Abstract
We have determined the genomic sequences of four virulent myophages, Bcep1, Bcep43, BcepB1A, and Bcep781, whose hosts are soil isolates of the Burkholderia cepacia complex. Despite temporal and spatial separations between initial isolations, three of the phages (Bcep1, Bcep43, and Bcep781, designated the Bcep781 group) exhibit 87% to 99% sequence identity to one another and most coding region differences are due to synonymous nucleotide substitutions, a hallmark of neutral genetic drift. Phage BcepB1A has a very different genome organization but is clearly a mosaic with respect to many of the genes of the Bcep781 group, as is a defective prophage element in Photorhabdus luminescens. Functions were assigned to 27 out of 71 predicted genes of Bcep1 despite extreme sequence divergence. Using a lambda repressor fusion technique, 10 Bcep781-encoded proteins were identified for their ability to support homotypic interactions. While head and tail morphogenesis genes have retained canonical gene order despite extreme sequence divergence, genes involved in DNA metabolism and host lysis are not organized as in other phages. This unusual genome arrangement may contribute to the ability of the Bcep781-like phages to maintain a unified genomic type. However, the Bcep781 group phages can also engage in lateral gene transfer events with otherwise unrelated phages, a process that contributes to the broader-scale genomic mosaicism prevalent among the tailed phages.
Phages typically exhibit a mosaic relationship to other phages, as both vertical and horizontal gene transfers play significant roles in phage evolution. Mosaic genomes share a modular relationship with regions of obvious homology interspersed among regions that are unrelated. Mosaicism was first described for the lambdoid phages that share common genome organization and size (3). Mosaicism is also detected among members of all three common morphotypes of double-stranded DNA phages: myophage (contractile tail), siphophage (flexible, noncontractile tail), and podophage (short tail). Moreover, mosaicism is found across phages of differing genome sizes, across phages with distinctly different genome organizations, and between phages that utilize distinctly different packaging and replication mechanisms (26). This has led to an evolutionary model in which phages participate in rampant lateral gene transfer and in which recombination is limited by physical access rather than degree of homology (26). One striking example of the phage capacity for mosaicism across what were initially considered to be distinct phage types was reported for phage D3112 (54). This phage resembles the myophage Mu in that it replicates its 40-kb genome by transposition, and yet its tail genes are related to the classic siphophage lambda. Mosaicism is not uniform across phages. Consequently, methods such as whole genome comparison generate phage groups comprised of members that share an overall high percentage of related genes (46). While members of a phage group might share more genes in common than they do with those of another phage group, the number and identity of the shared genes are generally not predictable. Members of any group can be more closely related to members of another group in terms of any aspect of phage growth cycle, including DNA replication, DNA packaging, and virion morphology, with the placement of podophages APSE-1 and P22 in the lambdoid siphophage group being obvious examples.
If phage mosaicism is limited mostly by physical access, then phages of hosts that occupy widely disparate ecological niches might be in a unique position to undergo mosaic exchange. The Burkholderia cepacia complex (Bcc) consists of heterogeneous members of the beta-Proteobacteria. Bcc members include opportunistic human pathogens, like B. cenocepacia, which account for the majority of infection for persons with cystic fibrosis, and phytopathogens, particularly B. cepacia, the causative agent of onion “sour skin” (5). Members of Bcc can also be recovered from the soil (22), water samples (52), and the rhizosphere of crop plants (12). Bcc isolates are not necessarily specific for one niche. For example, isolates of the B. cenocepacia electrophoretic type PHDC, a significant cause of cystic fibrosis Bcc infections, have been recovered from agricultural soils (34). We have found these soils to be a rich source of Bcc phages as well (C. F. Gonzalez, G. L. Mark, E. Mahenthiralingam, and J. J. LiPuma, Isolation of soilborne genomovar I, III and VII Burkholderia cepacia and lytic phages with intergenomovar host range, Int. B. cepacia Working Group 6th Annu. Meet., p. 115-117, 2001). In contrast with the distribution of phage morphotypes in the literature, our isolates are heavily biased towards myophages. Here, we describe the genomic organization of three Bcc-specific phages (Bcep phages) isolated from soil samples at disparate locations and times. The organization and relationships of these Bcep phages are discussed in respect to current models of phage genome evolution.
MATERIALS AND METHODS
Bacterial growth conditions and media.
Media and conditions for growth of Escherichia coli and Burkholderia spp. strains have been described previously (49). E. coli and Burkholderia spp. were grown at 37°C and 28°C, respectively.
Phage isolation and imaging.
For enrichment, 2 g soil was incubated for 30 min at room temperature with shaking in 20 ml of 0.1% peptone broth. After settling, the top 10 ml was removed, clarified twice by low-speed centrifugation, and filtered through a 0.2-μm filter to generate a cell-free phage suspension. Phages were isolated by inoculation of a 25-ml logarithmic (A550 of ∼0.4) Bcc culture with 1 ml of phage suspension and incubation for 20 m at room temperature without shaking, followed by overnight incubation with shaking (200 rpm) at 28°C. The culture was cleared of cells by centrifugation and filtration as above, generating a phage lysate. The titers of phage lysates were then determined with the host used for enrichment, and individual plaques were isolated. Pure phage stocks were obtained by amplification from a single plaque, followed by reisolation from a single plaque and reamplification. Preparation of high-titer lysates, determination of Bcep phage titers, and imaging by transmission electron microscopy were done as described previously (49).
Phage Bcep781 was isolated by S. Beer (Cornell) in 1978 from Orange County, NY, muck soils as a plaque former on Bcc strain 74-34, an onion pathogen provided by J. Lorbeer (Cornell) (20). Phage Bcep43 and its original host, Bcc43, were isolated from muck soil of Orange County, NY, obtained in 1999. Phage Bcep1 and its original host Bcc strain, HI2424 (34), were isolated from Oswego County, NY, soils in 1999. Phage BcepB1A and its original host, S198B1A, were isolated from soils obtained at a different site in Oswego County, NY, in 2000.
Phage infection parameters.
The eclipse period and burst size for bacteriophage Bcep781 were determined by conducting a one-step growth experiment, as described previously (49). The kinetics of phage adsorption was determined by infecting a logarithmic B. cepacia 74-34 strain, in the presence or absence of 0.01 M MgSO4 or 0.01 M CaCl2, at a multiplicity of infection of ∼10−3. Samples were taken at 5-min intervals, and titers were determined after removal of cells by filtration through a 0.2-μm filter (Nalgene). The rate of phage particle disappearance is defined as dP/dt = −kBP, where B is the concentration of bacteria, P is the concentration of free phage at any time (t), and k is the adsorption constant in ml cell−1 s−1 (48).
Genomic analysis.
Library preparation, shotgun sequencing, sequence assembly, and analysis were done essentially as described previously (49). The program Sequencher (Gene Codes Corporation) was used for sequence assembly from contigs. Areas of low-quality sequence were resequenced using primer walking. Protein coding regions were predicted initially using GeneMark.hmm (http://opal.biology.gatech.edu/GeneMark/) (2). Predicted coding regions were refined with Artemis (http://www.sanger.ac.uk/Software/Artemis/) (47). The predicted proteins were then compared to the NCBI protein database with BLASTP at the mirror site located at XBLAST (http://xBLAST.tamu.edu/pise/). Structural features (transmembrane helices and predicted molecular weights) of the proteins were determined with proteomic tools at ExPASy (http://us.expasy.org) (16). tRNAs were identified with the tRNAscan-SE search server (http://www.genetics.wustl.edu/eddy/tRNAscan-SE/). DNA pairwise comparisons were performed with the program Base by Base at the SARS Bioinformatics Suite (http://athena.bioc.uvic.ca/sars/index.php?page = tools). Phage genome maps were drawn utilizing the program DNA Master (http://cobamide2.bio.pitt.edu/computer.htm).
IST library construction and analysis.
An interactive sequence tag (IST) library of phage Bcep781 was produced and analyzed as previously described for BcepMu (49). Protocols can be found at http://oligomers.tamu.edu/doodle (41).
Bcep781 genome end cloning.
To clone the Bcep781 genomic end fragments, phosphorylated XbaI linkers (New England Biolabs) were ligated onto Bcep781 genomic DNA. The genomic DNA/linker ligation reaction product was digested to completion with XbaI and XhoI and ligated into XbaI/XhoI-digested pBluescript II SK (Stratagene). Transformants were picked at random and grown overnight in deep-well plates, with shaking (270 rpm). Plasmid DNA was isolated as described above and sequenced with the T3 primer (Stratagene). The positions of 47 independent end clones were determined with Sequencher (Gene Codes).
Amino-terminal sequencing of Bcep781 structural proteins.
Phage lysate proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrotransferred onto a polyvinylidene difluoride membrane. After the membrane was stained with Coomassie blue, the two predominant bands of 17 kDa and 33 kDa were excised from the blot and subjected to automated protein sequencing in the Texas A&M University Protein Chemistry Laboratory.
Cloning and assay of the Bcep43 endolysin.
To test for endolysin function, Bcep43 gene 27 was cloned into the expression vector pGemT-easy (Promega). (Bcep43 gp27 is identical in predicted amino acid sequence with gp27 of Bcep781 [see Table S2 in the supplemental material]). First, the coding sequence of gene 27 was amplified using Pfu polymerase and primers endo1 (ATAGGATCCCAGGAGGCCTGTAACATGGC) and 2endo2 (TCGGGCATTGTGTCAAGCTT). Following the manufacturer's guidelines, the resulting product was A-tailed with Taq polymerase, ligated into pGemT-easy, and transformed into E. coli JM109 cells. An insert with the correct orientation with respect to the T7 promoter in pGemT-easy was identified, designated pGemT-27, and transformed into XL1-Blue electrocompetent cells (Stratagene). For the cell lysis assay, overnight cultures containing pGemT-easy or pGemT-27 were diluted 250-fold into LB-ampicillin (ampicillin, 100 μg/ml) and aerated at 37°C. The cultures were induced at an A550 of ∼0.2 to 0.3 with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and the culture density was monitored at 10-min intervals for 1 h. CHCl3 was then added to 1% final concentration, and the A550 was determined at 5 and 10 min after addition.
Nucleotide sequence accession numbers.
The sequences of Bcep781, Bcep43, Bcep1, and BcepB1A have been entered as GenBank accession numbers AF543311 AY368235, AY369265 and NC_005886, respectively.
RESULTS
Phage isolation, morphology, and host range.
We have isolated four new Bcep phages, designated Bcep781, Bcep43, BcepB1A, and Bcep1. Phage Bcep781 was first isolated in 1978 by using a soil enrichment procedure with the onion pathogen Burkholderia cepacia strain 74-34NE. Although the original lysate had desiccated, it was found to still contain viable phage following hydration. BcepB1A, Bcep43, and Bcep1 were isolated using the same enrichment technique from soils obtained in 1999 and 2000.
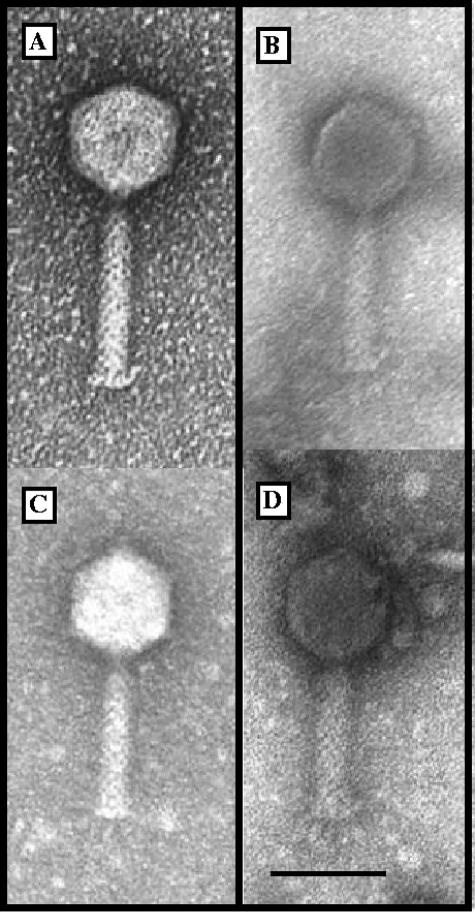
All four phages were found to have similar DNA sizes, of about 48 kb, based on pulsed-field gel electrophoresis (PFGE) (not shown). Transmission electron microscopy images revealed that all four phages had myophage morphologies, with collars, short appendages extruding from the baseplate, and isometric capsids decorated with knobs at the icosahedral vertices (Fig. 1). Bcep43 and Bcep781 plate efficiently on B. cepacia strains 74-34 and Bcc43, whereas phages Bcep1 and BcepB1A were restricted to the single isolates of B. cenocepacia Bcc1 and s198B1A, respectively. All four phages formed clear plaques on all susceptible hosts, suggesting that they were virulent; bioinformatics analysis of their genomes did not reveal any genes involved in lysogenization (see below).
FIG. 1.
Negatively stained transmission electron micrographs showing morphologies of phages of the Bcep781 group. (A) BcepB1A, (B) Bcep43, (C) Bcep1, and (D) Bcep781. Bar, 50 nm.
Physiological parameters of phage Bcep781 infection cycle.
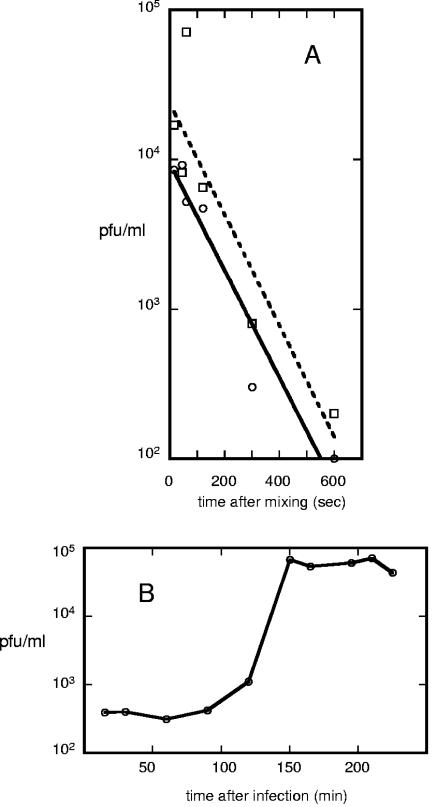
Bcep781 was chosen for a more detailed analysis of phage growth parameters. The adsorption of Bcep781 was characterized by measuring the disappearance of free phage after mixture with the susceptible host cells (Fig. 2A); the apparent adsorption rate constant was k = 1.6 × 10−11 ml cell−1 s−1 for the B. cepacia 74-34 cells. For comparison, this represents about 10-fold faster than for the adsorption of λPaPa, the commonly used variant of phage λ to E. coli K-12 (24). Addition of 0.01 M MgSO4 or 0.01 M CaCl2 had no effect on the adsorption rate. At 28°C, the average Bcep781 burst size was found to be 180 PFU/cell, with a latent period of ∼150 min (Fig. 2B).
FIG. 2.
Bcep781 adsorption rate and one-step growth curve. (A) Absorption of Bcep781 to B. cepacia 74-34NE in the presence (circles) or absence (squares) of 10 mM MgSO4. (B) One-step growth of Bcep781 in B. cepacia 74-34NE at 28°C.
Structures of the genomes of the virulent Bcep phages.
A combination of random shotgun sequencing and primer walking was used to determine the genomic sequences of these four phages. The ends of the original contig assemblages of all four were found to be overlapping, i.e., the sequences formed circular maps. When the duplicated end sequence was removed, the final genome assemblage resulted in unique coding region lengths of 48,247 bp (Bcep781), 48,177 bp (Bcep1) 48,024 bp (Bcep43), and 47,399 bp (BcepB1A).
The degree of circular permutation of Bcep781 was analyzed in more detail. Restriction maps of Bcep781 with several restriction enzymes with multiple recognition sites gave the pattern expected from a covalently closed circular molecule. However, the digests did not contain a submolar fragment that would indicate the presence of a pac site (57). In addition, digestion of Bcep781 genomic DNA independently with NdeI and NheI, with single cleavage sites 7.8 kb apart, resulted in a single band which resolved into a smear by PFGE (Fig. 3A). The close correlation in length as determined by PFGE (48.5 kb [Fig. 3A]) and the sequence length (48.2 kb) indicate the packaged DNA possesses little terminal redundancy. Bcep781 genomic DNA formed ladders following treatment with T4 DNA ligase (Fig. 3A), suggesting that the termini are not modified by proteins or dephosphorylated. A library enriched in phage genomic terminus clones was constructed by adding a linker with a unique restriction site (XbaI) to the end of purified genomic DNA and digesting the product with XbaI and an enzyme with multiple restriction sites (XhoI; 27 sites). The positions of 47 clones possessing XbaI and XhoI sites were mapped. The largest gap in the end clone library correlated to the largest gap in XhoI sites, which would produce fragments of a size likely to be underrepresented (Fig. 3B). The positions of the end clones were uniformly scattered throughout the length of the genome, which, along with the restriction digest analysis, indicates that Bcep781 has a highly circularly permuted genome similar to that of the classic coliphage T2 (50). These results are inconsistent with a packaging mechanism involving initial cleavage at or near a pac site, followed by subsequent rounds of headful packaging. However, the findings do not discriminate between random initiation, initiation at multiple pac sites, or terminase recognition of pac followed by movement and cleavage at distant sites (7, 33).
FIG. 3.
Bcep781 genome is highly circularly permuted. (A) PFGE of Bcep781 genomic DNA incubated with (A) NdeI, (B) NheI, (C) T4 DNA ligase, or (D) no pretreatment and visualized by ethidium bromide staining. Shown also are the relative positions of the NdeI and NheI restriction sites on the linearized Bcep781 map. (B) Bcep781 genomic DNA end cloning. A diagram of the end cloning strategy is shown. XbaI linkers (boxes) were ligated to Bcep781 genomic DNA. This was then digested to completion with XbaI and XhoI, which has 27 sites in the Bcep781 genomic DNA. The resulting end fragments were ligated into the XbaI/XhoI-digested vector and transformed into XL1-Blue cells. Below this are the relative positions of 47 independent end fragments, determined by sequencing randomly picked transformants and then aligning these fragments with the Bcep781 genomic sequence (bottom). Each horizontal dash represents an independent clone. Positions of the 27 XhoI sites in the Bcep781 genome are indicated with vertical lines. Alignment was made in Sequencher and converted to a line drawing.
Overview of the genomes.
The three phages Bcep781, Bcep43, and Bcep1 were found to be closely related and were designated the Bcep781 group. A combination of bioinformatics and experimental results suggests that Bcep781, Bcep43, and Bcep1 have 66, 65, and 71 protein-coding genes, respectively (Table 1 and Fig. 4A). The genes in these phages are arranged in a typically compact “head-to-tail” manner with little intergenic space and numerous overlapping or immediately adjoining start and stop codons. Bcep43 and Bcep781 additionally possess a tRNALeu and a tRNASer, respectively, while Bcep1 does not encode any tRNAs. BcepB1A was found to encode 72 proteins and exhibited a mosaic relationship, with 12 genes of detectable homology, with Bcep781 group genes (Fig. 5). In the Bcep781 group, the genes were found to be organized into four major transcription blocks, as judged by contiguous placement on one strand or the other (Fig. 4A). Phages Bcep781, Bcep43, and Bcep1 were found to be 63% GC. Although the GC contents of these phage host strains are not known, sequenced representatives of the Bcc, including B. cenocepacia strain J2315 and B. cepacia ATCC 17760, were found to be 66.9% (http://www.sanger.ac.uk/Projects/B_cenocepacia/) and 66.3% (http://genome.ornl.gov/microbial/bcep_18194) GC, respectively. In comparison, BcepB1A has an overall GC content of 53%. In the Bcep781 group, the GC content is about 30% in two regions that are devoid of genes. These regions have several predicted promoter sequences on both forward and reverse strands (not shown) and, since divergent transcriptional units emerge from both regions, presumably serve as transcriptional control regions (Fig. 4A). An exception to this simple transcriptional organization is a single open reading frame (orf16 in Bcep1) embedded within a cluster of head morphogenesis genes but on the opposite strand. This element has homology to a bacterial DNA polymerase subunit (see below), but it is highly degenerate in Bcep1. In Bcep781 and Bcep43, this reading frame possesses a legitimate Shine-Dalgarno sequence and is followed immediately by a strong rho-independent terminator (not shown). BcepB1A has an even simpler predicted transcriptional organization, which would require only one set of divergent promoters in a low-GC region between genes 65 and 66. However, there is a single opposite-strand gene, 37, within a region of small, novel genes that would be transcribed leftward (Fig. 5). In this case, the gene has no relatives in the database but it does have a consensus Shine-Dalgarno and is followed by a strong rho-independent terminator. These genes may be the equivalent of morons, which are typically found in temperate phages and are thought to provide selective advantage for the lysogenic host and thus help preserve the prophage sequences from systematic deletion (27).
TABLE 1.
Coding regions of Bcep781, Bcep43, and Bcep1d
| Gene(s)a,b | F/Rc | Size (kDa)b | Function | Organism, E value, gene code, and proteine |
|---|---|---|---|---|
| 781gp01, 43gp01, 1gp01 | R | 30.4 | Hypnovl | — |
| 781gp02, 43gp02, 1gp02 | R | 7.4 | Hypnovl | — |
| 781gp03, 43gp03, 1gp03 | R | 15.9 | Hypcons. | P. luminescens 2e-13, gi37527268, hyp. prt |
| 781gp04, 43gp04, 1gp04 | R | 52.3 | Phage | P. luminescens, 3e-59, gi37527269, hyp. prt; phage Aaφ23, 3e-06, gi31544040, hyp. prt; B1A gp17 |
| 781gp05, 43gp05, 1gp05 | R | 19.7 | Hypcons. | P. luminescens, 3e-12, gi37527270, hyp. prt |
| 781gp06, 43gp06, 1gp06 | R | 16.8 | Hypcons. | P. luminescens, 7e-17, gi37527271, hyp. prt |
| 781gp07, 43gp07, 1gp07 | R | 16.9 | Hypcons. | P. luminescens, 1e-4, gi37527272, hyp. prt |
| 781gp08, 43gp08, 1gp08 | R | 16.9 | Hypcons. | P. luminescens, 1e-16, gi37527273, hyp. prt |
| 781gp09, 43gp09, 1gp09 | R | 13.7 | Hypcons. | P. luminescens, 4e-12, gi37527274, hyp. prt |
| 781gp10, 43gp10, (1gp10) | R | 29.7 (26.1) | DNA methylase | C. crescentus 5e-13, gi1072869, DNA methyltransferase; phage Mx8, 2e-7, gi15320575, DNA methyltransferase; P. luminescens, 2e-5, gi37527291, hyp. prt |
| 781gp11, 43gp11, (1gp11) | R | 16.5 (16) | Hypnovl | — |
| 1gp12 | R | 20.1 | Phage | Phage RB49, 2e-10, gi33620550, hyp. prt |
| 781gp12, 43gp12, 1gp13 | R | 36.3 | Capsid | P. luminescens, 8e-82, gi37527275, hyp. prt; B1A gp23 |
| 781gp13, 43gp13, 1gp14 | R | 17.1 | Dec | P. luminescens, 1e-14, gi37527276, hyp. prt |
| 781gp14′, 43gp14′, 1gp15′ | R | 26.8 | Scaffold prt | —; IST |
| 781gp14, 43gp14, 1gp15 | R | 48.6 | Prohead protease | P. luminescens, 2e-41, gi37527277, hyp. prt; Xylella fastidiosa, 5e-11, gi28198879, hyp. prt; B1A gp26 IST |
| 781gp15, 43gp15, (1gp16) | F | 20.6 (4.7) | DNA Pol III | Yersinia pestis, 3e-9, gi16124205, DNA polymerase III, beta subunit |
| 781gp16, 43gp16, 1gp17 | R | 35.3 | Mu gp30 | P. luminescens, 2e-24, gi37527278, hyp. prt; phage Aaφ23, 3e-06, gi31544030, put. minor head prt; B1A gp27 |
| 781gp17, 43gp17, 1gp18 | R | 76.2 | Minor head protein | P. luminescens, 3e-86, gi37527279, hyp. prt; Magnetospirillum magnetotacticum; 6e-79, gi23011505, hyp. prt; X. fastidiosa, 1e-79, gi28198883, phage prt; B1A gp28 |
| 781gp18, 43gp18, 1gp19 | R | 55.5 | TerL | Phage Aaφ23, 1e-50, gi31544028, TerL |
| 781gp19, 43gp19, (1gp20) | R | 18.2 (17) | Hypnovl | —; IST |
| 781gp20, (43gp20), 1gp21 | F | 45.4 (46.1) | Phage | Bordetella phage BPP-1, 8e-16, gi41179398, Bbp38; Acyrthosiphon pisum phage APSE-1, 2e-7, gi9633598, hyp. prt; B1A gp67 |
| 781gp21, 43gp21, 1gp22 | F | 24.1 | Hypnovl | — |
| 781gp22, 43gp22, 1gp23 | F | 14.2 | Rus | Salmonella enterica serovar Typhimurium phage ST64T, 2e-8, gi24371573, Rus |
| 781gp23, 43gp23, 1gp24 | F | 10.9 | Hypnovl | — |
| 781gp24, 43gp24, 1gp25 | F | 10.9 | Rz | — |
| 781gp25, 43gp25, 1gp26 | F | 11.2 | Rz1 | — |
| 781gp26, 43gp26, (1gp27) | F | 17.2 (14.5) | Hypcons. | Mycobacterium avium, 9e-5, gi41408724, hyp. prt; IST |
| 781gp27, 43gp27, 1gp28 | F | 26.8 | Endolysin | Phage phiCTX, 3e-30, gi17313229, hyp. prt phiCTXp12 |
| 781gp28, 43gp28, (1gp29) | F | 21.2 (25) | Hypnovl | —; IST |
| 781gp30, 43gp30, 1gp31 | R | 12.1 | Holin | X. fastidiosa, 2e-6, gi28199004, cons. hyp. prt |
| 1gp32 | R | 6 | Hypnovl | — |
| 781gp31, 43gp31, (1gp33) | R | 44.6 (45.2) | P2 gpH equivalent | Bcep781 and Bcep43 homologues: S. flexneri, 2e-15, gi30063957, put. tail fiber prt; C. crescentus, 1e-7, gi16125259, S-layer prt RsaA pir. Bcep1 homology: phage GMSE-1, 7e-28, gi12276093, probable tail fiber prt; IST |
| 781gp32, 43gp32, 1gp34 | R | 24.8 | Hypcons. | S. flexneri, 6e-25, gi24113898, hyp. prt; P. luminescens, 6e-23, gi37527257, hyp. prt; B1A gp7 |
| 781gp33, 43gp33, 1gp35 | R | 43.5 | P2 gpW | P. luminescens, le-47, gi37527258, hyp. prt; IST B1A gp8 |
| 781gp34, 43gp34, 1gp36 | R | 6.6 | Hypnovl | — |
| 781gp35, 43gp35, 1gp37 | R | 6.9 | Hypnovl | — |
| 781gp36, 43gp36, 1gp38 | R | 25.7 | P2 gpV | P. luminescens, 4e-17, gi37527262, hyp. prt; B1A gp10 |
| 781gp37, 43gp37, 1gp39 | R | 6.9 | Hypnovl | — |
| 781gp38, 43gp38, 1gp40 | R | 33.4 | Hypcons. | P. luminescens, 1e-9, gi37527263, hyp. prt; B1A gp11 |
| 781gp39, | R | 11.8 | Hypnovl | — |
| 781gp40, 43gp39, 1gp41 | R | 18.8 | Hypnovl | — |
| 781gp41, 43gp40, 1gp42 | R | 5.6 | Hypnovl | — |
| 781gp42, 43gp41, 1gp43 | R | 7.8 | Hypnovl | — |
| 781gp43, [43gp42], (1gp44) | R | 7 [6.5] (9) | Hypnovl | — |
| 781gp44, 43gp43, 1gp45 | R | 64.8 | P2 gpT | P. luminescens, 6e-5, gi37527266, hyp. prt; B1A gp14 |
| 781gp45′, 43gp44′, (1gp46′) | R | 16 (13.6) | P2 E′ | — |
| 781gp45, 43gp44, 1gp46 | R | 21.4 | P2 E | P. luminescens, 4e-4, gi37527265, hyp. prt |
| 781gp46, 43gp45, 1gp47 | R | 14.6 | Hypnovl | —; IST |
| 781gp47, 43gp46, 1gp48 | R | 18.5 | Hypnovl | — |
| 1gp49 | R | 7.2 | Hypnovl | — |
| 781gp48, 43gp47, 1gp50 | R | 7.8 | Hypnovl | — |
| 781gp49, 43gp48, (1gp51) | R | 69.7 (51.60) | Tail spike | Bcep781, Bcep43: A. pisum, 4e-6, gi6752871, RTX prt; Sus scrofa; 2e-6, gi7460236, submaxillary mucin. Bcep1: Neurospora crassa, 2e-10, gi38567068, related to glucan 1,4-alpha-glucosidase |
| 781gp50, 43gp49, 1gp52 | R | 13.7 | Hypcons. | P. luminescens, 3e-8, gi37527264, hyp. prt |
| 781gp51, 43gp50, 1gp53 | R | 13.2 | Hypcons. | P. luminescens, 7e-12, gi37527259, hyp. prt |
| 781gp52, 43gp51, (1gp54) | R | 95.6 (90) | VirE | Phage APSE-1, 7e-18, gi9633552, hyp. prt |
| 781gp53, 43gp52, 1gp55 | F | 50.4 | Hypnovl | — |
| 781gp54, 43gp53, 1gp56 | F | 11.7 | Hypnovl | — |
| 781gp55, 43gp54, 1gp57 | F | 7.1 | Phage | Phage Bcep22, 3e-16, gi38640340, hyp. prt Bcep22p33; IST |
| 781gp56, 43gp55, 1gp58 | F | 5.8 | Hypnovl | — |
| 781gp57, 43gp56, 1gp59 | F | 7.4 | Hypnovl | — |
| 781gp58, 43gp57, 1gp60 | F | 69.2 | Uvs helicase | Phage Aeh1, 4e-11, gi38640174, UvsW; coliphage T4, 2e-8, gi9632837, UvsW; B1A gp65 |
| 781gp59, 43gp58, (1gp61) | F | 40 (14.5) | Hypnovl | —; IST B1A gp63 |
| 781gp60, 43gp59, 1gp62 | F | 8.2 | Hypnovl | — |
| 1gp63 | F | 7.5 | Txn factor | Bacillus cereus ATCC 14579, 3e-5, gi30019992, transcriptional regulator; B1A gp42 |
| 1gp64 | F | 7.9 | Hypnovl | — |
| 1gp65 | F | 7.2 | Hypnovl | — |
| 781gp61, 43gp60 | F | 8.5 | Hypnovl | — |
| 781gp62, 43gp61, 1gp66 | F | 72.5 | DNA Pol I | Phage VP16C, 9e-47, gi37626195, put. DNA polymerase; phage APSE-1, 2e-32, gi9633592, P45 |
| 781gp63, 43gp62, 1gp67 | F | 18.5 | Phage | Vibriophage VP2, 2e-10, gi40950045, hyp. prt |
| 781gp64, 43gp63, 1gp68 | F | 11.6 | Hypnovl | — |
| 781gp65, (43gp64), 1gp69 | F | 27.3 (28.7) | Hyp. | Many weak metazoan hits |
| (781gp66), 43gp65, 1gp70 | F | (11) 12.6 | Hypnovl | — |
| 1gp71 | F | 6.1 | Lar | Rac prophage, 2.4, gi16129309, modifies activity of EcoKI |
Gene designations are preceded by number indicating phage designation (i.e., 781 for Bcep781).
Gene designations and values in parentheses indicate phage homologues whose masses differ by more then 1 kDa.
F/R, coding region on forward or reverse strand, respectively.
prt, protein; cons., conserved; hyp., hypothetical; hypnovl, hypothetical novel protein; phage, conserved phage protein; hypcons., hypothetical conserved proteins; put., putative; txn, transcription.
—, no hits. Mycobacterium avium, M. avium subsp. paratuberculosis strain k10. P. luminescens, P. luminescens subsp. laumondii TTO. B. cenocepacia, B. cenocepacia phage BcepB1A.
FIG. 4.
(A) Alignment of Bcep781, Bcep43, and Bcep1 genomic maps showing amino acid similarities. The center line of each map is a DNA ruler with scaled representations of open reading frames (ORFs) in the forward direction drawn on top of the map and in the reverse direction indicated on the bottom of the map. Protein names are from phage P2 (H, W, V, T, and E’E), λ (Rz and Rz1), and E. coli (Rus and Lar). ORF boxes are color coded to indicate the degree of amino acid similarity with proteins in the public database as follows: blue, unique Bcep phage protein; yellow, conserved hypothetical protein; orange, homology to protein of known function; white, no homologue in database. Shading between maps corresponds to percent amino acid identity between these phages as follows: blue, greater than 90% identity; green, 50 to 89% identity; pink, less than 50% identity; white, protein is not present. Lavender boxes on the ruler correspond to 30% DNA GC content. The bottom ruler indicates nucleotide position in kb. Maps were created with DNAMaster. (B) Whole-genome nucleotide alignments of Bcep781, Bcep43, and Bcep1. Pairwise comparisons were performed with the program Base by Base. Shown are the alignments generated by comparing 1 (Bcep1 to Bcep781), 43 (Bcep43 to Bcep781), and 781 (Bcep781 to Bcep43). The label refers to the sequence used as the base sequence for the pairwise comparison; in this type of alignment, if two sequences have insertions or deletions relative to one another, the output looks different depending on which of the two sequences is used as the base sequence. White, perfect nucleotide homology; blue, SNP; red, deletions in the indicated phage; green, insertions in the indicated phage.
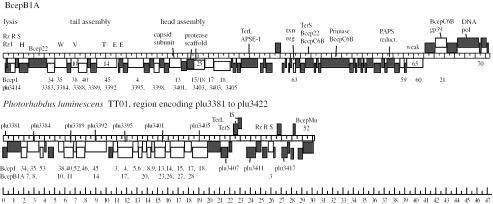
FIG. 5.
Mosaic relatives of Bcep781-like phages. Genomic maps of phages BcepB1A and a defective prophage in the P. luminescens TT01 genome (from plu3381 to plu3422). Maps were created with DNAMaster. The center line of each map is a ruler with scaled representations of open reading frames in the forward direction drawn on top of the map and in the reverse direction indicated on the bottom of the map. The functions of representative genes are indicated. Genes are color coded to indicate homology within this group of elements: white indicates homologues among these elements, and the corresponding gene(s) is indicated below the element; gray indicates genes without homologues among these elements. txn reg, transcription regulator; PAPS reduct., phosphoadenosine phosphosulfate reductase; IS, insertion sequence.
Functional assignments for the genes of the Bcep781 phage group.
In the following, except where significant differences exist among the three phages, only the relationship of Bcep781 proteins with database homologues will be treated in detail. The majority of Bcep781-encoded proteins lacked detectable homologues outside of this group of phages, BcepB1A, and the related Photorhabdus luminescens prophage (described below). Only five Bcep781 predicted proteins could be given a robust functional annotation based solely on primary sequence similarity (Table 1 and below). Protein sequencing was used to identify high-copy components of purified virions (see below), whereas functional analysis was used to identify the endolysin. A combination of gene position, size, and more sensitive PSI-BLAST analysis proved to be useful in annotation of the tail and head assembly cassettes. The classes of embedded genes found in many phages (the tail assembly frameshift gene [58], the protease-head scaffolding gene [9], and the Rz and Rz1 gene pair [62]) were annotated based on their unique gene architecture and the secretory signals of the encoded proteins. Finally, analysis of clones from an IST library provided experimental support for the existence of 10 Bcep781 genes encoding proteins capable of homotypic interactions, 6 of which were not assigned a functional annotation. This combination of bioinformatics and experimental results ultimately allowed for some degree of functional annotation of 25 Bcep781 genes, despite the great phylogenetic distance between these phages and more-characterized phages.
(i) DNA metabolism.
Most of the Bcep781 genes encoding proteins with robust functional homologues were involved in DNA metabolism. These included a DNA methyltransferase (gp10), a Holliday junction resolvase (gp22), a helicase (gp60), and a DNA polymerase (Pol) I homologue (gp66). Phage genomes typically show clustering of related genes. Replication and recombination genes are generally encoded in early transcriptional units, whereas morphogenesis and lysis genes are expressed in late transcripts. In terms of functional groupings, five Bcep781 genes showed similarity to or had motifs found in genes involved with DNA metabolism. Three of these genes are located within the two top-strand transcriptional blocks; no strong prediction of any morphogenesis genes is found in these blocks (but see “Lysis” below), suggesting that they are early transcriptional units and that the low GC regions have early, rightward promoters.
Bcep781 gene 22 encodes a homologue of RusA, a Holliday junction resolvase found in coliphage 82 and other lambdoid phages (37). Holliday junction resolvases are endonucleases that process the intermediate structure formed during homologous recombination events. The analogous but unrelated T4 gene product is gp49, which is responsible for cleaving branches prior to DNA packaging (13) and is functionally part of the packaging machinery (19). Bcep781 gene 58 encodes a homologue of the phage T4 DNA helicase, UvsW (Dar protein) (6). Like UvsW, Bcep781 gp58 contains imperfect ATP/GTP binding site and DEAH box ATP-dependent helicase signature motifs. Bcep781 gp62 shows significant homology to Bacillus subtilis phage SPO2 DNA-directed DNA polymerase gpL (44), and similar genes are found in phage APSE-1 (gene 45) and in numerous putative prophages (51). These phage and prophage DNA Pol I homologues are only weakly similar to authentic bacterial DNA Pol I subunits, primarily limited to the region around the DNA polymerase A signature domain.
The final two DNA metabolism gene assignments are more problematic. In the first transcriptional block, Bcep781 gene 10 encodes a weak homologue of Caulobacter crescentus and Agrobacterium tumefaciens cell cycle-regulated DNA adenine methyltransferase, CcrM, which is involved in methylation of DNA to effect recruitment of the replication complex (28). Significant homologues of CcrM are found in numerous bacteria and in archaea. There are distant homologues in phage genomes, including the mox gene of the myxococcal phage Mx8, which has been shown to encode DNA adenine methyltransferase activity but which had no nonsense phenotype in either lytic or lysogenic growth (36), leaving the role of this gene in phage DNA metabolism unknown. Although the Bcep1 gene 10 is also a homologue of CcrM, it is located in a cluster of three genes with no DNA sequence similarity with Bcep781 and is thus likely to have been acquired laterally.
As noted above, Bcep1 gene 16 encodes a highly truncated homologue, only 42 residues, of the bacterial DNA polymerase III β-clamp subunit. The Bcep781 and Bcep43 homologues are longer, at 193 residues, but still significantly smaller than the typical length (>300 residues) of authentic bacterial homologues. β-Clamp subunit homologues are not typically encoded by non-T4-like phages. In this phage group, Bcep781 gp15 appears to be a moron as it is located on the opposite strand within the head assembly gene cluster (Fig. 4). Pseudogenes are usually not detected in phage (31). Gene 43 (Bcep1 44, Bcep43 42), immediately downstream of the putative tape measure protein gene, is different in all three phages. Its size (60 to 88 residues) and the fact that it maintains its upstream gap with respect to the tape measure gene (67 bp in Bcep1 and Bcep43 and 68 bp in Bcep781) and the overlap of its stop codon with the downstream gene suggest that the three variants arose by different deletion events from the same original cistron.
(ii) Morphogenesis.
Even though individual similarities are low, Bcep781 gene 31 to gene 51 are likely to encode proteins involved in tail, baseplate, and tail fiber assembly (Table 1). Out of 20 predicted genes in this region, only 6 encode proteins with homologues outside of the Bcep781 group or the related prophage of Photorhabdus. However, these show weak or indirect similarity to tail and tail fiber structural proteins. The amino-terminal third of Bcep781 gp31 and Bcep1 gp33 are related both to each other and to the amino-terminal domain of a Shigella flexneri prophage tail fiber protein. In turn, the prophage gene is related to the P2 gpH tail fiber homologue over the C-terminal part of the tail fiber protein. Moreover, the C-terminal domain of the Bcep1 protein shows significant homology with the phage GMSE-1 tail fiber, also a P2 gpH homologue. In contrast, the carboxy terminus of Bcep781 (and Bcep43) gp31 exhibits sequence relationship to bacterial S-layer proteins and vertebrate mucins. PSI-BLAST results suggest that Bcep781 gp33 is related to Mu gp47, a probable homologue of P2 W, the base plate wedge (23). PSI-BLAST analysis also suggests that Bcep781 gp36 is related to P2 gpV and Mu gp45, a baseplate assembly protein. Based on indirect homology and length, Bcep781 gp44 is a candidate for the tape measure protein. Bcep781 gp44 shows weak similarity to Nocardia cryptic prophage protein, which in turn is related to the P2 T tape measure protein (10). While these homologies are weak, the additional compelling evidence for the annotation of Bcep781 genes 31, 33, 36, and 44 as functional homologues of P2 H, W, V, and T includes the similar gene order and sizes of the tail fiber and baseplate encoding genes of P2, Mu, and the Bcep781-like myophages. Our assignment of Bcep781 gene 44 as the tape measure protein gene suggests that the gene immediately preceding it would be predicted to encode the frameshift proteins, EE′, involved in tail assembly (10). A recent study identified such frameshift proteins in 49 out of 68 phages and prophages but, in the absence of an obvious candidate for the tape measure protein, did not detect a potential G/GT (the lambda equivalent of P2 EE′) equivalent in Bcep781 (58). A manual search of the coding region of Bcep781 gene 45 identified the “slippery sequence” GGCAAAC, which serves as the −1 frameshift motif that generates the alternate C terminus in the G/GT genes of Yersinia lambda (58). However, although this motif is conserved in Bcep43, it has a single base pair change in Bcep1, to GGCGAAC (with the change underlined), which should prevent the frameshift step. Since the frameshift is essential for tail morphogenesis, either this sequence is not the frameshift-inducing element in Bcep781/43 or Bcep1 has an alternative mechanism.
The most promoter-proximal gene in the tail morphogenesis transcription unit to which a function could be assigned is gp49, a tail spike protein. Particles formed by Bcep781 phage possessing point mutations in gp49 lack tail spikes in electron microscopy images (M. D. King, unpublished data). Bcep781 gp49 and Bcep43 gp48 are 99% identical but are only 27% identical to Bcep1 gp51. Moreover, more than a third of the amino acid residues in these proteins are glycine, serine, or threonine. The compositional bias of these proteins probably accounts for the significant homology to numerous bacterial and metazoan extracellular proteins, including mucins and S-layer proteins.
Capsid assembly and the terminase large subunit genes are found in the first leftward transcription unit, genes 19 to 13. Bcep781 gene 18 encodes a homologue of the phage terminase large subunit, TerL, from phages TP901-1 and Aaφ23. Like Bcep781, these phages use headful packaging mechanisms (4). Submolar fragments were detected in digests of TP901-1 and the TP901-1 pac site mapped upstream of terS (56). When TerL subunits from 114 phages were aligned, the clustering corresponded well to the structures of the ends of the packaged DNA (8). Thus, TerL subunits generating 5′-extended cohesive ends and 3′-extended cohesive ends fell into distinct groups. The TP901-1 and Aaφ23 TerL subunits, to which Bcep781 gp18 is related, formed a separate, poorly supported, and deeply branching group distinct from other TerL homologues (8). TerL typically provides the ATPase and DNA cleavage activity for the DNA packaging pathway, while the cognate terminase small subunit, TerS, is responsible for sequence recognition. No TerS homologue was identified in Bcep781, but TerS proteins are typically less conserved than TerL. Bcep781 gene 19 is likely to encode TerS because it is immediately upstream of TerL and the size of its predicted product, ∼18 kDa, is typical of TerS homologues. However, gp19 had no significant similarity to proteins in the database outside of the Bcep781 group.
Several lines of evidence indicate that the next two genes in this cassette, Bcep781 gp16 and gp17, encode minor head proteins. Bcep781 gp16 is a member of a cluster of orthologues (39), COG2369, which also contains the minor Mu head protein, gp30, and its homologues. Moreover, gp16 and gp17 are homologues of phage Aaφ23 proteins p32 and p31, respectively (45). Aaφ23p32 possesses an amino-terminal conserved domain (pfam04233.6) which includes SPP1 G7P. SPP1 G7P is the most well studied protein among members possessing this domain and has been shown to be present in low copy number in SPP1 phage heads (1). Homologues of Bcep781 gp16 and gp17 appear to segregate together, as out of 20 database homologues of gp16 with E values of <0.5, 13 are encoded by genes immediately adjacent to homologues of gp17 (identified with E values of <0.5) (Table S1 in the supplemental material). No other gene pair in Bcep781 shows such tight linkage of homologues. This suggests a functional interaction between the two proteins. The first step in double-stranded DNA head morphogenesis is assembly of a scaffold for the capsid protein subunits. The capsid and/or scaffolding proteins are then frequently processed by a prohead protease. Using a combination of computational and manual strategies, Cheng et al. identified Bcep1 gp15 (the Bcep781 gp14 orthologue) as one of 199 head maturation proteases in phage, prophage, and herpes viruses (9). Bcep1 gp15 was one of 17 sequences, of which 16 were from prophages and only 1 was from a phage, Aaφ23, forming the orthologue cluster, COG3566, which possesses conserved catalytic His and Ser residues and predicted secondary structure, despite great overall primary sequence distance. This annotation is particularly compelling in view of the location of this gene, immediately preceding the gene encoding the Dec (decoration or head stability protein [18]; see below). In many phages, there is conserved order of genes encoding essential capsid morphogenesis domains: capsid protease, scaffold, head stability (Dec), and major capsid proteins (9). In lambda, the protease and scaffold reading frames are fused, so that the gene for the scaffold Nu3 is constituted by a secondary downstream start codon in the protease gene C (25, 55). Accordingly, inspection of Bcep781 gene 14 revealed a consensus Shine-Dalgarno sequence (GGAGA) positioned 12 bases upstream of the AUG codon 199 and thus could serve as the site of initiation for the scaffolding protein gp14′.
The two predominant bands of 33 kDa and 17 kDa observed when purified Bcep781 phage particles were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (not shown) were subjected to N-terminal sequencing. The larger species had an N-terminal sequence of AADLS, corresponding to residues 35 to 39 of gp12, indicating that gp12 is proteolytically processed after residue 34. Moreover, although there was no significant homology with any phage major capsid protein, the predicted secondary structure of gp12, determined by use of the PredictProtein software suite, is very similar to that of a canonical major capsid protein, lambda E (not shown). This is consistent with the notion that the basic folds of major capsid proteins of phages and viruses with icosahedral-symmetry capsids are similar, despite the absence of sequence homology (14). In addition, gene 12 is perfectly positioned downstream of a strong Shine-Dalgarno sequence and is flanked by two very strong GC-rich stem-loop sequences; similar structures flank the lambda E gene and presumably facilitate the efficient translation required to produce high levels of the major capsid protein, relative to other cistrons on the late mRNA. The 17-kDa protein gave an N-terminal sequence of PFQKQVY, corresponding to residues 2 to 8 of gp13, which had a predicted molecular mass of 17.1 kDa. Given that the two proteins appear to be present in equimolar ratios (data not shown), it is possible that they represent the major capsid protein and the Dec protein, respectively. It has been found that with closely related pairs of phages, such as L and P22 or T2 and T4, one member may have a decorator protein while the other does not, suggesting an accessory role under certain conditions (18). Thus, despite the extremely limited primary sequence homology, the identified Bcep781-like phage DNA packaging and head assembly cassette gene order is as follows: terS (highly putative, implied by position only), terL, a minor head protein (possibly the portal gene) homologue, a Mu gene 30 homologue, the prohead protease gene with embedded scaffolding protein gene, decorator protein, and the major capsid protein.
(iii) Lysis.
Bcep781 gene 27 encodes a homologue of Pseudomonas aeruginosa phage phiCTX gp12, which was annotated as a possible lytic endolysin based on the presence of a peptidoglycan-binding motif (42). Bcep781 gp27 lacks a recognizable peptidoglycan-binding domain and has no primary sequence homology to known endolysins. To test for cell wall-degrading activity, gene 27 was cloned into an inducible expression vector. Cells expressing gene 27 rapidly lysed after membrane permeabilization with chloroform, while cells carrying the vector did not (Fig. 6A), demonstrating that gp27 is the authentic endolysin (60).
FIG. 6.
Lysis genes of Bcep781. (A) Induction of XL1-Blue cells carrying pGemT-27 (open squares) or the vector without insert (black circles). Chloroform (chl) was added at the indicated time. (B) Predicted membrane topology and charge distribution of the Bcep781 gp30 holin. Predicted transmembrane domains are underlined, and charges are indicated above the appropriate amino acid residues. (C) Organization of the Bcep781 Rz and Rz1 embedded gene pair. The presumptive Shine-Dalgarno sequences for the two start codons are in bold. The cysteine residue predicted by LipoP to be the site of processing by the signal peptidase II is bold and underlined.
Phage endolysins typically require a holin in order to be activated or to gain physical access to the bacterial cell wall (53, 59). Holins are small, hydrophobic proteins that form lesions in the bacterial membrane at a specified time during the infection cycle (53). Holins are extremely diverse, and thus it was not surprising that no Bcep781 predicted protein shared primary sequence homology with a known holin. However, gp30 shares significant structural similarities with known holins, making it an attractive candidate for the Bcep781 holin. Inspection and TMHMM analysis of Bcep781 gp30 suggest that it is a typical class I holin, with three transmembrane domains and a predicted N-out, C-in topology (Fig. 6B) (21).
Two additional genes present in the lysis cassette of gram-negative hosts are the nested genes Rz and Rz1, originally identified for phage λ as required for host lysis in the presence of cation concentrations that stabilize the outer membrane (61, 62). Rz encodes a secretory protein with a signal peptidase I leader peptide and has been proposed to encode an endopeptidase activity. The Rz1 gene is embedded in the +1 reading frame compared to Rz and encodes a short Pro-rich lipoprotein that has been localized to the outer membrane (29). A manual search of all Bcep781 genes predicted to contain a signal peptide revealed that gene 24 not only encodes a secretory protein of approximately the same size as λ Rz but also has an embedded reading frame, designated gene 25, that is served by a strong Shine-Dalgarno sequence (GGAGA) and encodes a predicted lipoprotein (Fig. 6B). Despite the lack of sequence similarity with the equivalent genes in other phages of gram-negative bacteria, we conclude that genes 24 and 25 are the Rz and Rz1 homologues in Bcep781.
The Bcep781 phage group lysis genes have an atypical organization. Unlike typical lysis cassettes as exemplified with lambdoid phages, in Bcep781 the Rz and Rz1 genes precede the endolysin gene and are separated from it by a gene of unknown function. Moreover, the holin gene is encoded on the opposite strand at the end with the tail assembly cassette (Fig. 6B).
IST analysis.
Overall, a minimal number of Bcep781 group genes could be assigned a function based solely on primary structure homology. To reinforce the identification of genes, an IST library was constructed. These libraries are based on the ability of expressed fusion proteins consisting of the N-terminal DNA-binding domain of the λ CI repressor and sequences encoded by randomly cloned fragments from the target genome to reconstitute repressor function (40). As CI requires separate DNA-binding and dimerization domains, immunity is conferred when the fragment of target DNA is in frame with CI and encodes a stretch of amino acid residues capable of homotypic interactions. A total of 77 immunity-conferring clones were isolated and sequenced. These were found to be in frame with 10 annotated Bcep781 genes (Tables 1 and 2). gp31, the putative P2 H homologue, had the most representatives in the IST library (26 representatives, corresponding to three domains). A similar result was found with the BcepMu IST library, where the P2 H homologue, BcepMu52, was the most abundant IST isolated (49). Hypothetical novel proteins Bcep781gp28 and gp46 were also represented multiple times in the library. Interestingly, Bcep781 hypothetical novel protein gp19, which is similar in size and genome location but not sequence homology to TerS subunits, was found in the IST library, just as BcepMu TerS was found in the BcepMu IST library. The λ TerS equivalent, Nu1, has been shown to form stable dimers (38). When the IST sequences were compared to the Prosite database (17), the most significant homology to known multimerization domains found was present in IST gp26, which exhibited 92% similarity with a myc-type, “helix-loop-helix” dimerization domain structure. The remaining ISTs did not exhibit similarity of over 75% to known motifs (data not shown).
TABLE 2.
Summary of Bcep781 IST analysis
| Bcep781 gene | No. of clones | Domain | No. of domain clones | No. of amino acid residues ata:
|
|
|---|---|---|---|---|---|
| Beginning | End | ||||
| gp14 | 7 | 1 | 6 | 179 | 210 |
| 2 | 1 | 206 | 340 | ||
| gp19 | 1 | 1 | 1 | 4 | 100 |
| gp26 | 1 | 1 | 1 | 3 | 120 |
| gp28 | 25 | 1 | 1 | 1 | 84 |
| 2 | 24 | 95 | 121 | ||
| gp31 | 26 | 1 | 10 | 142 | 192 |
| 2 | 10 | 274 | 286 | ||
| 3 | 6 | 363 | 392 | ||
| gp33 | 3 | 1 | 3 | 211 | 305 |
| gp36 | 1 | 1 | 1 | 1 | 95 |
| gp46 | 11 | 1 | 11 | 7 | 101 |
| gp55 | 1 | 1 | 1 | 1 | 55 |
| gp59 | 1 | 1 | 1 | 50 | 182 |
| Total | 77 | 14 | 77 | ||
Numbers of amino acid residues cloned as ISTs.
Relationships of Bcep781, Bcep43, and Bcep1 to each other.
A remarkable feature of the Bcep781 group of phages is the degree of DNA sequence identity exhibited between them. To our knowledge, these are the most closely related sequenced genomes from phages isolated from environmental samples. Whole-genome alignments indicate the phages exhibit overall DNA sequence identities of 97.6% (Bcep781 to Bcep43), 87.4% (Bcep781 to Bcep1), and 88.7% (Bcep43 to Bcep1) (Fig. 4B). Differences were primarily due to numerous single-nucleotide polymorphisms (SNP) and (<10-bp) insertions and deletions, as well as mosaic regions of recombinatorial origin (Table 3; see also Tables S2 and S3 in the supplemental material). Large (>5-kb) segments of the sequences exhibit only SNP.
TABLE 3.
Summary of coding region nucleotide differences between Bcep781, Bcep1, and Bcep43a
| Region | No. of aligned genes | Total CDS aligned (bp) | No. of NM genes | Aligned NM CDS (bp) | No of:
|
Ns | Nns | Ks | Kns | Kns/Ks | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dif. | Sub. | Inv. | Del. | ||||||||||
| Bcep781 to Bcep43 | 63 | 44,356 | 59 | 42,710 | 603 | 525 | 50 | 28 | 379 | 117 | 2.89 | 0.3 | 0.103 |
| Bcep781 to Bcep1 | 59 | 42,031 | 52 | 34,346 | 2,892 | 2,676 | 67 | 149 | 1,755 | 595 | 2.62 | 0.3 | 0.113 |
NM, nonmosaic; CDS, coding sequence; Dif., differences; Sub., substitutions; Ins., insertions; Del, deletions.
The relationships of the phages were analyzed in more detail. The obviously mosaic coding regions (regions encoding completely unrelated proteins or highly distant proteins that lacked recognizable DNA homology) of the sequence were eliminated. There were 42,710 base pairs of Bcep781 and Bcep43 coding sequences that aligned. Out of 603 nucleotide differences, 525 were due to SNP and 78 were due to insertions and deletions. Three hundred twenty-nine of the SNPs resulted in synonymous nucleotide substitutions (Ns) that maintained the same amino acid coding sequence, while 117 were nonsynonymous nucleotide substitutions (Nns) that resulted in the incorporation of a different amino acid. There were 34,346 base pairs of coding sequence unambiguously aligned between Bcep1 and Bcep781 (and Bcep43, data not shown). There were 2,892 total differences in this alignment. The majority of the differences were due to nucleotide substitutions (2,676). Synonymous substitutions (Ns = 1,755) were more numerous than nonsynonymous substitutions (Nns = 595). Two hundred sixteen insertions and deletions punctuated the regions.
Despite the high degree of identity, the complement of encoded proteins was not the same for the three phages. In some cases, there appear to be mosaic orthologues, as in the case of Bcep1 gp10, which is closely related at a protein level but not a DNA level to Bcep781 and Bcep43 gp10. At least nine genes between Bcep781 and Bcep1 and four genes between Bcep781 and Bcep43 appear to be the result of either extensive deletions or insertions or the acquisition of a distant yet still related homologue (Tables S2 and S3 in the supplemental material). Both Bcep781 and Bcep1 encode proteins not present in the other two phages. Five of the 71 predicted proteins of Bcep1 are unique, and 3 of these (gp12, gp63, and gp71) have identifiable database homologues (Table 1).
Relationship to BcepB1A and a P. luminescens prophage element.
At a protein level, two phages that share a significant number of genes with the Bcep781-like phages were identified. One is a prophage element present in the P. luminescens TT01 genomic sequence, consisting of 19 out of 41 genes (from plu3381 to plu3422) that are largely syntenic (albeit circularly permuted) with Bcep781 (Table 1 and Fig. 5) (11). These include the Dec and major capsid proteins, which have no other homologues in the database, and part of the tail and tail fiber cassette. Otherwise, this prophage element encodes lambdoid lysis proteins, terminase subunits, and tail fiber homologues. Thus, it appears to be a mosaic consisting of the structural genes of a Bcep781-like myophage and a temperate lambdoid siphophage. The Bcep781-related prophage element is about 30 kb and is immediately adjacent to another prophage, PhotoMu (a Mu-like prophage closely related to Burkholderia phage BcepMu), that extends from gi36786729 to gi36786769 (49).
The other phage related to the Bcep781-like phages at a predicted protein level is BcepB1A. Out of 72 predicted BcepB1A-encoded proteins, 14 show significant similarity to Bcep1 proteins and 12 to proteins encoded by the P. luminescens prophage described above (Table 1 and Fig. 5). These include numerous genes that lack appreciable homologues in the database. Overall, BcepB1A has a quite distinct genome arrangement from that of Bcep781-like phages. Although it is also circularly permuted and has one major divergent promoter region, BcepB1A has most of its genes on one strand. The BcepB1A endolysin is a “true lysozyme,” a homologue of the well-studied coliphage T4 lysozyme, but, like the endolysins R21 from lambdoid phage 21 and Lyz from coliphage P1, also has the additional feature of an N-terminal SAR domain, shown to direct holin-independent protein secretion (59). Despite disparate hosts, the Photorhabdus prophage is more similar to Bcep781 than Bcep781 is to BcepB1A (Fig. 5). BcepB1A is also mosaic to a lesser extent with other Burkholderia phages Bcep22 and BcepC6B (GenBank accession numbers NC_005262 and NC_005887). Although isolated using the same enrichment technique, these are quite distinct from the Bcep781-like phages. Bcep22 and BcepC6B are podophages with genome sizes of 64 kb and 42 kb, respectively (unpublished). BcepC6B is a temperate phage, mosaic to Bordetella podophage BPP-1 (35).
DISCUSSION
There are only limited data on the genomic stability of natural phage populations. Among 14 sequenced mycobacteriophage genomes, the two phages exhibiting the highest degree of synteny are L5 and D29 (43). Phage L5 was initially isolated as an induced lysogen, while phage D29 was isolated from soil samples (15). Although identified as a virulent phage, D29 appears instead to be derived from an L5-like temperate phage, having undergone a 3.6-kb deletion in the immunity region compared to L5. Phages L5 and D29 exhibit 80% nucleotide identity over the left ends of the genomes, but numerous insertions and deletions punctuate regions of otherwise very high identity. This can result in the insertion of new genes between homologous genes. Often, these insertions preserved the adjacent, “head-to-tail” arrangement of start/stop codons even if the reading frames of a few terminal codons were affected. There were six pairs of homologous genes with identities of less than 50%, implying recombinatorial substitution (15). Within the mycobacteriophage genomes surveyed, where very high levels of mosaicism and diversity predominated, this degree of synteny was found to be the exception (43). A recent analysis of the genomic sequence of 27 Staphylococcus aureus bacteriophage also revealed extremes of relatedness, with the two most related phages, G1 and K, having 90% identity (30).
Because it is so widespread, lateral gene transfer is obviously advantageous to phage. What then is the contribution of divergence due to positive selection for random mutations relative to this mosaicism? The high degree of DNA sequence identity exhibited by Bcep781, Bcep43, and Bcep1 (87% to 99%) made it possible to generate values for synonymous and nonsynonymous nucleotide substitutions for the majority of the genes. These data can be interpreted in terms of the selective pressures on the phages. If it is assumed that 25% of random substitutions result in synonymous changes and 75% in nonsynonymous substitutions (32), then the number of synonymous substitutions per potential synonymous substitution site (Ks) and the number of nonsynonymous substitutions per nonsynonymous site (Kns) can be estimated. For Bcep781 compared to Bcep43, the values over all nonmosaic protein coding genes are Ks = 2.89 and Kns = 0.3. Thus, the Kns/Ks ratio is 0.103. For Bcep781 compared to Bcep1, the values for Kns and Ks are 2.62 and 0.3, respectively, making the Kns/Ks ratio 0.113. When Kns is less than Ks (Kns/Ks < 1), the simplest interpretation is that the selection pressure is purifying, i.e., natural selection is acting to decrease the frequency of deleterious alleles (32). These results were remarkably uniform across all genes determined to be nonmosaic (Tables S2 and S3 in the supplemental material). Because of the high sequence identity between Bcep43 and Bcep781, when values are assessed on an individual gene basis, only 22 of the 59 aligned genes possessed enough nucleotide differences to perform the analysis (Table S2 in the supplemental material). Of these genes, only one, gene 11, showed Kns/Ks to be >1. As Bcep781 and Bcep1 are more distant, more genes could be analyzed on an individual basis when Bcep781 was compared to Bcep1. Out of 52 aligned coding regions, 44 had enough nucleotide differences to perform the calculation (Table S3 in the supplemental material). Again, only one, Bcep781 gene 28 (compared to Bcep1 gene 29), showed Kns/Ks to be >1. The simplest interpretation of this observation is that for 32,150 bases of aligned Bcep1 and Bcep781 DNA sequence (corresponding to 66% of the genome), the overwhelming majority of nucleotide differences observed reflect an evolutionary path for purifying selection against, rather than positive selection for, amino acid changes.
A similar bias towards neutral genetic drift was found with a comparison of lambdoid phages Sf6 and HK620 (7). Sf6 and HK620 exhibit greater than 83% nucleotide identity over 42.9% of their genomic sequence. These regions were distributed across 20 homology regions encoding 24 proteins. Of these proteins, only one that possessed more nonsynonymous substitutions per nonsynonymous site than synonymous substitutions per synonymous site was identified.
Despite this high identity, the phages exhibit some mosaicism in relation to one another. Similarly to what was observed with Sf6 and HK620, mosaicism was not limited to the acquisition of completely unrelated sequences but also applied to the acquisition of close homologues of the same gene (7). This type of mosaicism is not obvious at a protein sequence level, and thus the degree of mosaicism among phages is probably underestimated. An example of this is Bcep1 gp10, which is 40% identical at an amino acid level to Bcep781 gp10, despite there being no DNA sequence homology. Among the Bcep781 group of phages, therefore, it appears that mosaicism is a dominant mechanism for protein sequence level changes. Given the immensity and diversity of the phage population, it is likely that optimized genes are already available for most conditions. One interpretation of these data is that for proteins under selection to change, adaptive mosaicism is more successful than selection for adaptive divergence among phages.
Supplementary Material
Acknowledgments
Support for this work was provided primarily from a grant from the National Science Foundation, MCB-0135653, to establish a research and instruction program in phage genomics for undergraduate students. T.C., L.M., A.E., A.M.K., M.B., and W.C.M. were participants in the program and conducted all of the genomic sequencing and the primary annotation in partial fulfillment of the requirements of this program. J.L. was funded by an Office of Naval Research/National Science Foundation summer undergraduate internship program. Other support for this work was derived from funding provided by the U.S. Army Medical Research and Material Command Disaster Relief and Emergency Medical Services Program (DREAMS), the Texas Agricultural Experiment Station, PHS grant 27099 to R.Y., and grant GONZALOO3GO from the Cystic Fibrosis Foundation to C.F.G. and J.J.L.
The assistance of Jim Hu and Leonardo Mariño-Ramirez in generating the IST library was essential. We thank Chris Upton, Rachel Roper, and Vasily Tcherepanov for access and help with the SARS Bioinformatics Suite programs. We are grateful for sequencing and robotics facilities provided to this program through the cooperation of John Mullet and Eun G. No (Center for Plant Genomics and Biotechnology) and Robert Klein (Southern Plains Agricultural Research Center, USDA-ARS). Electron microscope imaging was done by Cristos Savva at the Microscopy and Imaging Center of the Department of Biology at Texas A&M University.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Becker, B., N. de la Fuente, M. Gassel, D. Gunther, P. Tavares, R. Lurz, T. A. Trautner, and J. C. Alonso. 1997. Head morphogenesis genes of the Bacillus subtilis bacteriophage SPP1. J. Mol. Biol. 268:822-839. [DOI] [PubMed] [Google Scholar]
- 2.Besemer, J., and M. Borodovsky. 1999. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 27:3911-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Botstein, D. 1980. A theory of modular evolution for bacteriophages. Ann. N. Y. Acad. Sci. 354:484-490. [DOI] [PubMed] [Google Scholar]
- 4.Brondsted, L., S. Ostergaard, M. Pedersen, K. Hammer, and F. K. Vogensen. 2001. Analysis of the complete DNA sequence of the temperate bacteriophage TP901-1: evolution, structure, and genome organization of lactococcal bacteriophages. Virology 283:93-109. [DOI] [PubMed] [Google Scholar]
- 5.Burkholder, W. H. 1950. Sour skin, a bacterial rot of onion bulbs. Phytopathology 40:115-117. [Google Scholar]
- 6.Carles-Kinch, K., J. W. George, and K. N. Kreuzer. 1997. Bacteriophage T4 UvsW protein is a helicase involved in recombination, repair and the regulation of DNA replication origins. EMBO J. 16:4142-4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casjens, S., D. A. Winn-Stapley, E. B. Gilcrease, R. Morona, C. Kuhlewein, J. E. Chua, P. A. Manning, W. Inwood, and A. J. Clark. 2004. The chromosome of Shigella flexneri bacteriophage Sf6: complete nucleotide sequence, genetic mosaicism, and DNA packaging. J. Mol. Biol. 339:379-394. [DOI] [PubMed] [Google Scholar]
- 8.Casjens, S. R., E. B. Gilcrease, D. A. Winn-Stapley, P. Schicklmaier, H. Schmieger, M. L. Pedulla, M. E. Ford, J. M. Houtz, G. F. Hatfull, and R. W. Hendrix. 2005. The generalized transducing Salmonella bacteriophage ES18: complete genome sequence and DNA packaging strategy. J. Bacteriol. 187:1091-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng, H., N. Shen, J. Pei, and N. V. Grishin. 2004. Double-stranded DNA bacteriophage prohead protease is homologous to herpesvirus protease. Protein Sci. 13:2260-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christie, G. E., L. M. Temple, B. A. Bartlett, and T. S. Goodwin. 2002. Programmed translational frameshift in the bacteriophage P2 FETUD tail gene operon. J. Bacteriol. 184:6522-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duchaud, E., C. Rusniok, L. Frangeul, C. Buchrieser, A. Givaudan, S. Taourit, S. Bocs, C. Boursaux-Eude, M. Chandler, J. F. Charles, E. Dassa, R. Derose, S. Derzelle, G. Freyssinet, S. Gaudriault, C. Medigue, A. Lanois, K. Powell, P. Siguier, R. Vincent, V. Wingate, M. Zouine, P. Glaser, N. Boemare, A. Danchin, and F. Kunst. 2003. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 21:1307-1313. [DOI] [PubMed] [Google Scholar]
- 12.Fiore, A., S. Laevens, A. Bevivino, C. Dalmastri, S. Tabacchioni, P. Vandamme, and L. Chiarini. 2001. Burkholderia cepacia complex: distribution of genomovars among isolates from the maize rhizosphere in Italy. Environ. Microbiol. 3:137-143. [DOI] [PubMed] [Google Scholar]
- 13.Flemming, M., B. Deumling, and B. Kemper. 1993. Function of gene 49 of bacteriophage T4 III. Isolation of Holliday structures from very fast-sedimenting DNA. Virology 196:910-913. [DOI] [PubMed] [Google Scholar]
- 14.Fokine, A., P. G. Leiman, M. M. Shneider, B. Ahvazi, K. M. Boeshans, A. C. Steven, L. W. Black, V. V. Mesyanzhinov, and M. G. Rossmann. 2005. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc. Natl. Acad. Sci. USA 102:7163-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ford, M. E., G. J. Sarkis, A. E. Belanger, R. W. Hendrix, and G. F. Hatfull. 1998. Genome structure of mycobacteriophage D29: implications for phage evolution. J. Mol. Biol. 279:143-164. [DOI] [PubMed] [Google Scholar]
- 16.Gasteiger, E., A. Gattiker, C. Hoogland, I. Ivanyi, R. D. Appel, and A. Bairoch. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31:3784-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gattiker, A., E. Gasteiger, and A. Bairoch. 2002. ScanProsite: a reference implementation of a PROSITE scanning tool. Appl. Bioinformatics 1:107-108. [PubMed] [Google Scholar]
- 18.Gilcrease, E. B., D. A. Winn-Stapley, F. C. Hewitt, L. Joss, and S. R. Casjens. 2005. Nucleotide sequence of the head assembly gene cluster of bacteriophage L and decoration protein characterization. J. Bacteriol. 187:2050-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Golz, S., and B. Kemper. 1999. Association of Holliday-structure resolving endonuclease VII with gp20 from the packaging machine of phage T4. J. Mol. Biol. 285:1131-1144. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez, C. F., and A. K. Vidaver. 1979. Bacteriocin, plasmid and pectolytic diversity in Pseudomonas cepacia of clinical and plant origin. J. Gen. Microbiol. 110:161-170. [DOI] [PubMed] [Google Scholar]
- 21.Grundling, A., U. Blasi, and R. Young. 2000. Biochemical and genetic evidence for three transmembrane domains in the class I holin, lambda S. J. Biol. Chem. 275:769-776. [DOI] [PubMed] [Google Scholar]
- 22.Hagedorn, C., W. D. Gould, T. R. Bardinelli, and D. R. Gustavson. 1987. A selective medium for enumeration and recovery of Pseudomonas cepacia biotypes from soil. Appl. Environ. Microbiol. 53:2265-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haggard-Ljungquist, E., E. Jacobsen, S. Rishovd, E. W. Six, O. Nilssen, M. G. Sunshine, B. H. Lindqvist, K. J. Kim, V. Barreiro, E. V. Koonin, et al. 1995. Bacteriophage P2: genes involved in baseplate assembly. Virology 213:109-121. [DOI] [PubMed] [Google Scholar]
- 24.Hendrix, R. W., and R. L. Duda. 1992. Bacteriophage lambda PaPa: not the mother of all lambda phages. Science 258:1145-1148. [DOI] [PubMed] [Google Scholar]
- 25.Hendrix, R. W., and R. L. Duda. 1998. Bacteriophage HK97 head assembly: a protein ballet. Adv. Virus Res. 50:235-288. [DOI] [PubMed] [Google Scholar]
- 26.Hendrix, R. W., M. C. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juhala, R. J., M. E. Ford, R. L. Duda, A. Youlton, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 299:27-51. [DOI] [PubMed] [Google Scholar]
- 28.Kahng, L. S., and L. Shapiro. 2001. The CcrM DNA methyltransferase of Agrobacterium tumefaciens is essential, and its activity is cell cycle regulated. J. Bacteriol. 183:3065-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kedzierska, S., A. Wawrzynow, and A. Taylor. 1996. The Rz1 gene product of bacteriophage lambda is a lipoprotein localized in the outer membrane of Escherichia coli. Gene 168:1-8. [DOI] [PubMed] [Google Scholar]
- 30.Kwan, T., J. Liu, M. Dubow, P. Gros, and J. Pelletier. 2005. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc. Natl. Acad. Sci. USA 102:5174-5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawrence, J. G., R. W. Hendrix, and S. Casjens. 2001. Where are the pseudogenes in bacterial genomes? Trends Microbiol. 9:535-540. [DOI] [PubMed] [Google Scholar]
- 32.Li, W.-H. 1987. Molecular evolution. Sinauer Associates, Inc., Sunderland, Mass.
- 33.Lin, H., and L. W. Black. 1998. DNA requirements in vivo for phage T4 packaging. Virology 242:118-127. [DOI] [PubMed] [Google Scholar]
- 34.LiPuma, J. J., T. Spilker, T. Coenye, and C. F. Gonzalez. 2002. An epidemic Burkholderia cepacia complex strain identified in soil. Lancet 359:2002-2003. [DOI] [PubMed] [Google Scholar]
- 35.Liu, M., M. Gingery, S. R. Doulatov, Y. Liu, A. Hodes, S. Baker, P. Davis, M. Simmonds, C. Churcher, K. Mungall, M. A. Quail, A. Preston, E. T. Harvill, D. J. Maskell, F. A. Eiserling, J. Parkhill, and J. F. Miller. 2004. Genomic and genetic analysis of Bordetella bacteriophages encoding reverse transcriptase-mediated tropism-switching cassettes. J. Bacteriol. 186:1503-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magrini, V., D. Salmi, D. Thomas, S. K. Herbert, P. L. Hartzell, and P. Youderian. 1997. Temperate Myxococcus xanthus phage Mx8 encodes a DNA adenine methylase, Mox. J. Bacteriol. 179:4254-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahdi, A. A., G. J. Sharples, T. N. Mandal, and R. G. Lloyd. 1996. Holliday junction resolvases encoded by homologous rusA genes in Escherichia coli K-12 and phage 82. J. Mol. Biol. 257:561-573. [DOI] [PubMed] [Google Scholar]
- 38.Maluf, N. K., Q. Yang, and C. E. Catalano. 2005. Self-association properties of the bacteriophage lambda terminase holoenzyme: implications for the DNA packaging motor. J. Mol. Biol. 347:523-542. [DOI] [PubMed] [Google Scholar]
- 39.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33:D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marino-Ramirez, L., L. Campbell, and J. C. Hu. 2003. Screening peptide/protein libraries fused to the lambda repressor DNA-binding domain in E. coli cells. Methods Mol. Biol. 205:235-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marino-Ramirez, L., and J. C. Hu. 2002. Isolation and mapping of self-assembling protein domains encoded by the Saccharomyces cerevisiae genome using lambda repressor fusions. Yeast 19:641-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakayama, K., S. Kanaya, M. Ohnishi, Y. Terawaki, and T. Hayashi. 1999. The complete nucleotide sequence of phi CTX, a cytotoxin-converting phage of Pseudomonas aeruginosa: implications for phage evolution and horizontal gene transfer via bacteriophages. Mol. Microbiol. 31:399-419. [DOI] [PubMed] [Google Scholar]
- 43.Pedulla, M. L., M. E. Ford, J. M. Houtz, T. Karthikeyan, C. Wadsworth, J. A. Lewis, D. Jacobs-Sera, J. Falbo, J. Gross, N. R. Pannunzio, W. Brucker, V. Kumar, J. Kandasamy, L. Keenan, S. Bardarov, J. Kriakov, J. G. Lawrence, W. R. Jacobs, Jr., R. W. Hendrix, and G. F. Hatfull. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171-182. [DOI] [PubMed] [Google Scholar]
- 44.Raden, B., and L. Rutberg. 1984. Nucleotide sequence of the temperate Bacillus subtilis bacteriophage SPO2 DNA polymerase gene L. J. Virol. 52:9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Resch, G., E. M. Kulik, F. S. Dietrich, and J. Meyer. 2004. Complete genomic nucleotide sequence of the temperate bacteriophage AaΦ23 of Actinobacillus actinomycetemcomitans. J. Bacteriol. 186:5523-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rohwer, F., and R. Edwards. 2002. The phage proteomic tree: a genome-based taxonomy for phage. J. Bacteriol. 184:4529-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 48.Schwartz, M. 1975. Reversible interaction between coliphage lambda and its receptor protein. J. Mol. Biol. 99:185-201. [DOI] [PubMed] [Google Scholar]
- 49.Summer, E. J., C. F. Gonzalez, T. Carlisle, L. M. Mebane, A. M. Cass, C. G. Savva, J. LiPuma, and R. Young. 2004. Burkholderia cenocepacia phage BcepMu and a family of Mu-like phages encoding potential pathogenesis factors. J. Mol. Biol. 340:49-65. [DOI] [PubMed] [Google Scholar]
- 50.Thomas, C. A., Jr., and L. A. MacHattie. 1964. Circular T2 DNA molecules. Proc. Natl. Acad. Sci. USA 52:1297-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van der Wilk, F., A. M. Dullemans, M. Verbeek, and J. F. van den Heuvel. 1999. Isolation and characterization of APSE-1, a bacteriophage infecting the secondary endosymbiont of Acyrthosiphon pisum. Virology 262:104-113. [DOI] [PubMed] [Google Scholar]
- 52.Vermis, K., M. Brachkova, P. Vandamme, and H. Nelis. 2003. Isolation of Burkholderia cepacia complex genomovars from waters. Syst. Appl. Microbiol. 26:595-600. [DOI] [PubMed] [Google Scholar]
- 53.Wang, I. N., D. L. Smith, and R. Young. 2000. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54:799-825. [DOI] [PubMed] [Google Scholar]
- 54.Wang, P. W., L. Chu, and D. S. Guttman. 2004. Complete sequence and evolutionary genomic analysis of the Pseudomonas aeruginosa transposable bacteriophage D3112. J. Bacteriol. 186:400-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weisberg, R. A., M. E. Gottesmann, R. W. Hendrix, and J. W. Little. 1999. Family values in the age of genomics: comparative analyses of temperate bacteriophage HK022. Annu. Rev. Genet. 33:565-602. [DOI] [PubMed] [Google Scholar]
- 56.Willi, K., and J. Meyer. 1998. DNA analysis of temperate bacteriophage Aaφ23 isolated from Actinobacillus actinomycetemcomitans. Mol. Gen. Genet. 258:323-325. [DOI] [PubMed] [Google Scholar]
- 57.Wu, H., L. Sampson, R. Parr, and S. Casjens. 2002. The DNA site utilized by bacteriophage P22 for initiation of DNA packaging. Mol. Microbiol. 45:1631-1646. [DOI] [PubMed] [Google Scholar]
- 58.Xu, J., R. W. Hendrix, and R. L. Duda. 2004. Conserved translational frameshift in dsDNA bacteriophage tail assembly genes. Mol. Cell 16:11-21. [DOI] [PubMed] [Google Scholar]
- 59.Xu, M., A. Arulandu, D. K. Struck, S. Swanson, J. C. Sacchettini, and R. Young. 2005. Disulfide isomerization after membrane release of its SAR domain activates P1 lysozyme. Science 307:113-117. [DOI] [PubMed] [Google Scholar]
- 60.Young, R. 1992. Bacteriophage lysis: mechanism and regulation. Microbiol. Rev. 56:430-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Young, R., J. Way, S. Way, J. Yin, and M. Syvanen. 1979. Transposition mutagenesis of bacteriophage lambda: a new gene affecting cell lysis. J. Mol. Biol. 132:307-322. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, N., and R. Young. 1999. Complementation and characterization of the nested Rz and Rz1 reading frames in the genome of bacteriophage lambda. Mol. Gen. Genet. 262:659-667. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.