (77). The isolated 1C domain can be artificially driven to the poles (by a fusion to the B. subtilis DivIVA protein (20), where it is able to recruit some of the downstream division proteins, indicating that it must be sufficient for their localization. Most strikingly, when fused to FtsZ, it could even partially complement a temperature-sensitive mutation in ftsA, indicating that FtsA might be the major protein connecting the FtsZ ring and the downstream proteins. Deletion of the tip of domain 2B (S12 and S13 β-strands), at the opposite side, induces formation of misplaced rings (some even locate near the pole), suggesting that this domain interacts with FtsZ and that the mutation somehow increases the stability of the ring, which is then able to partially overcome the actions of the Min and Noc systems (77), supporting a regulatory role for FtsA. A point mutation in the tip of domain 2B of FtsA can bypass completely the function of ZipA (30). This indicates that the functions of both proteins, FtsA and ZipA, are related, and that ZipA, besides being absent from many bacteria, is dispensable for the recruitment of the downstream proteins.
A fusion of FtsQ to ZapA has been found both to be efficiently recruited to the Z-ring independently of FtsA and FtsK (32) and to direct the recruitment of other downstream proteins, from FtsK to FtsI. This indicates that the recruitment of these division proteins by FtsA does not depend on a modification of the Z-ring catalyzed by FtsA but rather on the physical interaction between the proteins. The function of FtsA can be bypassed only partially, and the rings formed are not functional for division, probably because they lack FtsN and perhaps some other activity of FtsA.
FtsA has been shown to interact with itself (95, 96), with the cytoplasmic proteins FtsZ and ZapA, and with the membrane proteins FtsQ, FtsI, and FtsN (25, 46). Contrary to this, ZipA has been found to interact only with FtsZ and with itself (25). This is surprising because under normal conditions ZipA is essential for the localization of the downstream proteins, suggesting that there might exist some additional undetected interactions or otherwise that ZipA has some activity on the structure or with the dynamics of the Z-ring that is necessary for the recruitment of the downstream proteins. ZipA, like FtsN, is present only in the γ-proteobacteria, suggesting that these two proteins might interact or might have related functions (59).
ASSEMBLY OF THE DOWNSTREAM PROTEINS
After FtsA and ZipA, the linear assembly sequence proceeds with FtsE and FtsX, two proteins that are related to the family of ABC transporters and are essential in salt-free media (82). The function of the FtsEX complex in division is not known, but it is more likely to be related to the assembly of downstream proteins than to transport. Its dispensability in high-salt media underlines the flexibility of the architecture of the divisome.
Next comes FtsK, a very large multifunctional membrane protein that has three cytoplasmic domains. The amino-terminal domain is essential for cell division, the intermediate linker domain also has some role in division, and the carboxy-terminal domain is an ATP-dependent DNA translocase that has two separate activities involved in chromosome segregation and chromosome dimer resolution (10).
In E. coli, FtsQ assembles with FtsL and FtsB into a trimeric complex before their localization (14), while in S. pneumoniae, their homologs form a transient complex during septation (69). Assembly of this complex is essential in E. coli for the localization of FtsL and FtsB, which are codependent, but not for the localization of FtsQ. FtsQ is the protein that establishes the higher number of interactions with the other components of the ring. It has a polypeptide transport-associated (POTRA) domain which is functionally associated with protein interactions or chaperon function (80), suggesting that it might be a connector protein able to nucleate or regulate the assembly of the other membrane proteins.
The next proteins in the assembly process are FtsI and FtsW, both of which are involved in peptidoglycan synthesis during division. FtsI is a class B penicillin-binding protein (PBP3) with transpeptidase activity, and FtsW belongs to the shape, elongation, division, and sporulation (SEDS) family of polytopic membrane proteins (41). Each SEDS protein is usually associated with a single class B PBP (57), so FtsW is proposed to act in concert with PBP3 (FtsI) in cell division (44), as RodA and PBP2 do in cell elongation, although their exact molecular roles are not known.
The last membrane protein in the linear sequence is FtsN, which spans the periplasm and has a peptidoglycan-binding domain. The function of FtsN is unknown but is likely to be required for both early and late phases of assembly (2, 20, 32). The sequence of known division ring elements closes with two periplasmic proteins, the AmiC amidase and the EnvC hydrolase (7, 8).
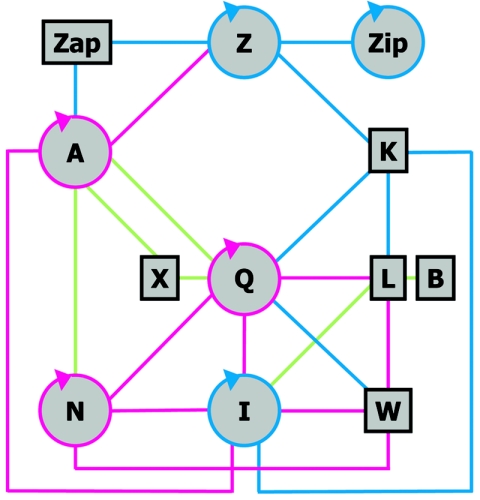
Data from two-hybrid and other interaction assays indicate the existence of interactions between the E. coli cell division proteins (Fig. 5) (16, 20, 25, 32, 46). Strikingly, most of the proteins interact with multiple partners. FtsZ binds to ZipA, FtsA, and FtsK. Then FtsA and FtsK bind to slightly different subsets of the group of downstream membrane proteins, and these are themselves connected by multiple interactions. The ability of FtsQ to establish numerous interactions is also supported by these assays. The existence of a complex network of interactions fits well with the cooperative or concerted assembly of the division proteins described to occur in B. subtilis (27) but is difficult to reconcile with the linearity of the assembly pathway described for E. coli (13). Possibly the linear sequence reveals a dominant set of strong interactions under physiological conditions, while the rest are weaker interactions that can cooperatively strengthen the complex in vivo but are detected individually only in experimental set-ups in which the proteins are overproduced. This issue should be addressed by studying the interactions by other, different methods to produce a quantitative description of the network that takes into account both the amounts of the different division proteins in the cell (Table 1) and the affinities of at least some of their interactions.
FIG. 5.
Network of interactions between division proteins. Schematic drawing of the protein-protein interactions among the E. coli cell division proteins found in systematic functional bacterial two-hybrid assays. The circular arrows indicate self-interactions. Blue lines indicate only those interactions described by Di Lallo et al. (25), green lines indicate only those described by Karimova et al. (46), while magenta lines are those detected in both studies. Note that the report of Karimova et al. (46) did not include FtsK. The FtsA self-interaction (96) and the ZipA-FtsZ interaction (38) are also found in functional yeast two-hybrid assays.
TIMING THE ASSEMBLY
The timing for the assembly of the division proteins has been studied by immunofluorescence microscopy in steady-state cultures of E. coli by two different methods (1, 22, 78). The first method makes use of the distribution of cell ages in batch cultures growing under steady-state conditions to infer, from the fraction of cells with rings, the fraction of the cell cycle in which the rings are present and from this the age at which theproteins localize in the division ring (22). This age at assembly corresponds to an ideal population in which all the cells haveidentical life cycles; i.e., all the cells in a given age class are identical, and all the cells divide at a critical age into two identical daughter cells (47). The second method measures the fraction of cells with detectable rings at different times along the cell cycle in synchronous cultures (78). In both studies it was found that the distribution of the frequency of rings by cell age classes (cell lengths) was rather broad, indicating that the cells were heterogeneous with respect to the ring assembly even in steady-state populations. Pooling the FtsZ data from the synchronous cultures of Ref (78) to calculate the assembly timing from the frequency of cells with rings (22) yields an estimate of approximately 21 min for the period of the cell cycle in which the ring is present. This indicates that the Z-ring assembled at minute 28 in a 49-min cell cycle, that is to say at 57% of the full cycle. The maximum frequency of cells with FtsZ rings in the synchronous cultures was reached at 40 min, which is 81% of the cycle. Therefore, the period of FtsZ ring assembly in the population since the appearance of the first rings until most cells had rings spanned about 24% of the cell cycle. This means that some cells maintain a ring assembled for a significant part of the cell cycle but that others assemble the ring shortly before division. Another possibility is that the ring formed by the upstream proteins can disassemble and reassemble, being a transient structure until the downstream proteins localize into it at a late cell cycle stage (see below). Whatever the case may be, this heterogeneity of the cells with respect to ring assembly is surprising and should be taken into account for a better understanding of the processes of assembly of the different components of the division ring.
Using the age distribution method, it was found that the age at assembly was the same for FtsZ and FtsA, and this assembly occurs around the time of termination of DNA replication and before chromosome segregation, suggesting that termination of DNA replication could provide a signal to initiate cell division. In the synchronous cultures, the first cells with FtsZ, FtsA, or ZipA rings appear also around the end of chromosome replication, and there were no significant differences in the timings of localization of these three early proteins.
The age distribution method has been applied to determine the timing of assembly of the downstream proteins FtsQ, FtsW, FtsI, and FtsN (1). Surprisingly, it was found that these proteins localize into the ring nearly at the same time and 14 to 21 min after FtsZ, FtsA, and ZipA (in cell cycles of 40 to 140 min). This indicates that assembly of division proteins into the ring occurs in two steps (Fig. 3 and 4) that are chronologically separated and raises the question of what the early proteins are doing at midcell during all this time, which is a significant fraction of the cell cycle. This is not a trivial question because maintenance of the Z-ring consumes energy (Fig. 3) and is therefore unlikely to gratuitously be assembled before it is needed.
It has been shown that in the E. coli cell envelope there are two different types of peptidoglycan. Both are well separated into topological domains: the peptidoglycan of the lateral wall, which grows by diffuse insertion of new material, and an inert and very stable peptidoglycan that is synthesized at midcell to be finally placed at the poles of the daughter cells. The synthesis of the latter type is initiated by a process that depends on the previous localization of FtsZ, but it does not depend on FtsA or, surprisingly, on FtsI, which is the PBP involved in the synthesis of the septal peptidoglycan (23, 24). This suggests that the early assembly of the FtsZ ring might be the signal to start the processes of murein differentiation and segregation of the topological domains of the cell wall, two processes that might require some time to operate before the wall is mature for septation. Compartmentalization of the periplasm (29) or the cytoplasm (45, 90) is a process that might also be under way once the assembly of the early upstream division proteins has occurred and before the downstream proteins reach the division ring.
SUMMARY AND OUTLOOK
The main components of the E. coli cell division machinery have been identified, and their relations are being studied. Several of the genes identified are widely conserved, suggesting that the core of the division engine is common to most bacterial groups. Two-hybrid and other gene fusion approaches have shown that the division proteins are related by a small but complex interaction network, in which FtsQ and FtsA may be the most ubiquitous partners. Most of the downstream proteins, which comprise the late assembly group, form a membrane complex connected to the Z-ring through FtsA and possibly FtsK. Issues that remain unresolved are the heterogeneity between cells in the timing of ring assembly and the continuity and role of the Z-ring during the part of the cell cycle in which the late proteins are not yet assembled. The functions of several division genes are still poorly understood (or unknown); therefore, to obtain a comprehensive functional description of the division ring, we need to obtain the detailed structural and biochemical characterization of the individual components as well as their role in the partial or complete division complexes. Another important future challenge ahead is to determine the fine structure of the cell wall and in particular the structural and functional differences between the lateral and polar/septal peptidoglycans.
Research in bacterial cell division has surpassed its own field, providing some fundamental ideas to develop a new bacterial cell biology that recognizes that bacteria do have internal cytoskeletons and a dynamic subcellular structure (31, 53). Our current knowledge of the assembly of the bacterial septum has reached a point in which we can envisage its application to enlighten other fields. Among them, synthetic biology and nanotechnology appear to be interesting challenges. More-mundane applications, from the discovery of new medicines to the design of nanoengines, will hopefully help to brighten our future.
Acknowledgments
We thank Pilar Palacios for the raw images used to produce Fig. 1.
Work in M.V.'s laboratory was supported by grant QLK3-2000-00079 from the European Commission, grants BIO2000-0451-P4-02 and BIO2001-1542 from the Ministerio de Ciencia y Tecnología, and grant GR/SAL/0642/2004 from the Consejería de Educación de la Comunidad de Madrid. J.M. is recipient of a CSIC I3P fellowship financed by the European Social Fund, and R.M.-A. received a fellowship from Heskuntza, Unibertsitate Eta Ikerketa Saila, Eusko Jaurlaritza.
REFERENCES
- 1.Aarsman, M. E., A. Piette, C. Fraipont, T. M. Vinkenvleugel, M. Nguyen-Distéche, and T. den Blaauwen. 2005. Maturation of the Escherichia coli divisome occurs in two steps. Mol. Microbiol. 55:1631-1645. [DOI] [PubMed] [Google Scholar]
- 2.Addinall, S. G., C. Cao, and J. Lutkenhaus. 1997. FtsN, a late recruit to the septum in Escherichia coli. Mol. Microbiol. 25:303-309. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, D. E., F. J. Gueiros-Filho, and H. P. Erickson. 2004. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J. Bacteriol. 186:5775-5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayala, J. A., T. Garrido, M. A. de Pedro, and M. Vicente. 1994. Molecular biology of bacterial septation. .In J. M. Ghuysen and R. Habenneck (ed.), Bacterial cell wall. Elsevier Science, Amsterdam, The Netherlands.
- 5.Barondess, J. J., M. Carson, L. M. Guzmán Verduzco, and J. Beckwith. 1991. Alkaline phosphatase fusions in the study of cell division genes. Res. Microbiol. 142:295-299. [DOI] [PubMed] [Google Scholar]
- 6.Ben-Yehuda, S., and R. Losick. 2002. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 109:257-266. [DOI] [PubMed] [Google Scholar]
- 7.Bernhardt, T. G., and P. A. de Boer. 2003. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol. Microbiol. 48:1171-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernhardt, T. G., and P. A. de Boer. 2004. Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol. Microbiol. 52:1255-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernhardt, T. G., and P. A. de Boer. 2005. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol. Cell 18:555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bigot, S., J. Corre, J. M. Louarn, F. Cornet, and F. X. Barre. 2004. FtsK activities in Xer recombination, DNA mobilization and cell division involve overlapping and separate domains of the protein. Mol. Microbiol. 54:876-886. [DOI] [PubMed] [Google Scholar]
- 11.Bork, P., C. Sander, and A. Valencia. 1992. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. USA 89:7290-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyle, D. S., M. M. Khattar, S. G. Addinall, J. Lutkenhaus, and W. D. Donachie. 1997. ftsW is an essential cell-division gene in Escherichia coli. Mol. Microbiol. 24:1263-1273. [DOI] [PubMed] [Google Scholar]
- 13.Buddelmeijer, N., and J. Beckwith. 2002. Assembly of cell division proteins at the E. coli cell center. Curr. Opin. Microbiol. 5:553-557. [DOI] [PubMed] [Google Scholar]
- 14.Buddelmeijer, N., and J. Beckwith. 2004. A complex of the Escherichia coli cell division proteins FtsL, FtsB and FtsQ forms independently of its localization to the septal region. Mol. Microbiol. 52:1315-1327. [DOI] [PubMed] [Google Scholar]
- 15.Buddelmeijer, N., N. Judson, D. Boyd, J. J. Mekalanos, and J. Beckwith. 2002. YgbQ, a cell division protein in Escherichia coli and Vibrio cholerae, localizes in codependent fashion with FtsL to the division site. Proc. Natl. Acad. Sci. USA 99:6316-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butland, G., J. M. Peregrin-Alvarez, J. Li, W. Yang, X. Yang, V. Canadien, A. Starostine, D. Richards, B. Beattie, N. Krogan, M. Davey, J. Parkinson, J. Greenblatt, and A. Emili. 2005. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433:531-537. [DOI] [PubMed] [Google Scholar]
- 17.Caldas, T., E. Binet, P. Bouloc, A. Costa, J. Desgres, and G. Richarme. 2000. The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S ribosomal RNA methyltransferase. J. Biol. Chem. 275:16414-16419. [DOI] [PubMed] [Google Scholar]
- 18.Carrión, M., M. J. Gómez, R. Merchante-Schubert, S. Dongarra, and J. A. Ayala. 1999. mraW, an essential gene at the dcw cluster of Escherichia coli codes for a cytoplasmic protein with methyltransferase activity. Biochimie 81:879-888. [DOI] [PubMed] [Google Scholar]
- 19.Chen, J. C., and J. Beckwith. 2001. FtsQ, FtsL and FtsI require FtsK, but not FtsN, for co-localization with FtsZ during Escherichia coli cell division. Mol. Microbiol. 42:395-413. [DOI] [PubMed] [Google Scholar]
- 20.Corbin, B. D., B. Geissler, M. Sadasivam, and W. Margolin. 2004. Z-ring-independent interaction between a subdomain of FtsA and late septation proteins as revealed by a polar recruitment assay. J. Bacteriol. 186:7736-7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Leeuw, E., B. Graham, G. J. Phillips, C. M. ten Hagen-Jongman, B. Oudega, and J. Luirink. 1999. Molecular characterization of Escherichia coli FtsE and FtsX. Mol. Microbiol. 31:983-993. [DOI] [PubMed] [Google Scholar]
- 22.Den Blaauwen, T., N. Buddelmeijer, M. E. Aarsman, C. M. Hameete, and N. Nanninga. 1999. Timing of FtsZ assembly in Escherichia coli. J. Bacteriol. 181:5167-5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Pedro, M. A., J. V. Höltje, and H. Schwarz. 2002. Fast lysis of Escherichia coli filament cells requires differentiation of potential division sites. Microbiology 148:79-86. [DOI] [PubMed] [Google Scholar]
- 24.de Pedro, M. A., J. C. Quintela, J. V. Höltje, and H. Schwarz. 1997. Murein segregation in Escherichia coli. J. Bacteriol. 179:2823-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Lallo, G., M. Fagioli, D. Barionovi, P. Ghelardini, and L. Paolozzi. 2003. Use of a two-hybrid assay to study the assembly of a complex multicomponent protein machinery: bacterial septosome differentiation. Microbiology 149:3353-3359. [DOI] [PubMed] [Google Scholar]
- 26.Erickson, H. P., D. W. Taylor, K. A. Taylor, and D. Bramhill. 1996. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc. Natl. Acad. Sci. USA 93:519-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Errington, J., R. A. Daniel, and D. J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feucht, A., I. Lucet, M. D. Yudkin, and J. Errington. 2001. Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis. Mol. Microbiol. 40:115-125. [DOI] [PubMed] [Google Scholar]
- 29.Foley, M., J. M. Brass, J. Birmingham, W. R. Cook, P. B. Garland, C. F. Higgins, and L. I. Rothfield. 1989. Compartmentalization of the periplasm at cell division sites in Escherichia coli as shown by fluorescence photobleaching experiments. Mol. Microbiol. 3:1329-1336. [DOI] [PubMed] [Google Scholar]
- 30.Geissler, B., D. Elraheb, and W. Margolin. 2003. A gain-of-function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc. Natl. Acad. Sci. USA 100:4197-4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gitai, Z. 2005. The new bacterial cell biology: moving parts and subcellular architecture. Cell 120:577-586. [DOI] [PubMed] [Google Scholar]
- 32.Goehring, N. W., F. Gueiros-Filho, and J. Beckwith. 2005. Premature targeting of a cell division protein to midcell allows dissection of divisome assembly in Escherichia coli. Genes Dev. 19:127-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.González, J. M., M. Vélez, M. Jiménez, C. Alfonso, P. Schuck, J. Mingorance, M. Vicente, A. P. Minton, and G. Rivas. 2005. Cooperative behavior of Escherichia coli cell-division protein FtsZ assembly involves the preferential cyclization of long single-stranded fibrils. Proc. Natl. Acad. Sci. USA 102:1895-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gueiros-Filho, F. J., and R. Losick. 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16:2544-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guzmán, L. M., J. J. Barondess, and J. Beckwith. 1992. FtsL, an essential cytoplasmic membrane protein involved in cell division in Escherichia coli. J. Bacteriol. 174:7716-7728. [PMC free article] [PubMed] [Google Scholar]
- 36.Hale, C. A., and P. A. de Boer. 1997. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88:175-185. [DOI] [PubMed] [Google Scholar]
- 37.Hale, C. A., and P. A. de Boer. 1999. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J. Bacteriol. 181:167-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haney, S. A., E. Glasfeld, C. Hale, D. Keeney, Z. He, and P. de Boer. 2001. Genetic analysis of the Escherichia coli FtsZ · ZipA interaction in the yeast two-hybrid system. Characterization of FtsZ residues essential for the interactions with ZipA and with FtsA. J. Biol. Chem. 276:11980-11987. [DOI] [PubMed] [Google Scholar]
- 39.Harry, E. J. 2001. Bacterial cell division: regulating Z-ring formation. Mol. Microbiol. 40:795-803. [DOI] [PubMed] [Google Scholar]
- 40.Hayashi, S., H. Hara, H. Suzuki, and Y. Hirota. 1988. Lipid modification of Escherichia coli penicillin-binding protein 3. J. Bacteriol. 170:5392-5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henriques, A. O., P. Glaser, P. J. Piggot, and C. P. Moran, Jr. 1998. Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol. Microbiol. 28:235-247. [DOI] [PubMed] [Google Scholar]
- 42.Höltje, J. V., and C. Heidrich. 2001. Enzymology of elongation and constriction of the murein sacculus of Escherichia coli. Biochimie 83:103-108. [DOI] [PubMed] [Google Scholar]
- 43.Hu, Z., A. Mukherjee, S. Pichoff, and J. Lutkenhaus. 1999. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. USA 96:14819-14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ikeda, M., T. Sato, M. Wachi, H. K. Jung, F. Ishino, Y. Kobayashi, and M. Matsuhashi. 1989. Structural similarity among Escherichia coli FtsW and RodA proteins and Bacillus subtilis SpoVE protein, which function in cell division, cell elongation, and spore formation, respectively. J. Bacteriol. 171:6375-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Judd, E. M., K. R. Ryan, W. E. Moerner, L. Shapiro, and H. H. McAdams. 2003. Fluorescence bleaching reveals asymmetric compartment formation prior to cell division in Caulobacter. Proc. Natl. Acad. Sci. USA 100:8235-8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karimova, G., N. Dautin, and D. Ladant. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 187:2233-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koch, A. L. 1993. Similarities and differences of individual bacteria within a clone, p. 1640-1651. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 2. ASM Press, Washington, D.C. [Google Scholar]
- 48.Koch, A. L. 2003. Were Gram-positive rods the first bacteria? Trends Microbiol. 11:166-170. [DOI] [PubMed] [Google Scholar]
- 49.Lara, B., A. I. Rico, S. Petruzzelli, A. Santona, J. Dumas, J. Biton, M. Vicente, J. Mingorance, and O. Massidda. 2005. Cell division in cocci: localization and properties of the Streptococcus pneumoniae FtsA protein. Mol. Microbiol. 55:699-711. [DOI] [PubMed] [Google Scholar]
- 50.Levin, P. A., I. G. Kurtser, and A. D. Grossman. 1999. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 96:9642-9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Low, H. H., M. C. Moncrieffe, and J. Löwe. 2004. The crystal structure of ZapA and its modulation of FtsZ polymerisation. J. Mol. Biol. 341:839-852. [DOI] [PubMed] [Google Scholar]
- 52.Löwe, J. 1998. Crystal structure determination of FtsZ from Methanococcus jannaschii. J. Struct. Biol. 124:235-243. [DOI] [PubMed] [Google Scholar]
- 53.Löwe, J., F. van den Ent, and L. A. Amos. 2004. Molecules of the bacterial cytoskeleton. Annu. Rev. Biophys. Biomol. Struct. 33:177-198. [DOI] [PubMed] [Google Scholar]
- 54.Lu, C., J. Stricker, and H. P. Erickson. 1998. FtsZ from Escherichia coli, Azotobacter vinelandii, and Thermotoga maritima—quantitation, GTP hydrolysis, and assembly. Cell Motil. Cytoskelet. 40:71-86. [DOI] [PubMed] [Google Scholar]
- 55.Ma, X., and W. Margolin. 1999. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol. 181:7531-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Margolin, W. 2004. The assembly of proteins at the cell division site, p.79-102. In M. Vicente, J. Tamames, A. Valencia, and J. Mingorance (ed.), Molecules in time and space. Bacterial shape, division and phylogeny. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 57.Matsuhashi, M., M. Wachi, and F. Ishino. 1990. Machinery for cell growth and division: penicillin-binding proteins and other proteins. Res. Microbiol. 141:89-103. [DOI] [PubMed] [Google Scholar]
- 58.Meinhardt, H. 2004. Models for pattern formation in bacteria applied to bacterial morphogenesis, p. 59-78. In M. Vicente, J. Tamames, A. Valencia, and J. Mingorance (ed.), Molecules in time and space. Bacterial shape, division and phylogeny. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 59.Mingorance, J., A. I. Rico, and P. Gómez-Puertas. 2004. Bacterial morphogenes, p. 173-194. In M. Vicente, J. Tamames, A. Valencia, and J. Mingorance (ed.), Molecules in time and space. Bacterial shape, division and phylogeny. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 60.Mingorance, J., S. Rueda, P. Gomez-Puertas, A. Valencia, and M. Vicente. 2001. Escherichia coli FtsZ polymers contain mostly GTP and have a high nucleotide turnover. Mol. Microbiol. 41:83-91. [DOI] [PubMed] [Google Scholar]
- 61.Mingorance, J., M. Tadros, M. Vicente, J. M. González, G. Rivas, and M. Vélez. 2005. Visualization of single Escherichia coli FtsZ filament dynamics with atomic force microscopy. J. Biol. Chem. 280:20909-20914. [DOI] [PubMed] [Google Scholar]
- 62.Mingorance, J., and J. Tamames. 2004. The bacterial dcw gene cluster: an island in the genome?, p. 249-272. In M. Vicente, J. Tamames, A. Valencia, and J. Mingorance (ed.), Molecules in time and space. Bacterial shape, division and phylogeny. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 63.Mingorance, J., J. Tamames, and M. Vicente. 2004. Genomic channeling in bacterial cell division. J. Mol. Recognit. 17:481-487. [DOI] [PubMed] [Google Scholar]
- 64.Morlot, C., M. Noirclerc-Savoye, A. Zapun, O. Dideberg, and T. Vernet. 2004. The d,d-carboxypeptidase PBP3 organizes the division process of Streptococcus pneumoniae. Mol. Microbiol. 51:1641-1648. [DOI] [PubMed] [Google Scholar]
- 65.Mosyak, L., Y. Zhang, E. Glasfeld, S. Haney, M. Stahl, J. Seehra, and W. S. Somers. 2000. The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 19:3179-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mukherjee, A., K. Dai, and J. Lutkenhaus. 1993. Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. Proc. Natl. Acad. Sci. USA 90:1053-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nagasawa, H., Y. Sakagami, A. Suzuki, H. Suzuki, H. Hara, and Y. Hirota. 1989. Determination of the cleavage site involved in C-terminal processing of penicillin-binding protein 3 of Escherichia coli. J. Bacteriol. 171:5890-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nanninga, N. 1991. Cell division and peptidoglycan assembly in Escherichia coli. Mol. Microbiol. 5:791-795. [DOI] [PubMed] [Google Scholar]
- 69.Noirclerc-Savoye, M., A. Le Gouellec, C. Morlot, O. Dideberg, T. Vernet, and A. Zapun. 2005. In vitro reconstitution of a trimeric complex of DivIB, DivIC and FtsL, and their transient co-localization at the division site in Streptococcus pneumoniae. Mol. Microbiol. 55:413-424. [DOI] [PubMed] [Google Scholar]
- 70.Ogino, H., M. Wachi, A. Ishii, N. Iwai, T. Nishida, S. Yamada, K. Nagai, and M. Sugai. 2004. FtsZ-dependent localization of GroEL protein at possible division sites. Genes Cells 9:765-771. [DOI] [PubMed] [Google Scholar]
- 71.Ohashi, T., C. A. Hale, P. A. de Boer, and H. P. Erickson. 2002. Structural evidence that the P/Q domain of ZipA is an unstructured, flexible tether between the membrane and the C-terminal FtsZ-binding domain. J. Bacteriol. 184:4313-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oliva, M. A., S. Huecas, J. M. Palacios, J. Martín-Benito, J. M. Valpuesta, and J. M. Andreu. 2003. Assembly of archaeal cell division protein FtsZ and a GTPase-inactive mutant into double-stranded filaments. J. Biol. Chem. 278:33562-33570. [DOI] [PubMed] [Google Scholar]
- 73.Paradis-Bleau, C., F. Sanschagrin, and R. C. Levesque. 2005. Peptide inhibitors of the essential cell division protein FtsA. Protein Eng. Des. Sel. 18:85-91. [DOI] [PubMed] [Google Scholar]
- 74.Pichoff, S., and J. Lutkenhaus. 2005. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol. Microbiol. 55:1722-1734. [DOI] [PubMed] [Google Scholar]
- 75.Pichoff, S., and J. Lutkenhaus. 2002. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 21:685-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pla, J., M. Sánchez, P. Palacios, M. Vicente, and M. Aldea. 1991. Preferential cytoplasmic location of FtsZ, a protein essential for Escherichia coli septation. Mol. Microbiol. 5:1681-1686. [DOI] [PubMed] [Google Scholar]
- 77.Rico, A. I., M. García-Ovalle, J. Mingorance, and M. Vicente. 2004. Role of two essential domains of Escherichia coli FtsA in localization and progression of the division ring. Mol. Microbiol. 53:1359-1371. [DOI] [PubMed] [Google Scholar]
- 78.Rueda, S., M. Vicente, and J. Mingorance. 2003. Concentration and assembly of the division ring proteins FtsZ, FtsA, and ZipA during the Escherichia coli cell cycle. J. Bacteriol. 185:3344-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sánchez, M., A. Valencia, M. J. Ferrándiz, C. Sander, and M. Vicente. 1994. Correlation between the structure and biochemical activities of FtsA, an essential cell division protein of the actin family. EMBO J. 13:4919-4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sánchez-Pulido, L., D. Devos, S. Genevrois, M. Vicente, and A. Valencia. 2003. POTRA: a conserved domain in the FtsQ family and a class of beta-barrel outer membrane proteins. Trends Biochem. Sci. 28:523-526. [DOI] [PubMed] [Google Scholar]
- 81.Sayed, A., S. Matsuyama, K. Inoue, J. Alsina, F. Cai, J. Chen, and M. Inouye. 2000. ATPase and GTPase activities copurifying with GTP-binding proteins in E. coli. J. Mol. Microbiol. Biotechnol. 2:261-263. [PubMed] [Google Scholar]
- 82.Schmidt, K. L., N. D. Peterson, R. J. Kustusch, M. C. Wissel, B. Graham, G. J. Phillips, and D. S. Weiss. 2004. A predicted ABC transporter, FtsEX, is needed for cell division in Escherichia coli. J. Bacteriol. 186:785-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stricker, J., P. Maddox, E. D. Salmon, and H. P. Erickson. 2002. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc. Natl. Acad. Sci. USA 99:3171-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tamames, J., M. González-Moreno, J. Mingorance, A. Valencia, and M. Vicente. 2001. Bringing gene order into bacterial shape. Trends Genet. 17:124-126. [DOI] [PubMed] [Google Scholar]
- 85.Tomoyasu, T., T. Yuki, S. Morimura, H. Mori, K. Yamanaka, H. Niki, S. Hiraga, and T. Ogura. 1993. The Escherichia coli FtsH protein is a prokaryotic member of a protein family of putative ATPases involved in membrane functions, cell cycle control, and gene expression. J. Bacteriol. 175:1344-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Uehara, T., H. Matsuzawa, and A. Nishimura. 2001. HscA is involved in the dynamics of FtsZ-ring formation in Escherichia coli K12. Genes Cells 6:803-814. [DOI] [PubMed] [Google Scholar]
- 87.Ursinus, A., F. van den Ent, S. Brechtel, M. de Pedro, J. V. Holtje, J. Lowe, and W. Vollmer. 2004. Murein (peptidoglycan) binding property of the essential cell division protein FtsN from Escherichia coli. J. Bacteriol. 186:6728-6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.van den Ent, F., and J. Löwe. 2000. Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO J. 19:5300-5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vicente, M., and J. Löwe. 2003. Ring, helix, sphere and cylinder: the basic geometry of prokaryotic cell division. EMBO Rep. 4:655-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vicente, M., P. Palacios, A. Dopazo, T. Garrido, J. Pla, and M. Aldea. 1991. On the chronology and topography of bacterial cell division. Res. Microbiol. 142:253-257. [DOI] [PubMed] [Google Scholar]
- 91.Wang, H., and R. C. Gayda. 1992. Quantitative determination of FtsA at different growth rates in Escherichia coli using monoclonal antibodies. Mol. Microbiol. 6:2517-2524. [DOI] [PubMed] [Google Scholar]
- 92.Weiss, D. S. 2004. Bacterial cell division and the septal ring. Mol. Microbiol. 54:588-597. [DOI] [PubMed] [Google Scholar]
- 93.Weiss, D. S., K. Pogliano, M. Carson, L. M. Guzmán, C. Fraipont, M. Nguyen-Distéche, R. Losick, and J. Beckwith. 1997. Localization of the Escherichia coli cell division protein Ftsl (PBP3) to the division site and cell pole. Mol. Microbiol. 25:671-681. [DOI] [PubMed] [Google Scholar]
- 94.Witte, A., E. Brand, P. Mayrhofer, F. Narendja, and W. Lubitz. 1998. Mutations in cell division proteins FtsZ and FtsA inhibit phiX174 protein-E-mediated lysis of Escherichia coli. Arch. Microbiol. 170:259-268. [DOI] [PubMed] [Google Scholar]
- 95.Yan, K., K. H. Pearce, and D. J. Payne. 2000. A conserved residue at the extreme C-terminus of FtsZ is critical for the FtsA-FtsZ interaction in Staphylococcus aureus. Biochem. Biophys. Res. Commun. 270:387-392. [DOI] [PubMed] [Google Scholar]
- 96.Yim, L., G. Vandenbussche, J. Mingorance, S. Rueda, M. Casanova, J. M. Ruysschaert, and M. Vicente. 2000. Role of the carboxy terminus of Escherichia coli FtsA in self-interaction and cell division. J. Bacteriol. 182:6366-6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yu, X. C., and W. Margolin. 1999. FtsZ ring clusters in min and partition mutants: role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol. Microbiol. 32:315-326. [DOI] [PubMed] [Google Scholar]