Abstract
Trp-rich antimicrobial peptides play important roles in the host innate defense mechanisms of many plants, insects, and mammals. A new type of Trp-rich peptide, Ac-KWRRWVRWI-NH2, designated Pac-525, was found to possess improved activity against both gram-positive and -negative bacteria. We have determined that the solution structures of Pac-525 bound to membrane-mimetic sodium dodecyl sulfate (SDS) micelles. The SDS micelle-bound structure of Pac-525 adopts an α-helical segment at residues Trp2, Arg3, and Arg4. The positively charged residues are clustered together to form a hydrophilic patch. The three hydrophobic residues Trp2, Val6, and Ile9 form a hydrophobic core. The surface electrostatic potential map indicates the three tryptophan indole rings are packed against the peptide backbone and form an amphipathic structure. Moreover, the reverse sequence of Pac-525, Ac-IWRVWRRWK-NH2, designated Pac-525rev, also demonstrates similar antimicrobial activity and structure in membrane-mimetic micelles and vesicles. A variety of biophysical and biochemical methods, including circular dichroism, fluorescence spectroscopy, and microcalorimetry, were used to show that Pac-525 interacted strongly with negatively charged phospholipid vesicles and induced efficient dye release from these vesicles, suggesting that the antimicrobial activity of Pac-525 may be due to interactions with bacterial membranes.
Antimicrobial peptides play important roles in the host innate defense mechanisms of many plants, insects, and mammals (2, 3, 8, 11). Most antimicrobial peptides do not target specific molecular receptors of the pathogens but rather interact and permeabilize microbial membranes (36). With an increase of antibiotic resistance within bacteria, the potential for the development of antimicrobial peptides as novel therapeutic agents could overcome the problem of resistance (13).
To date, several hundred antimicrobial peptides have been characterized (2, 31). The naturally derived antimicrobial peptides are generally 12 to 50 amino acids in length and are folded into several structural groups, including α-helices, β-sheets, extended peptides, and looped peptides. Although these peptides show a marked degree of variability, they share two common and functionally important characteristics, namely, a net cationicity that facilitates interaction with negatively charged microbial surfaces and the ability to assume amphipathic structures that permit incorporation into microbial membranes (36).
Tryptophan has a strong ability to insert into membranes and to cause Trp-rich peptides to partition into membranes, which affects lipid polymorphism (9, 27). Recent studies by Killian and coworkers also indicate that Trp residues are located in the interfacial region of lipid bilayers (19). Due to these important features of Trp residues in the antimicrobial and hemolytic activities, the structures and mechanisms of action of several Trp-rich cationic antimicrobial peptides have been studied recently (10, 15, 17, 18, 26, 28-30). A well-studied example of a Trp-rich antimicrobial peptide is indolicidin (26). Indolicidin, with the amino acid sequence ILPWKWPWWPWRR-NH2, is a short Trp-rich peptide isolated from the cytoplasmic granules of bovine neutrophils. Indolicidin has an extended boat-shaped structure and a wide-spectrum antimicrobial activity against gram-positive and -negative bacteria, protozoa, fungi, and the enveloped virus human immunodeficiency virus type 1 (26). Antimicrobial activity has also been proposed for other Trp-rich peptides, such as tritrpticin (29), puroA (17), Lfcin B (28), and PW2 (30).
In addition to the large-scale screening of individual natural products and synthetic compounds, combinatorial chemistry has the potential to greatly advance the development of biologically active antimicrobial peptides (4). Several antimicrobial and/or antifungal peptides of various lengths were identified using non-support-bound, mixture-based synthetic combinatorial libraries (4). In an effort to develop more-effective low-molecular-mass antimicrobial peptides, we have designed and synthesized a linear library of peptides ranging in length from 3 to 9 amino acid residues, using tryptophan as a template at various positions. Of the newly designed peptides, the 9-amino-acid-residue peptide Ac-KWRRWVRWI-NH2, designated Pac-525, demonstrates improved activity against both gram-positive and -negative bacteria and reduced hemolytic activity. We have found that a decrease in the number of positively charged amino acids abates the antimicrobial activities of Pac-525. Alternatively, the replacement of the tryptophan residues of Pac-525 causes minimal or no effect on the antimicrobial activities. However, it is not known how such a short peptide possesses antimicrobial activity. In this paper, we have used various biophysical and biochemical methods, including circular dichroism (CD), fluorescence spectroscopy, and microcalorimetry to study the properties of Pac-525 through its interaction with various bacteria and model membranes. In addition, we have determined the solution structure of Pac-525 bound to membrane-mimetic sodium dodecyl sulfate (SDS) micelles by two-dimensional nuclear magnetic resonance (NMR) methods. Our results provide a molecular basis for elucidating the mechanism of Trp-rich peptide-membrane interactions and highlighting several important structural features of these interactions.
MATERIALS AND METHODS
Materials.
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (POPG), and calcein were purchased from Sigma (St Louis, MO). Acrylamide was purchased from Bio-Rad Laboratories Inc., and Escherichia coli (ATCC 25922 and ATCC 10536), Staphylococcus aureus strains (ATCC 29213 and ATCC 33591 [methicillin resistant]), and Pseudomonas aeruginosa (ATCC 27853) strains were obtained from the Bioresource Collection and Research Center in Taiwan. Perdeuterated sodium dodecyl sulfate and D2O were purchased from Cambridge Isotope Laboratories, Inc.
Peptide synthesis, purification, and analysis.
Peptides were synthesized by a solid-phase method using the 9-fluorenylmethoxy carbonyl (Fmoc) protocol. 1-hydroxybenzotriazole and diisopropylethylamine were used as coupling reagents, and fourfold excess Fmoc amino acids were added during every coupling cycle. Removal of the N-terminal Fmoc protecting group was accomplished by gentle stirring with 20% piperidine in dimethylformamide for 2 h at room temperature. The acetylation of the peptides was achieved by adding 10-fold acetic anhydride and 20-fold diisopropylethylamine in dimethylformamide and stirring for 2 h at room temperature. The cleavage of peptides from the resin was carried out by using reagent K following the procedure of King et al. (20). The crude peptides were then purified by high-performance liquid chromatography using a Vydac Cl8 reversed-phase column with two solvent systems of 0.1% trifluoroacetic acid/H2O and 0.1% trifluoroacetic acid/acetonitrile (24). The identity of the peptides was checked by electrospray mass spectroscopy, and the purity (>95%) was assessed by high-performance liquid chromatography.
Antimicrobial activity.
E. coli strains (ATCC 25922 and ATCC 10536), S. aureus strains (ATCC 29213 and ATCC 33591 [methicillin resistant]), and Pseudomonas aeruginosa (ATCC 27853) were used to test the antimicrobial activity of the peptides. MICs were determined by the standard broth microdilution method of the National Committee for Clinical Laboratory Standards (24a) with Mueller-Hinton broth. Bacteria were cultivated in Luria broth at 37°C to stationary phase. Overnight cultures of the test organisms were diluted to 5 × 105 CFU/ml in Mueller-Hinton broth (Difco). Peptides in 100-μl volumes were put in a 96-well microtiter plate, and all wells were subsequently inoculated with the diluted culture of the test organisms. After 18 h of incubation at 37°C, the results were assayed for turbidity (cell growth) by measuring the absorbance at 600 nm in a microplate reader (Thermo Max; Molecular Devices). The MIC is the lowest concentration of peptide at which there was no change in optical density. All peptides were tested in triplicate.
Hemolytic activity.
The hemolytic activities of the peptides were determined based on hemolysis against human red blood cells (hRBC). The hRBCs were isolated from heparinized human blood by centrifugation after being washed three times with phosphate-buffered saline (PBS) buffer. The hRBCs were diluted in a solution of 10% PBS. The peptides dissolved in PBS buffer were added to 50 μl of 10% solution of hRBCs and incubated for 1 hour at 37°C (final hRBC concentration, 5% [vol/vol]). The samples were centrifuged for 10 min. Various concentrations of peptides were incubated with pretreated hRBC, and the percentage of hemolysis was determined by measuring the optical density of the supernatant at 540 nm. Zero hemolysis (blank) and 100% hemolysis were determined in PBS buffer and 1% Triton X-100, respectively.
Preparation of unilamellar vesicles.
Large unilamellar vesicles (LUVs) were prepared by the extrusion method using an Avanti small-volume extrusion apparatus (Avanti Polar Lipids, Inc., Alabaster, AL). After a final trace amount of organic solvent was evaporated, the dried lipid film was suspended by vortexing it in PBS buffer (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4). After freezing and thawing cycles, the lipid suspensions were extruded by using an extruder (Avanti Polar Lipids, Inc., Alabaster, AL) 10 times through two stacked 0.4-μm-pore-size polycarbonate filters (Avanti Polar Lipids, Inc., Alabaster, AL), followed by 10 times through two stacked 0.1-μm-pore-size filters.
Calcein-filled vesicles were prepared by the use of a calcein-containing buffer (70 mM calcein and 10 mM Tris at pH 7.4) with the extrusion method (14) as foregoing. Unencapsulated calcein was removed by centrifugation (10 min at 10,000 rpm) three times using isosmotic buffer (10 mM Tris and 100 mM NaCl, pH 7.4).
Dye leakage experiments.
Peptide-induced calcein leakage (1), reflected by an increase in fluorescence, was investigated by using a Perkin-Elmer luminescence spectrofluorimeter (model LS55) at excitation and emission wavelengths of 496 and 515 nm, respectively. Measurements of calcein-entrapped LUVs were made when the lipid concentration was 30 μM. One hundred percent leakage was induced in 5 min by the addition of 0.1% Triton X-100. Using this as a reference, the degree of leakage induced by various concentrations of peptides was calculated using the following equation: % leakage = [(F − F0)/(Fr − F0)] × 100, where F0 and Fr are the initial fluorescence intensities observed without peptide and after treatment with 20 μl 10% Triton X-100, respectively.
ITC.
Isothermal titration calorimetry (ITC) experiments were performed using a VP-ITC isothermal titration calorimeter (MicroCal Inc., Northhampton, MA) (33). All the ITC experiments were conducted in sodium phosphate buffer with 150 mM NaCl at pH 7.4. The solutions were degassed under a vacuum prior to use. The peptide solution was placed in the 1.4-ml reaction cell and titrated with the LUVs delivered from a 250-μl motor-driven syringe. The cell contents were stirred continuously at 300 rpm during the experiment. In control experiments, the corresponding vesicle suspension was injected into buffer without peptide. The heats of dilution of lipid vesicles were subtracted from the heats determined in the corresponding peptide-lipid binding experiments. Data were analyzed with the Origin version 5.0 software. For calculation of the isotherm, only the outer layer of the lipid vesicles (60% of total lipid) was considered.
Fluorescence spectroscopy.
Fluorescence emission spectra were recorded on a model LS55 spectrofluorimeter (Perkin-Elmer). Measurements were performed between 300 and 450 nm within 0.5-nm increments by using a 4- by 10-mm quartz cell at 25°C. The excitation wavelength was set to 295 nm with both the excitation and emission slit widths set to 5 nm. The concentration of peptide samples was 1 μM in phosphate buffer (pH 7.4) in the presence or absence of 25 mM SDS or 1 mM phospholipid vesicles. Spectra were baseline corrected by subtracting blank spectra of the corresponding solutions without peptide. For fluorescence quenching experiments, acrylamide was used as the quencher. Acrylamide was added to ensure that final concentrations were between 2 and 135 mM. The quenched samples were excited at 295 nm, and the emission was monitored at the peak determined from the wavelength scan in the absence of quencher. The effect of acrylamide on the fluorescence of the peptide was analyzed using the modified Stern-Volmer equation (6): F0/F = 1 + KSV × Q, where KSV is the Stern-Volmer quenching constant and Q is the quencher concentration.
CD spectroscopy.
CD spectra were recorded on a AVIV 202 spectropolarimeter after calibration with d-10-camphorsulfonic acid. All the measurements reported were carried out in buffer, 25 mM SDS, or 1 mM POPG and POPC vesicles at 25°C using a l-mm-path-length cuvette. Three scans were averaged for each spectrum with a 0.2-nm step size. Peptide concentrations were 60 μM and were obtained by a quantitative serial dilution of a stock solution. Spectra were baseline corrected by subtracting blank spectra of the corresponding solutions without peptide, and ellipticities were converted to mean residue molar ellipticities in degrees · cm2 · dmol−1.
NMR spectroscopy.
All NMR experiments were performed on a Bruker Avance 600-MHz spectrometer at 37°C using a 2 mM sample. NMR spectra were collected in the phase-sensitive mode with time-proportional phase incrementation. The acquired data were transferred to a Silicon Graphics extreme Indigo2 workstation and processed using XWINNMR (Bruker). Nuclear Overhauser effect spectroscopy (NOESY) spectra were acquired at three different mixing times: 60, 120, and 250 ms. Solvent suppression was done by presaturation in the relaxation and mixing period. These phase-sensitive NOESY spectra were collected into 2,048 points in the t2 dimension and 300 points in the tl dimension with 48 scans per tl increment and a relaxation delay of 1.5 s between each scan. The NOESY data sets were apodized with a shifted-sine-bell window function and zero-filled to 2,048 and 1,024 points in the t2 and tl dimensions, respectively. Double-quantum filtered correlation spectroscopy spectra were recorded into 2,048 points in t2 and 256 increments in tl with 48 scans per tl experiment; 6-Hz Gaussian broadening was applied in the t2 domain, and a skewed-sine-bell window function was used in the tl domain. Total correlation spectroscopy (TOCSY) spectra were recorded using the MLEV17 pulse sequence with mixing times of 35 and 70 ms at 2,048 points in t2 and 300 points in tl. All chemical shifts were referenced to internal 2,2-dimethyl-2-silapentane-5-sulfonate.
Structural calculations.
Distance restraints were acquired from NOEs assigned in a two-dimensional NOESY spectrum with 60 ms of mixing time. The NOE cross-peak intensities were classified as strong, medium, and weak, corresponding to distance limits of 1.8 to 2.8 Å, 1.8 to 3.4 Å, and 1.8 to 5.0 Å, respectively. Structural calculations were carried out with the programs InsightII 950 (MSI Inc.) and X-PLOR 3.851 (A. T. Brünger, Yale University) on a Silicon Graphics Indigo2 workstation. The starting structure was generated as an extended form by using the program Insight II and then was calculated and refined with the simulated annealing protocol in X-PLOR 3.851. All force constants and molecular parameters were set to their default values, as in the original sa.inp and refine.inp protocols, with the exception of the time step, which was decreased to 0.001 ps throughout the calculations. Simulated annealing was performed using 10,000 steps at 1,000 K and 15,000 steps as the molecule was gradually cooled to 100 K. The structures generated by the SA procedure were refined using SA (simulated annealing protocol) refinement. The temperature in refine.inp was set to 1,000 K and was gradually cooled to 100 K in 25,000 steps. Finally these structures were energy minimized using 500 steps of Powell energy minimization. The final 20 structures contained no distance constraint violations greater than 0.3 Å and no dihedral angle constraint violations greater than 5 degrees. An average structure was calculated by averaging the 20 structures, which was refined using the refine.inp protocol as mentioned above. The structures were analyzed with MOLMOL and PROCHECK-NMR (23).
RESULTS
Antibacterial and hemolytic activities.
E. coli (ATCC 25922 and ATCC 10536), S. aureus (ATCC 29213 and ATCC 33591 [methicillin resistant]), and Pseudomonas aeruginosa (ATCC 27853) strains were used to determine the antimicrobial activities of Pac-525. The MICs were 2 μM for E. coli and Pseudomonas aeruginosa and 4 μM for S. aureus. These MICs indicate that Pac-525 exhibits antimicrobial activity against gram-positive, gram-negative, and anaerobic bacteria. Compared to the MICs (2 to 4 μM), the hemolytic activity of Pac-525 is very low. It only gives 50% lysis of human red blood cells at a 350 μM peptide concentration.
Peptide-induced dye leakage.
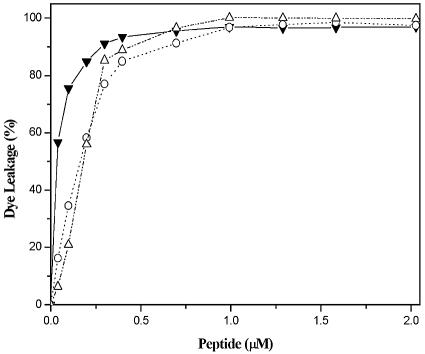
The membrane-permeabilizing abilities of Pac-525 were investigated by studying calcein release from phospholipid vesicles of different surface charge densities. The electrically neutral POPC LUVs were used to mimic the eukaryotic membranes, and the highly negatively charged POPG and the negatively charged mixed POPC/POPG (3:1) LUVs were used to mimic the bacterial membranes. Upon addition of Pac-525 to these LUVs, the entrapped calcein was released into the buffer due to lysis or leakage induced by the peptide (Fig. 1). The calcein leakage percentages in POPG, POPC/POPG (3:1), and POPC LUVs all approached 100% at very low peptide concentrations (∼1 μM) with only small differences. These results indicate that Pac-525 has a very strong membrane-disruptive activity.
FIG. 1.
Calcein leakage experiments for Pac-525 in POPG (▵), POPC/POPG (3:1) (○), and POPC (▾) LUVs at 25°C.
ITC.
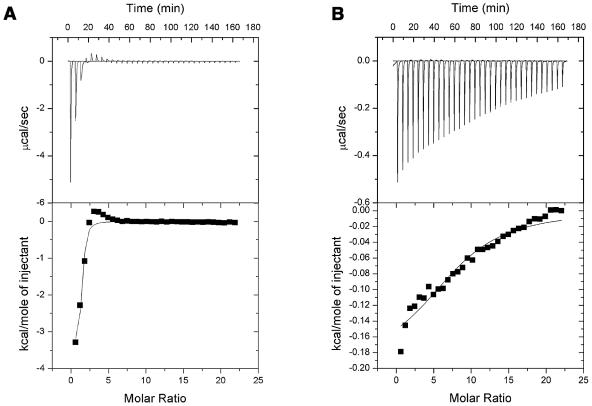
Figure 2 shows the titrations determined when Pac-525 was placed in the reaction cell and titrated with LUVs delivered from a motor-driven syringe. The ITC data were used to compare the affinities of Pac-525 to the POPG and POPC LUVs. The binding constants determined for Pac-525 bound to POPG and POPC LUVs were 2.4 × 105 and 4.8 × 103 M−1, respectively. These data clearly indicate that Pac-525 binds much more tightly to the negatively charged POPG vesicles than to the electrically neutral POPC vesicles.
FIG.2.
Titration calorimetry of Pac-525 with POPG LUVs (A) and POPC LUVs (B) in PBS buffer (pH 7.4) at 25°C.
Tryptophan fluorescence spectroscopy.
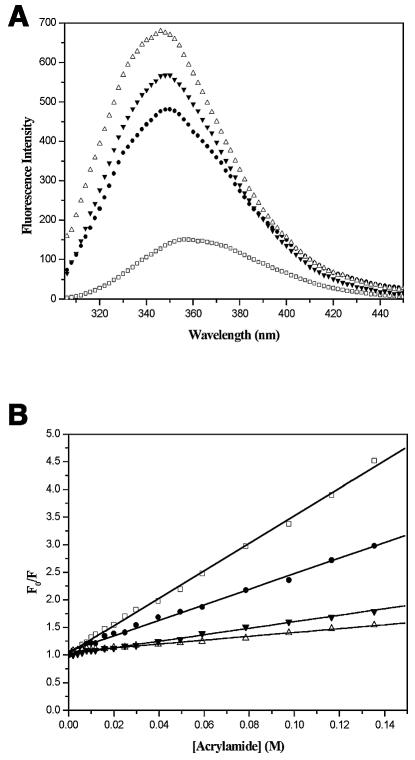
When excited at 295 nm, Trp residues in Pac-525 in buffer gave rise to an emission peak centered at 357 nm (Fig. 3A). When Pac-525 bound to SDS micelles, POPC LUVs, and POPG LUVs, the more hydrophobic environment and the decreased flexibility of Trp residues caused a shift to the lower wavelength (“blue shift”) and an increase in the fluorescence intensity (Fig. 3A). The blue shifts of Pac-525 in SDS micelles, POPC LUVs, and POPG LUVs were found to be 9, 10, and 11 nm, respectively. These results indicate that Trp side chains of Pac-525 were positioned in a more hydrophobic environment and were more sterically confined in SDS micelles, POPC LUVs, and POPG LUVs (22).
FIG. 3.
(A) Fluorescence emission spectra of 1 μM Pac-525 in PBS buffer (□), SDS (•), POPG LUVs (▵), and POPC LUVs (▾) at 25°C. (B) Stern-Volmer plots of 1 μM Pac-525 in PBS buffer (□), SDS (•), POPG LUVs (▵), and POPC LUVs (▾) at 25°C.
To determine the extent to which the Trp residues were sequestered in the hydrophobic cores of the micelles and vesicles, fluorescence quenching experiments were performed using acrylamide (34). The effective KSV values for Pac-525 were calculated to be 25.3, 14.8, 6.1, and 4.2 M−1 in buffer, SDS micelles, POPC LUVs, and POPG LUVs (Fig. 3B). The more extensively Trp residues are shielded from acrylamide, the smaller the value for KSV. These values confirm that when Pac-525 is free in solution, it is more accessible to the quencher than when it is in the presence of micelles and vesicles. The order of protection of Trp residues by micelles and vesicles from acrylamide is as follows: POPG > POPC > SDS > buffer.
NMR spectroscopy.
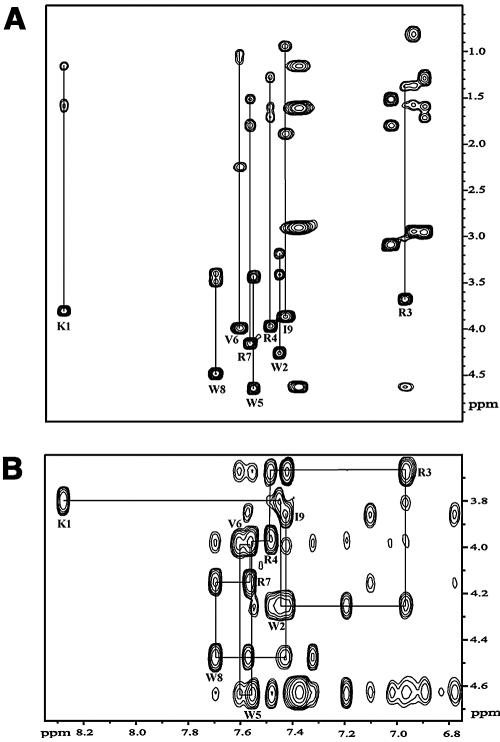
The CD spectra of Pac-525 recorded in buffer presented a negative band at 200 nm, indicating a typical random coil conformation. The addition of SDS micelles, POPC LUVs, and POPG LUVs led to a dramatic structure change resulting in a more ordered structure (data not shown). In addition to the CD studies, we have used two-dimensional 1H NMR to study the conformational change of Pac-525 in SDS micelles. Use of micelles rather than phospholipid vesicles was necessary to acquire high-resolution solution state NMR data due to the unfavorable large rotational correlation time of the vesicles. The chemical shift assignments of Pac-525 (2 mM) in 20 mM sodium phosphate buffer (pH 4.5) and in SDS (200 mM) at 37°C were performed by using the standard sequential assignment procedures (35). Assignment of all the proton chemical shifts was accomplished by using the TOCSY and NOESY spectra (Fig. 4). DQF-COSY was used to assign the hydrogen protons in the delta and gamma positions in residues and to obtain the backbone dihedral angle constraints. Many of the chemical shifts observed deviated significantly from the random coil values.
FIG. 4.
(A) A 600-MHz TOCSY spectrum, recorded at 70 ms of mixing time of Pac-525 (2 mM) in 200 mM SDS at 37°C, showing the NH (F2)-aliphatic (F1) region. (B) NH-CαH region of the 600-MHz NOESY spectrum recorded at 120 ms of mixing time for Pac-525 (2 mM) in 200 mM SDS at 37°C. Peaks are labeled at the positions of the NH-CαH cross-peaks.
Structure calculation and description.
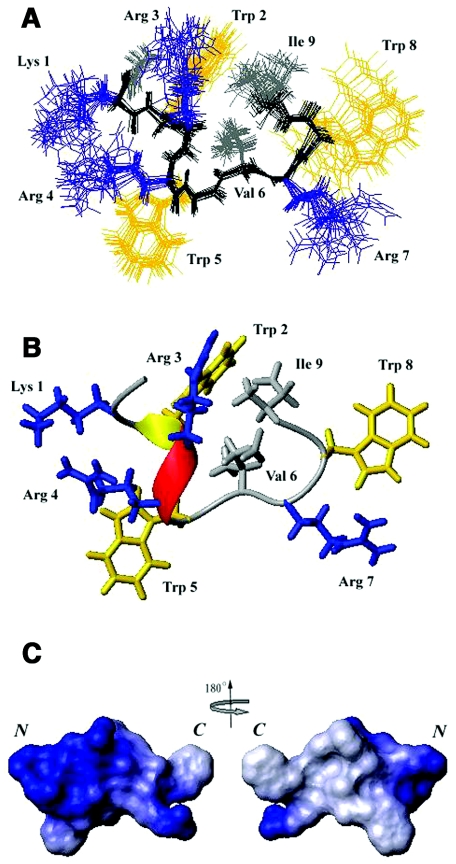
When working with such a short peptide in solution, highly interconverting conformational ensembles are normal. Based on the single set of NMR peaks observed in the NOESY and TOCSY spectra, the interconverting conformations are concluded in the fast-exchange region. Thus, the solution structure of Pac-525 calculated based on NMR data reflects the ensemble average of the interconverting conformations. A total of 154 NOE-derived distance constraints, including 100 intraresidue, 31 sequential, and 23 medium-range distance restraints in conjunction with eight backbone dihedral angles were used in the structure calculations. Some of the observed NOEs were not used due to their weak intensities and/or overlaps in the NOESY spectra collected at the 60- and 120-ms mixing times. No hydrogen bond was detected in the two-dimensional TOCSY experiments. The overlay of the backbone atoms for the 20 lowest energy structures of Pac-525 in SDS is shown in Fig. 5A. Figure 5B shows the final refined averaged structure of Pac-525. Although calculating the surface electrostatic potential for such a short and flexible peptide is deceptive, the electrostatic surface plots of Pac-525 computed using MOLMOL is shown in Fig. 5C to illustrate the charge distribution of Pac-525. The energetic and structural statistics are listed in Table 1. The root mean square deviation (RMSD) calculated from the averaged coordinates for Pac-525 is 0.297 Å for the backbone heavy atoms (N, C, and Cα) and 1.033 Å for all heavy atoms.
FIG. 5.
Calculated structures of Pac-525. (A) Twenty lowest-energy structures calculated for Pac-525 (2 mM in 20 mM sodium phosphate buffer at pH 4.5 and 37°C with 200 mM SDS). (B) The final refined average structure for Pac-525 bound to SDS. The positively charged residues are shown in blue, the tryptophan residues are shown in yellow, and the hydrophobic residues are shown in gray. (C) The electrostatic surface plot of Pac-525. Positive charge is indicated by blue and neutral charge is indicated by white. The figure was generated using the program MOLMOL (21).
TABLE 1.
Summary of structural constrains and structure statistics
| Parameter | Value |
|---|---|
| Total no. of NOE restraints | 154 |
| Intraresidue (|i − j| = 0) | 100 |
| Sequential (|i − j| = 1) | 31 |
| Medium range (2 ≦ |i − j| ≦ 4) | 23 |
| Dihedral angle | 8 |
| RMSD for geometrical analysis | |
| Bond length (Å) | 0.0014 ± 0.00005 |
| Bond angle (degrees) | 0.1663 ± 0.01127 |
| Improper (degrees) | 0.0926 ± 0.01109 |
| RMSD from experimental restraints | |
| Distance (Å) | 0.0155 ± 0.00112 |
| Dihedral angle (degrees) | 0.00003 ± 0.00015 |
| Pairwise atomic RMSD (Å) | |
| All heavy atoms | 1.033 |
| Backbone heavy atoms | 0.297 |
| Energy statistics (kcal/mol) | |
| Overall | 4.45 ± 0.31 |
| Bond | 0.43 ± 0.028 |
| Angle | 1.62 ± 0.22 |
| Improper | 0.18 ± 0.046 |
| van der Waals | 0.36 ± 0.17 |
| NOE | 1.86 ± 0.27 |
| Dihedral angle | 0.00 ± 0.00 |
| Ramachandran statistics (%) | |
| Most favored region | 82.9 |
| Additionally allowed | 17.1 |
| Generously allowed | 0.0 |
| Disallowed | 0.0 |
The SDS micelle-bound structure of Pac-525 had a helical segment at residues Trp2, Arg3, and Arg4 (Fig. 5). The positively charged residues were clustered together to form a hydrophilic patch. The three hydrophobic residues Trp2, Val6, and Ile9 formed a hydrophobic core. The surface electrostatic potential map indicated the three tryptophan indole rings were packed against the peptide backbone to form an amphipathic structure. A cationic amphipathic structure would be best suited for maximizing both electrostatic and hydrophobic interactions with a membrane.
Bidirectional antimicrobial activity of Pac-525.
There are many examples that the reversed analogs of antimicrobial peptides posses equal or enhanced antimicrobial activities (16). While viewing the amino acid sequence and the solution structure of Pac-525, it was noticed that the reversed sequence of Pac-525, Ac-IWRVWRRWK-NH2, designated Pac-525rev, may also possess the antimicrobial activity. Indeed, the MICs of Pac-525rev against E. coli and S. aureus were found to be 4 μM and 8 μM, respectively. The hemolytic activity of Pac-525rev is very low and similar to that of Pac-525 (50% lysis against human red blood cells at 880 μM). In addition, Pac-525rev also caused a similar extent of calcein release from POPC and POPG LUVs and induced a blue shift to the lower wavelength and an increase in the fluorescence intensity while bound to the micelles and vesicles (data not shown).
DISCUSSION
In this paper, we report that the 9-amino-acid peptide Pac-525 (Ac-KWRRWVRWI-NH2), derived from a linear peptide library using tryptophan as a template, exhibits antimicrobial activity against both gram-positive and gram-negative bacteria. Based on dye leakage and ITC experiments, Pac-525 was found to bind strongly to negatively charged phospholipid vesicles and efficiently induce dye leakage from the vesicles. These results suggest that the antimicrobial activity of Pac-525 may act through binding to and destabilization of the microbial membrane rather than through a specific protein receptor (17). This hypothesis was further confirmed by the fact that an all-d-amino-acid Pac-525 possesses similar antimicrobial activities against E. coli and S. aureus (data not shown) (12).
Pac-525 has a very strong membrane-disruptive activity as can be seen from the calcein leakage experiments in POPG, POPC/POPG (3:1), and POPC LUVs. In contrast to other antimicrobial peptides studied (25, 32), Pac-525 induced 100% dye leakage at very low peptide concentrations (∼1 μM). These results may explain the strong antimicrobial activity of Pac-525 against gram-negative and gram-positive bacteria. However, the outer surface of gram-negative bacteria contains lipopolysaccharides, and that of gram-positive bacteria contains acidic polysaccharides (teichoic acids), conferring a net negative charge to the surface of both gram-positive and gram-negative bacteria (5). Furthermore, the phospholipids composing the inner membrane of gram-negative bacteria and the single membrane of gram-positive bacteria are negatively charged. It is expected that the MIC will be higher (2 to 4 μM) than the peptide concentration required for membrane disruption of the POPG and POPC vesicles (∼1 μM) due to the differences between the outer polysaccharides and phospholipids of bacteria and the POPG and POPC vesicles. On the other hand, the ITC experiments indicated that Pac-525 binds much more strongly to the negatively charged POPG LUVs than to the neutral POPC LUVs. This preference may explain the selective activity of Pac-525 against negatively charged bacterial membranes rather than mammalian cells.
Membrane leakage can be induced either by perturbation of lipid packing or by formation of peptide pores (7, 8). The fluorescence quenching experiments were performed to determine the depth of penetration of Pac-525 into micelles and vesicles. The burial of Trp residues in the acyl core of the micelles or LUVs protects the fluorescence of Trp residues from acrylamide quenching. The results indicated that Trp residues of Pac-525 were not fully sequestered in the hydrophobic interior to form stable channels or pores since partial quenching of Trp fluorescence was still detected.
A common feature found in most of the antimicrobial peptides is that the distribution and the amount of the net charge correlate with biological function. The NMR structures of indolicidin have been determined in both dodecylphosphorylcholine and SDS micelles (26). Although the structures of the two types of micelles differ, indolicidin was found to have a similar arrangement of hydrophobic and cationic regions in SDS and dodecylphosphorylcholine micelles. The NMR structure of another Trp-rich antimicrobial peptide, tritrpticin, with the amino acid sequence VRRFPWWWPFLRR, shows an amphipathic turn-turn structure with the Trp residues clustered together and inserted in the hydrophobic core of the SDS micelle (29). Residues 4 to 9 of the lactoferricin peptide (LfcinB4-9) with the sequence RRWQWR-NH2, has been shown to display antimicrobial activity which compares favorably to that of LfcinB. LfcinB4-9 has been found to form a stable amphipathic structure in SDS micelles, with the Trp side chains located deeper within the micelle than the Arg and Gln residues (28). Table 2 shows the antimicrobial activities of Pac-525 and its derivatives as well as the above-mentioned antimicrobial peptides. It was found that the decrease in positively charged amino acids (Pac-521 and Pac-529) abates the peptides' antimicrobial activities. Alternatively, the replacement of the tryptophan residues of Pac-525 causes minimal or no effect on their antimicrobial activities (Pac-527 and Pac-528). For Pac-525 and other tryptophan-rich antimicrobial peptides, the rules guiding structure-activity relationships tend to be somewhat dependent on the parent peptide structure and the distribution of the charged amino acids. In the case of Pac-525, four of the nine residues are positively charged amino acids and distributed in an amphipathic structure. This may explain the broad antimicrobial activity and low hemolytic activity of Pac-525.
TABLE 2.
MIC values for Pac-525 and its derivatives as well as other Trp-rich peptides against E. coli and S. aureus
| Peptidea | Sequence | MIC (μM) for:
|
|
|---|---|---|---|
| E. coli | S. aureus | ||
| Pac-525 | Ac-KWRRWVRWI-NH2 | 2 | 4 |
| Pac-525rev | Ac-IWRVWRRWK-NH2 | 4 | 4 |
| Pac-521 | Ac-KWIKWIKWI-NH2 | 8 | 16 |
| Pac-529 | Ac-KWIRWVRWI-NH2 | 8 | 4 |
| Pac-527 | Ac-KFRRFVRFI-NH2 | 4 | 4 |
| Pac-528 | Ac-KPRRPVRPI-NH2 | 2 | 4 |
| Indolicidin | ILPWKWPWWPWRR-NH2 | 10 | 1 |
| Tritrpticin | VRRFPWWWPFLRR | 10 | 10 |
| PuroA | FPVTWRWWKWWKG-NH2 | 7 | 16 |
| LfcinB4-14 | RRWQWRMKKLG | 20 | 20 |
| PW2 | HPLKQYWWRPSI | >125 | >125 |
In conclusion, we have determined the solution structure of the antimicrobial peptide Pac-525 in membrane-mimetic SDS micelles. It was found that Pac-525 forms a marked amphipathic structure in SDS with hydrophobic core flanked by positively charged residues. A variety of biophysical and biochemical experiments, including fluorescence spectroscopy and microcalorimetry, were used to show that Pac-525 interacted strongly with negatively charged phospholipid vesicles and induced efficient dye release from these vesicles, suggesting that the antimicrobial activity of Pac-525 may be due to lipid interactions with bacterial membranes.
Acknowledgments
We thank Feng-Di Lung and Jia-Yin Tsai for synthesizing some of the peptides for us in the early stage of this project. This study was carried out on the 600-MHz NMR spectrometer at the Regional Instrument Center at Hsinchu, National Tsing Hua University, Taiwan.
This work is supported by grants from the NTHU-Pacgen-commissioned research project. Structural studies are supported by grants from National Science Council, ROC (NSC93-2113-M-007-002 and NSC93-3112-B-007-012), and the Program for Promoting Academic Excellence of Universities from the Ministry of Education, ROC (89-B-FA04-1-4).
REFERENCES
- 1.Allen, T. M. (ed.). 1984. Calcein as a tool in liposome methodology. CRC Press, Boca Raton, Fla.
- 2.Andreu, D., and L. Rivas. 1998. Animal antimicrobial peptides: an overview. Biopolymers 47:415-433. [DOI] [PubMed] [Google Scholar]
- 3.Bals, R., and J. M. Wilson. 2003. Cathelicidins—a family of multifunctional antimicrobial peptides. Cell. Mol. Life Sci. 60:711-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blondelle, S., and K. Lohner. 2000. Combinatorial libraries: a tool to design antimicrobial and antifungal peptide analogues having lytic specificities for structure-activity relationship studies. Biopolymers 55:74-87. [DOI] [PubMed] [Google Scholar]
- 5.Brock, T. D. 1974. Biology of microorganisms, 2nd ed. Prentice-Hall Inc., Englewood Cliffs, N.J.
- 6.Eftink, M. R., and C. A. Ghiron. 1976. Exposure of tryptophanyl residues in proteins. Quantitative determination by fluorescence quenching studies. Biochemistry 15:672-680. [DOI] [PubMed] [Google Scholar]
- 7.El Jastimi, R., K. Edwards, and M. Lafleur. 1999. Characterization of permeability and morphological perturbations induced by nisin on phosphatidylcholine membranes. Biophys. J. 77:842-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Epand, R. M., and H. J. Vogel. 1999. Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta 1462:11-28. [DOI] [PubMed] [Google Scholar]
- 9.Fimland, G., V. G. Eijsink, and J. Nissen-Meyer. 2002. Mutational analysis of the role of tryptophan residues in an antimicrobial peptide. Biochemistry 41:9508-9515. [DOI] [PubMed] [Google Scholar]
- 10.Friedrich, C. L., A. Rozek, A. Patrzykat, and R. E. W. Hancock. 2001. Structure and mechanism of action of an indolicidin peptide derivative with improved activity against gram-positive bacteria. J. Biol. Chem. 276:24015-24022. [DOI] [PubMed] [Google Scholar]
- 11.Ganz, T. 2003. Defensins: antimicrobial peptides of innate immunity. Nature Rev. Immunol. 3:710-720. [DOI] [PubMed] [Google Scholar]
- 12.Hancock, R. E. W., and R. Lehrer. 1998. Cationic peptides: a new source of antibiotics. Trends Biotechnol. 16:82-88. [DOI] [PubMed] [Google Scholar]
- 13.Hancock, R. E. W., and A. Patrzykat. 2002. Clinical development of cationic antimicrobial peptides: from natural to novel antibiotics. Curr. Drug Targets Infect. Disorders 2:79-83. [DOI] [PubMed] [Google Scholar]
- 14.Hope, M. J., M. B. Bally, G. Webb, and P. R. Cullis. 1985. Production of large unilamellar vesicles by a rapid extrusion procedure. Characterization of size distribution, trapped volume and ability to maintain a membrane potential. Biochim. Biophys. Acta 812:55-66. [DOI] [PubMed] [Google Scholar]
- 15.Hwang, P. M., N. Zhou, X. Shan, C. H. Arrowsmith, and H. J. Vogel. 1998. Three-dimensional solution structure of lactoferricin B, an antimicrobial peptide derived from bovine lactoferrin. Biochemistry 37:4288-4298. [DOI] [PubMed] [Google Scholar]
- 16.Iwahori, A., Y. Hirota, R. Sampe, S. Miyano, and N. Numao. 1997. Synthesis of reversed magainin 2 analogs enhanced antibacterial activity. Biol. Pharm. Bull. 20:267-270. [DOI] [PubMed] [Google Scholar]
- 17.Jing, W., A. R. Demcoe, and H. J. Vogel. 2003. Conformation of a bactericidal domain of puroindoline a: structure and mechanism of action of a 13-residue antimicrobial peptide. J. Bacteriol. 185:4938-4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jing, W., H. N. Hunter, J. Hagel, and H. J. Vogel. 2003. The structure of the antimicrobial peptide Ac-RRWWRF-NH2 bound to micelles and its interactions with phospholipid bilayers. J. Pept. Res. 61:219-229. [DOI] [PubMed] [Google Scholar]
- 19.Killian, J. A., and G. von Heijne. 2000. How proteins adapt to a membrane-water interface. Trends Biochem. Sci. 25:429-434. [DOI] [PubMed] [Google Scholar]
- 20.King, D. S., C. G. Fields, and G. B. Fields. 1990. A cleavage method which minimizes side reactions following Fmoc solid phase peptide synthesis. Int. J. Pept. Protein Res. 36:255-266. [DOI] [PubMed] [Google Scholar]
- 21.Koradi, R., M. Billeter, and K. Wuthrich. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14:51-55. [DOI] [PubMed] [Google Scholar]
- 22.Lakowicz, J. R. 1999. Principles of fluorescence spectroscopy. Plenum Press, New York, N.Y.
- 23.Laskowski, R. A., J. A. Rullmannn, M. W. MacArthur, R. Kaptein, and J. M. Thornton. 1996. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8:477-486. [DOI] [PubMed] [Google Scholar]
- 24.Lin, I. J., Y. C. Lou, M. T. Pai, H. N. Wu, and J. W. Cheng. 1999. Solution structure and RNA binding activity of the N-terminal leucine repeat region of hepatitis delta antigen. Proteins 37:121-129. [DOI] [PubMed] [Google Scholar]
- 24a.National Committee for Clinical Laboratory Standards. 1993. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 3rd ed. Approved standard. NCCLS document M7-A3. National Committee for Clinical Laboratory Standards, Villanova, Pa.
- 25.Prenner, E. J., M. Kiricsi, M. Jelokhani-Niaraki, R. N. A. H. Lewis, R. S. Hodges, and R. N. McElhaney. 2005. Structure-activity relationships of diastereomeric lysine ring size analogs of the antimicrobial peptide gramicidin S. J. Biol. Chem. 280:2002-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rozek, A., C. L. Friedrich, and R. E. W. Hancock. 2000. Structure of the bovine antimicrobial peptide indolicidin bound to dodecylphosphocholine and sodium dodecyl sulfate micelles. Biochemistry 39:15765-15774. [PubMed] [Google Scholar]
- 27.Schibli, D. J., R. F. Epand, H. J. Vogel, and R. M. Epand. 2002. Tryptophan-rich antimicrobial peptides: comparative properties and membrane interactions. Biochem. Cell Biol. 80:667-677. [DOI] [PubMed] [Google Scholar]
- 28.Schibli, D. J., P. M. Hwang, and H. J. Vogel. 1999. The structure of the antimicrobial active center of lactoferricin B bound to sodium dodecyl sulfate micelles. FEBS Lett. 446:213-217. [DOI] [PubMed] [Google Scholar]
- 29.Schibli, D. J., P. M. Hwang, and H. J. Vogel. 1999. Structure of the antimicrobial peptide tritrpticin bound to micelles: a distinct membrane-bound peptide fold. Biochemistry 38:16749-16755. [DOI] [PubMed] [Google Scholar]
- 30.Tinoco, L. W., A. da Silva, Jr., A. Leite, A. P. Valente, and F. C. L. Almeida. 2002. NMR structure of PW2 bound to SDS micelles. J. Biol. Chem. 277:36351-36356. [DOI] [PubMed] [Google Scholar]
- 31.Tossi, A., L. Sandri, and A. Giangaspero. 2000. Amphipathic, a-helical antimicrobial peptides. Biopolymers 55:4-30. [DOI] [PubMed] [Google Scholar]
- 32.Wieprecht, T., O. Apostolov, M. Beyermann, and J. Seelig. 2000. Membrane binding and pore formation of the antibacterial peptide PGLa: thermodynamic and mechanistic aspects. Biochemistry 39:442-452. [DOI] [PubMed] [Google Scholar]
- 33.Wiseman, T., S. Williston, J. F. Brandts, and L. N. Lin. 1989. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 179:131-137. [DOI] [PubMed] [Google Scholar]
- 34.Woody, R. W. 1994. Contributions of tryptophan side chains to the far-ultraviolet circular dichroism of proteins. Eur. Biophys. J. 23:253-262. [DOI] [PubMed] [Google Scholar]
- 35.Wuthrich, K. 1986. NMR of proteins and nucleic acids. John Wiley & Sons, New York, N.Y.
- 36.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389-395. [DOI] [PubMed] [Google Scholar]