Abstract
In Salmonella enterica serovar Typhimurium, σ28 and anti-sigma factor FlgM are regulatory proteins crucial for flagellar biogenesis and motility. In this study, we used S. enterica serovar Typhimurium as an in vivo heterologous system to study σ28 and anti-σ28 interactions in organisms where genetic manipulation poses a significant challenge due to special growth requirements. The chromosomal copy of the S. enterica serovar Typhimurium σ28 structural gene fliA was exchanged with homologs of Aquifex aeolicus (an extreme thermophile) and Chlamydia trachomatis (an obligate intracellular pathogen) by targeted replacement of a tetRA element in the fliA gene location using λ-Red-mediated recombination. The S. enterica serovar Typhimurium hybrid strains showed σ28-dependent gene expression, suggesting that σ28 activities from diverse species are preserved in the heterologous host system. A. aeolicus mutants defective for σ28/FlgM interactions were also isolated in S. enterica serovar Typhimurium. These studies highlight a general strategy for analysis of protein function in species that are otherwise genetically intractable and a straightforward method of chromosome restructuring using λ-Red-mediated recombination.
Many organisms possess flagellar organelles that enable them to move about in liquid environments. The cells are able to direct their movement through chemical gradients as a result of intricate interactions between individual flagella and a chemosensory system, a process known as chemotaxis (3, 46). Motility is advantageous, since it allows cells to seek and acquire nutrients and to escape from adverse environmental conditions (3, 46). In addition, in many pathogenic organisms, flagella are important for adhesion, invasion, colonization, and biofilm formation on host cells and thereby contribute to virulence (17, 19). Flagellin monomers that constitute the flagella have also been shown to induce immune responses in mammalian hosts and the activation of plant defense mechanisms (33).
Flagellar gene regulation and assembly have been extensively characterized in Salmonella enterica serovar Typhimurium (1, 25). These studies revealed important regulatory mechanisms evolved by the organism to build these complex supramolecular structures. The assembly of the flagellum coincides with the cocoordinated regulation and expression of over 50 genes (25). The flagellar genes are expressed in a transcriptional hierarchy involving three promoter classes, classes 1, 2, and 3 (21). The class 1 operon responds to global regulatory signals and encodes transcriptional activators that interact with the housekeeping sigma factor σ70 and RNA polymerase (RNAP) to transcribe class 2 promoters. The class 2 genes encode proteins required for the structure and assembly of the hook-basal body (HBB) and regulatory proteins including FliA (σ28), required for class 3 promoter transcription, and the anti-σ28 factor FlgM. The class 3 genes encode proteins that are required for filament assembly, motor force generation, and chemotaxis. The HBB anchors the flagellum to the bacterial membranes and includes a channel that enables the flagellar type III secretion system to secrete flagellar protein subunits out of the cytoplasm (26). Prior to completion of the HBB structure, FlgM binds σ28, and class 3 promoters are not transcribed. When the HBB is completed, FlgM is secreted from the cell; this frees σ28, and class 3 genes are expressed (18). This σ28/FlgM regulatory checkpoint is thought to prevent production of class 3 gene products until they can be assembled into the flagellar structure (15).
Genetic and biochemical studies of σ28/FlgM interactions have contributed to the understanding of flagellar regulation. Nuclear magnetic resonance studies with FlgM revealed that the protein was unstructured in solution but that, upon binding to σ28, the C-terminal domain gained structure (9). These studies showed that the C-terminal domain of FlgM is important for interactions with σ28 (9). Mutations in σ28 that were insensitive to FlgM inhibition mapped throughout the protein. Based on these studies, a model suggesting that FlgM binds to σ28 in a multipartite manner to inhibit σ28 interaction with RNAP was proposed (4). In addition, it was shown that FlgM could inhibit σ28 activity by destabilizing the σ28-RNAP holoenzyme complex (4, 5).
Recently, a cocrystal structure of A. aeolicus σ28/FlgM has been solved, and these studies defined the important domains in σ28 and FlgM interactions (41). Further, these studies provide insights into the mechanism of FlgM inhibition through destabilization of the σ28-RNAP holoenzyme complex (41). Because the overall amino acid homologies of A. aeolicus σ28 and FlgM to S. enterica serovar Typhimurium are low, it is difficult to extrapolate whether the important interacting amino acids identified in S. enterica serovar Typhimurium are significant in A. aeolicus σ28/FlgM interactions. Hence, we sought to isolate mutants that specifically affect A. aeolicus σ28/FlgM interactions. A. aeolicus is a motile extreme thermophile, and unlike S. enterica serovar Typhimurium, it is difficult to propagate and requires special growth conditions (11). As a result, genetic studies with A. aeolicus are challenging. To corroborate the structural studies of A. aeolicus σ28/FlgM interaction with an in vivo analysis, we utilized S. enterica serovar Typhimurium as a host cell system to genetically characterize the flagellar regulatory proteins of A. aeolicus. The availability of genetic and molecular tools and the extensive characterization of the flagellar regulon in S. enterica serovar Typhimurium present this organism as an ideal model system.
We also sought to test whether S. enterica serovar Typhimurium could be used as a general host to study σ28 proteins from organisms that are genetically intractable. For this reason, we included the σ28 homolog rspD from Chlamydia trachomatis in these studies. C. trachomatis is an obligate intracellular pathogen and is the causal agent of different human diseases such as trachoma, genital infections, and sexually transmitted diseases (37). Chlamydia are also difficult to propagate, and construction of stable transformants has been difficult (43). Although C. trachomatis is nonmotile, its σ28 homolog was shown to recognize the fliC promoter of Escherichia coli in vitro (39). In this study, we present a genetic characterization of σ28 from A. aeolicus and C. trachomatis in S. enterica serovar Typhimurium and an isolation of A. aeolicus σ28/FlgM through complementation and mutagenesis experiments.
MATERIALS AND METHODS
Strains.
Bacterial strains, plasmids, and primers used in this study are listed in Table 1.
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Genotype description, or sequence (5′→3′)a | Source or referenceb |
|---|---|---|
| Strains | ||
| S. enterica serovar Typhimurium | ||
| TH6238 | motA5461::MudJ | |
| TH8059 | motA5461::MudJ fliA6088 (A. aeolicus GTG-fliA+) | |
| TH8120 | motA5461::MudJ flgM6095 (A. aeolicus flgM+) | |
| TH8122 | motA5461::MudJ fliA6088 flgM6095 | |
| TH8194 | motA5461::MudJ ΔflgM5794::FRTc | |
| TH8196 | motA5461::MudJ fliA6088 ΔflgM5794::FRT | |
| TH8197 | motA5461::MudJ ΔflgM5794::FRT ΔfliA5805::tetRA | |
| TH4098 | fliC5050::MudJ fljBe,n,xvh2 | |
| TH8061 | fliC5050::MudJ fljBe,n,xvh2 fliA6090 (A. aeolicus GTG-fliA+) | |
| TH8123 | fliC5050::MudJ fljBe,n,xvh2 flgM6095 | |
| TH8125 | fliC5050::MudJ fljBe,n,xvh2 fliA6090 flgM6095 | |
| TH8198 | fliC5050::MudJ fljBe,n,xvh2 ΔflgM5794::FRT | |
| TH8200 | fliC5050::MudJ fljBe,n,xvh2 fliA6090 ΔflgM5794::FRT | |
| TH8201 | fliC5050::MudJ fljBe,n,xvh2 ΔflgM5794::FRT ΔfliA5805::tetRA | |
| MST4190 | leuBCD485 trp::[Spcr PlacT7-RNAP lacI] | S. Maloy |
| TH9031 | leuBCD485 trp::[Spcr PlacT7-RNAP lacI] motA5461::MudJ fliA6325 (C. trachomatis L2 GTG-fliA+) ΔflgM5794::FRT/pJK627 | |
| TH9032 | leuBCD485 trp::[Spcr PlacT7-RNAP lacI] motA5461::MudJ fliA6325 ΔflgM5794::FRT/pET15b | |
| TH9033 | leuBCD485 trp::[Spcr PlacT7-RNAP lacI] motA5461::MudJ fliA6325/pET15b | |
| TH9040 | leuBCD485 trp::[Spcr PlacT7-RNAP lacI] fliC5050::MudJ fljBe,n,xvh2 fliA6325 ΔflgM5794::FRT/pJK627 | |
| TH9041 | leuBCD485 trp::[Spcr PlacT7-RNAP lacI] fliC5050::MudJ fljBe,n,xvh2 fliA6325 ΔflgM5794::FRT/pET15b | |
| TH9042 | leuBCD485 trp::[Spcr PlacT7-RNAP lacI] fliC5050::MudJ fljBe,n,xvh2 fliA6325/pET15b | |
| TH8604 | fliC5050::MudJ fljBe,n,xvh2 fliA6090 flgM6237 (A. aeolicus flgM A→G −6 bp from ATG) | |
| TH8605 | fliC5050::MudJ fljBe,n,xvh2 fliA6090 flgM6238 (A. aeolicus flgM A→C −5 bp from ATG) | |
| TH8606 | fliC5050::MudJ fljBe,n,xvh2 fliA6090 flgM6239 (A. aeolicus flgM A→G −4 bp from ATG) | |
| TH8607 | fliC5050::MudJ fljBe,n,xvh2 fliA6090 flgM6240 (A. aeolicus flgM ATG→GTG M1V) | |
| TH8608 | fliC5050::MudJ fljBe,n,xvh2 fliA6090 flgM6241 (A. aeolicus flgM AAG→AGG K19R) | |
| TH8609 | fliC5050::MudJ fljBe,n,xvh2 fliA6090 flgM6242 (A.aeolicus flgM Δ bp +55-56) | |
| TH8610 | fliC5050::MudJ fljBe,n,xvh2 fliA6090 flgM6243 (A. aeolicus flgM ΔA bp +177 from ATG) | |
| TH8611 | fliC5050::MudJ fljBe,n,xvh2 fliA6090 flgM6244 (A. aeolicus flgM AAA→AAG K64K, ΔA bp +199 from ATG) | |
| TH8612 | fliC5050::MudJ fljBe,n,xvh2 flgM6095 fliA6245 (A. aeolicus fliA CCT→TCT P4S) | |
| TH8613 | fliC5050::MudJ fljBe,n,xvh2 flgM6095 fliA6246 (A. aeolicus fliA ACA→TCA T27S) | |
| TH8614 | fliC5050::MudJ fljBe,n,xvh2 flgM6095 fliA6247 (A. aeolicus fliA ATA→ATG 129M) | |
| TH8615 | fliC5050::MudJ fljBe,n,xvh2 flgM6095 fliA6248 (A. aeolicus fliA AGA→GGA R149G) | |
| TH8616 | fliC5050::MudJ fljBe,n,xvh2 flgM6095 fliA6249 (A. aeolicus fliA ACG→ATG 1175M) | |
| TH8617 | fliC5050::MudJ fljBe,n,xvh2 flgM6095 fliA6250 (A. aeolicus fliA GAA→GGA E189G) | |
| TH8618 | fliC5050::MudJ fljBe,n,xvh2 flgM6095 fliA6251 (A. aeolicus fliA GAA→GGA E199G) | |
| TH9607 | fliC5050::MudJ fljBe,n,xvh2 flgM6095 fliA6245 ΔflgM5794::FRT | |
| TH9608 | fliC5050::MudJ fljBe,n,xvh2 flgM6095 fliA6246 ΔflgM5794::FRT | |
| TH9609 | fliC5050::MudJ fljBe,n,xvh2 flgM6095 fliA6247 ΔflgM5794::FRT | |
| TH9610 | fliC5050::MudJ fljBe,n,xvh2 flgM6095 fliA6248 ΔflgM5794::FRT | |
| TH9611 | fliC5050::MudJ fljBe,n,xvh2 flgM6095 fliA6249 ΔflgM5794::FRT | |
| TH9612 | fliC5050::MudJ fljBe,n,xvh2 flgM6095 fliA6250 ΔflgM5794::FRT | |
| TH9647 | fliC5050::MudJ fljBe,n,xvh2 flgM6095 fliA6251 ΔflgM5794::FRT | |
| TH3467 | proAB47/F′128 (pro-lac) zzf-3833::Tn10dTc[del-25] (T-POP) | 34 |
| TH4387 | fliA5059::Tn10dTc fljBe,n,xvh2 | |
| TH7953 | flgM6085::tetRA/pKD46 | |
| TH8009 | ΔfliA5805::tetRA/pKD46 | |
| TH8010 | ΔfliA5805::tetRA motA5461::MudJ/pKD46 | |
| TH8011 | ΔfliA5805::tetRA fliC5050::MudJ fljBe,n,xvh2/pKD46 | |
| TH8349 | fliC5050::MudJ fljBe,n,xvh2 fliA6090 flgM6085::tetRA/pKD46 | |
| TH8350 | fliC5050::MudJ fljBe,n,xvh2 flgM6095 ΔfliA5805::tetRA/pKD46 | |
| TH8142 | fliA6088 (A. aeolicus GTG-fliA+) | |
| TH8982 | fliA6325 (C. trachomatis L2 GTG fliA+) | |
| TH9065 | ΔfliA5647::FRT/pBAD24 | |
| TH9066 | ΔfliA5647::FRT/pMC147 | |
| TH9067 | ΔfliA5647::FRT/pJK558 | |
| TH9068 | ΔfliA5647::FRT/pJK629 | |
| E. coli | ||
| DH5α | φ80dlacZΔM15 endA1 hsdR17 (rK−mK−) supE44 thi-1 recA1 gyrA (Nalr (lacZYA-argF) U169) | New England Biolabs |
| Plasmids | ||
| pMC147 | (pBAD24-fliA S. enterica serovar Typhimurium) | |
| pJK558 | (pBAD24-fliA A. aeolicus) | |
| pJK629 | (pBAD24-rspD C. trachomatis) | |
| pJK627 | (pET15b-rsbW C. trachomatis L2) | |
| pKD46 | (pBAD- λ-Red (γ β exo) | 8 |
| pBAD24 | (Para cloning vector pBR322 ori, Apr) | 14 |
| pET15b | Novagen | |
| pMS531 | (S. enterica serovar Typhimurium fliA) | 42 |
| pLF28 | (PBAD-rspD C. trachomatis) | 39 |
| pET28a-rsbW | C. trachomatis L2 | T. P. Hatch |
| Primers | ||
| FliAdeltetR | CTCATTTAACGCAGGGCTGTTTATCGTGAATATAGGTCGCGCATGATCGCACCC GAAAAGTttaagacccactttcaca | |
| FliAdeltetA | CTTTTCGGGTGCGATCATGCGCGACCTATATTCACGATAAACAGCCCTGCGTTA AATGAGTctaagcacttgtctcctg | |
| FlgMAUGtetR | AGCTGGCCGCTACAACGTAACCCTCGATGAGGATAAATAAttaagacccactttcacatt | |
| FlgMAUGtetA | TGCTAACGGGTTTCAAAGGTGAGGTACGGTCAATGCTCATctaagcacttgtctcctg | |
| AAGTG | TAATCATGCCGATAACTCATTTAACGCAGGGCTGTTTATCgtgaaaaacccttacagcaacc | |
| AA28R | CGTTGTGCGGCACTTTTCGGGTGCGATCATGCGCGACCTAtagaggattagagagcatttc | |
| AAFLGM1 | AGCTGGCCGCTACAACGTAACCCTCGATGAGGATAAATAAatggtgaacaggattcaactc | |
| AAFLGM2 | CATCTGGTCAAGTATTTCTGACAAACGAGTCATACGCTTAcgtaaaaaactctatcagtcc | |
| CTFLIAGTG | TAATCATGCCGATAACTCATTTAACGCAGGGCTGTTTATCgtgaagactcacgatctcgc | |
| CTFLIASTOP | CGTTGTGCGGCACTTTTCGGGTGCGATCATGCGCGACCTAaagcagactggacaatgtac | |
| AQAA1 | AAGAATTCATGAAAAACCCTTACAGCAACC | |
| AQAA2 | ACAAAGCTTATAGAGGATTAGAGAGCATTTC | |
| FliA#3 | GGCGCTACAGGTTACATAAG | |
| FliA#4 | TAGTCTATACGTTGTGCGGC | |
| FlgM5′UP | GAACCGTCGATTCTGATG | |
| FlgN reverse | GTCTTCAGGTCATTCAG |
In primer sequences, lowercase indicates homology to the tetRA element.
All strains were constructed for this study unless otherwise noted.
FRT, FLP recognition target scar (8).
Media and standard genetic and molecular techniques.
Media, growth conditions, phage P22 transduction methods, and motility assays were as described previously (10, 13). Tetracycline-sensitive (Tcs) plates were made as described by Maloy (28). Antibiotics in rich media were used at the following concentrations (in μg/ml): ampicillin (Ap; 100), kanamycin (50), tetracycline (15), and spectinomycin (100). PCR and DNA cloning were performed as described previously (35). DNA primers (Integrated DNA Technologies, Coralville, IA) used in this study are listed in Table 1. PCRs were performed with Taq (Promega, Madison, WI), Accuzyme (Bioline, Randolph, MA), or ThermalAce (Invitrogen, Carlsbad, CA). All enzymes used for DNA cloning were from New England Biolabs (Beverly, MA). DNA sequencing was performed using BigDye v3.1 (Applied Biosystems, Foster City, CA), and the reactions were analyzed using a 3730XL sequencer at the DNA sequencing facility of the Department of Biochemistry, University of Washington, Seattle, WA.
λ-Red-mediated insertion of the tetRA element.
The fliA coding region in S. enterica serovar Typhimurium was replaced by allelic exchange with a tetRA element using λ-Red-mediated recombination (8, 32, 48). In addition, a tetRA element was inserted just before the ATG start codon of flgM. This tetRA element includes the coding sequences of tetR and tetA from transposon Tn10dTc and confers tetracycline resistance (47). The PCR primers for amplification of the tetRA element were flanked by 40-bp sequences of homology for recombination on the chromosome. Primers FliAdeltetR, FliAdeltetA, FlgMAUGtetR, and FlgMAUGtetAwere used for the construction of ΔfliA5805::tetRA and flgM6085::tetRA, respectively. The PCRs containing genomic DNA of TH3467, DNA primers, dNTPs, and Taq polymerase were amplified as follows; 3 min at 95°C for 1 cycle, 30 s at 95°C, 30 s at 49°C, 2 min at 72°C for 30 cycles, and 10 min at 72°C for 1 cycle. A 1,990-bp product was purified using a QIAquick PCR purification kit (QIAGEN, Valencia, CA). S. enterica serovar Typhimurium strain TH4702 containing a λ-Red plasmid pKD46 that has a temperature-sensitive replicon (8) was grown under inducing conditions in LB plus Ap (100 μg/ml) plus arabinose at 0.2% at 30°C until the optical density at 600 nm reached 0.6 to 0.8. The cells were washed twice in equal volumes of cold sterile water and concentrated 250-fold. Fifty μl of cells was electroporated with 100 to 200 ng of purified PCR fragment using 0.1-cm cuvettes at 200 Ω, 1.6 kV, and 25 μF. Subsequently, one ml of LB was added, and cells were incubated for 1 hour at 37°C. Approximately 0.5 ml of cells was plated on LB plates containing tetracycline and incubated at 37°C. Tcr colonies were purified on LB plates without antibiotics and incubated at 42°C. Tcr colonies were screened for Aps to confirm the loss of the pKD46 plasmid. To confirm that the tetRA integrated into the correct region on the chromosome, Tcr and Aps colonies were screened by PCR using primers within the tetRA element and in sequences flanking fliA or flgM, respectively.
λ-Red-mediated replacement of the tetRA element.
The fliA and flgM genes were replaced in S. enterica serovar Typhimurium with homologs from A. aeolicus and C. trachomatis using λ-Red-mediated replacement of the tetRA element as described below. The fliA homologs from A. aeolicus (fliA) and C. trachomatis (rspD) were PCR amplified using genomic DNA from A. aeolicus (kind gift from K. O. Stetter) and pLF28 (39), respectively. Primer sets to amplify the fliA homologs are as follows: for A. aeolicus, AAGTG and AA28R; for C. trachomatis, CTFLIAGTG and CTFLIASTOP. PCR amplification was performed as described above, except the annealing temperature used was 45°C. The PCR products were purified, electroporated into strain TH8350 (ΔfliA::tetRA) as described above except for subsequent plating of the cells onto Tcs medium (28), and incubated at 42°C overnight. Colonies were purified once on Tcs medium at 42°C and then on LB plates at 37°C. The constructs were screened for the loss of the tetRA element by PCR, and the region was subsequently PCR amplified for DNA sequencing to confirm the replacement of S. enterica serovar Typhimurium fliA with the fliA homologs of A. aeolicus and C. trachomatis. The coding region of flgM in S. enterica serovar Typhimurium was replaced with the homolog from A. aeolicus as described for fliA using A. aeolicus genomic DNA, primers AAFLGM1 and AAFLGM2, and TH8349.
Isolation of A. aeolicus fliA and flgM mutants using error-prone PCR and λ-Red-mediated replacement of the tetRA element.
The coding regions of fliA and flgM from A. aeolicus were mutagenized by error-prone PCR with genomic DNA from TH8125, primers FliA#3 and FliA#4 (for σ28), FlgM5′UP and FlgN reverse (for flgM), dNTPs, and 5 units of Promega Taq per reaction. The PCR amplifications were performed as described above. Electrocompetent cells of strains TH8350 (for fliA) and TH8349 (for flgM) were prepared as described above, and transformants were selected on Tcs medium and incubated at 42°C. Colonies were further purified on Tcs medium at 42°C and then on LB medium at 37°C. The colonies were screened for Lac+ on MacConkey medium. The fliA or flgM regions from the Lac+ isolates were DNA sequenced.
Plasmid constructions.
The fliA coding sequences from S. enterica serovar Typhimurium, A. aeolicus, and C. trachomatis were cloned under the arabinose promoter to construct pMC147, pJK558, and pJK629 as follows. A 770-bp EcoRI fragment blunted with Klenow fragment containing S. enterica serovar Typhimurium fliA from pMS531 was ligated into pBAD24 digested with NcoI and blunted with Klenow fragment to generate pMC147. A. aeolicus fliA was PCR amplified using A. aeolicus genomic DNA and primers AQAA1 and AQAA2. The PCR product was digested with BspHI and HindIII and ligated into pBAD24 digested with NcoI and HindIII to generate pJK588. A 515-bp NcoI-PstI fragment from pLF28 containing C. trachomatis rspD was ligated into pBAD24 digested with NcoI-PstI to generate pJK629. A 500-bp NcoI-HindIII fragment from pET-rsbW C. trachomatis L2 containing His-tagged rsbW from C. trachomatis (gift from T. P. Hatch) was ligated into pET15b digested with NcoI-XhoI to generate pJK627.
β-Galactosidase assays.
β-Galactosidase assays were performed as described by Maloy (27). Cells were grown in LB or LB plus Ap (100 μg/ml) plus 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) (in strains containing plasmids) at 37°C until mid-log phase. Assays were performed in triplicate, and the values were normalized as percentages of the wild-type (WT) value.
Western blot analysis.
Cells were grown in LB until mid-log phase, and 1 ml of cells was centrifuged and resuspended in sodium dodecyl sulfate sample buffer. The samples were run on a 10% Tricine-sodium dodecyl sulfate polyacrylamide gel and electrotransferred in CAPS buffer onto polyvinylidene difluoride membranes (29, 38). The membrane was probed with rabbit anti-FliC antibodies (Salmonella H antiserum i rabbit, catalog number 228241; Difco) purified as described previously (15) and developed using an ECL-Plus kit (Amersham Biosciences, Piscataway, NJ).
Software programs.
Amino acid alignments were performed using Clustal W (6). The sequence alignments were edited using Jalview alignment editor (7). Aquifex aeolicus σ28/FlgM cocrystal analysis used coordinates PDB ID 1SC5 from the RCSB protein database (www.rcsb.org/pdb/) and RasMol v. 2.7 (2, 36). The analysis of helix interactions was done with PDBsum (23). The analysis of residue contacts was determined on PDB ID 1SC5 using CSU (contact of structural units) software (40).
RESULTS
A. aeolicus and C. trachomatis σ28 restore motility to an S. enterica serovar Typhimurium σ28 mutant.
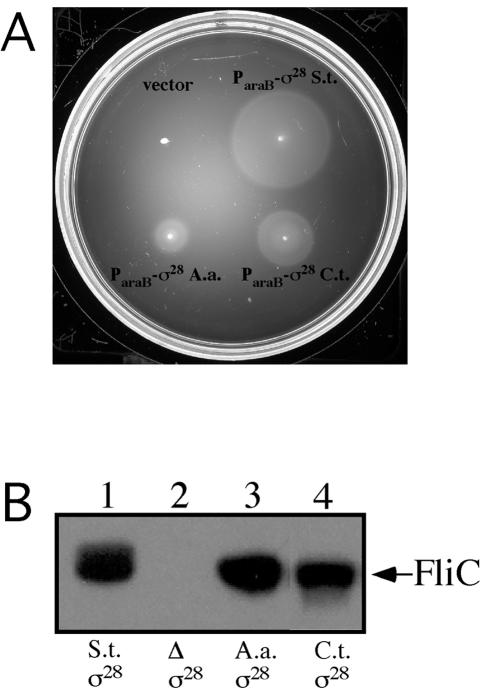
Given the similarities in the critical domains of σ28 (see alignments in Fig. 1), we tested whether σ28 homologs of A. aeolicus and C. trachomatis would complement an S. enterica serovar Typhimurium σ28 mutant. The genes encoding σ28 of S. enterica serovar Typhimurium (fliASeT), A. aeolicus (fliAAa), and C. trachomatis (rspDCt) were cloned under the arabinose-inducible promoter (ParaB) and introduced into the S. enterica serovar Typhimurium fliA mutant strain TH4387. The plasmid-containing strains were assessed for motility on plates containing 0.2% arabinose (Fig. 2A). Levels of complementation for motility by ParaB-A. aeolicus fliA+ andParaB-C. trachomatis rspD+ were 39% and 58%, respectively, of the level for ParaB-S. enterica serovar Typhimurium fliA+ (Fig. 2A). Similar results were previously observed with A. aeolicus fliA+ from E. coli. (44). Interestingly, a ParaB-C. trachomatis rspD+ plasmid did not restore motility in E. coli with a fliA deletion (39). Although E. coli and S. enterica serovar Typhimurium possess nearly identical flagellar systems (25), the dramatic differences in complementation for motility with ParaB-C. trachomatis rspD+ demonstrates that differences in the flagellar regulons between the two species can be uncovered.
FIG. 1.
(A) Clustal W amino acid alignments of σ28 homologs. The GenInfo Identifier numbers of the σ28s used in the alignments are as follows: Yersinia pestis KIM (gi22126347), Salmonella enterica serovar Typhimurium LT2 (gi16765294), Yersinia enterocolitica (gi1706832), Proteus mirabilis (gi6959882), Escherichia coli K-12 (gi33347603), Ralstonia solanacearum (gi17549609), Pseudomonas syringae (gi28869183), Bordetella pertussis (gi33571813), Vibrio cholerae (gi15642066), Aquifex aeolicus (gi15606452), Bacillus subtilis (gi1350863), Chlamydia trachomatis (gi15604780), and Helicobacter pylori (gi15645646). Conserved amino acid regions as described by Lonetto et al. (24) are marked in black at the top of the alignments. The σ28 domains from previous structural studies by Sorenson et al. (41) are noted below. (B) Clustal W amino acid alignments of FlgM homologs. The GenInfo Identifier numbers of the FlgM used in the alignments are as follows: Yersinia enterocolitica (gi6166171), Escherichia coli K-12 (gi1651526), Yersinia pestis KIM (gi22126396), Salmonella enterica serovar Typhimurium LT2 (gi16764528), Proteus mirabilis (gi1857441), Bordetella pertussis (gi33572120), Bacillus subtilis (gi729520), Vibrio cholerae (gi15642203), Pseudomonas syringae (gi28869129), Ralstonia solanacearum (gi17548561), Helicobacter pylori (gi18075990), and Aquifex aeolicus (gi15605866). The σ28 binding domain in S. enterica serovar Typhimurium FlgM is highlighted at the bottom of the alignments. The shading scheme for amino acid homologies was determined using Blosum62 (7).
FIG. 2.
(A) Motility assay of S. enterica serovar Typhimurium fliA-defective strain TH4387 with the following plasmids: vector (pBAD24), ParaB-σ28 S. enterica serovar Typhimurium (S.t.) (pMC147), ParaB-σ28 A. aeolicus (A.a.) (pJK558), and ParaB-σ28 C. trachomatis (C.t.) (pJK629). (B) Western analysis of total cellular proteins from S. enterica serovar Typhimurium hybrid strains fliA (σ28) from A. aeolicus and C. trachomatis probing with anti-FliC antiserum. Lane 1, TH437 (S. enterica serovar Typhimurium wild-type strain LT2); lane 2, TH6827 (ΔfliA5805::tetRA); lane 3, TH8142 ((fliA6068 [A. aeolicus fliA]); lane 4, TH8982 ((fliA6325 [C. trachomatis rspD]). Fourfold more cellular lysate was loaded in lanes 2 to 4 than in lane 1.
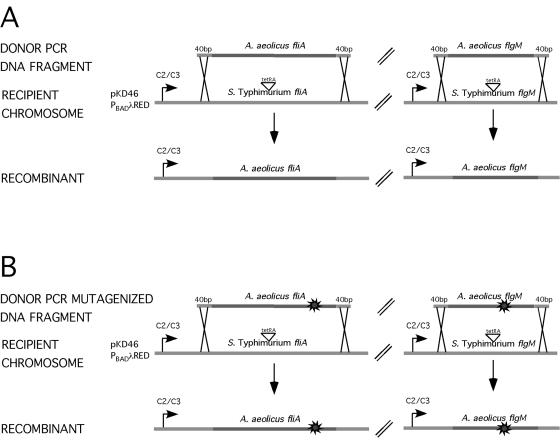
The coding region of chromosomal S. enterica serovar Typhimurium fliA was replaced with either the fliA from A. aeolicus or the rspD from C. trachomatis by targeted replacement of a tetRA element using a λ-Red recombination system (Fig. 3A; also see reference 8 and Materials and Methods). Therefore, σ28-mediated gene expression in these hybrid strains remained under the regulation of the S. enterica serovar Typhimurium flagellar regulon. These σ28 hybrid strains were weakly motile (data not shown). Because motility was weak, we analyzed whether the hybrid strains expressed the σ28-dependent flagellar filament protein FliC. Western blot analysis using anti-FliC serum on cellular lysates showed the presence of FliC protein in S. enterica serovar Typhimurium hybrid strains with A. aeolicus σ28 or C. trachomatis σ28 (Fig. 2B, lanes 3 and 4, respectively). However, the levels of FliC protein were reduced in the σ28 hybrid strains by fourfold relative to the level for WT S. enterica serovar Typhimurium (Fig. 2B). Although motility was reduced in the σ28 hybrid strains, the presence of FliC protein and the ability to transcribe class 3 flagellar promoters (see below) indicate the conservation of σ28 function. Together, these results indicate that σ28 from diverse species can interact with the heterologous host transcriptional machinery for flagellar biogenesis.
FIG. 3.
(A) λ-Red-mediated replacement of the tetRA element. Donor PCR products from coding sequences of fliA (σ28) or flgM from A. aeolicus were flanked by 40-bp sequences of homology for recombination in the chromosome. The donor PCR products were electroporated into S. enterica serovar Typhimurium strains with tetRA elements inserted in the fliA or flgM genes. The tetRA element includes the coding sequences of tetR and tetA from transposon Tn10dTc and confers tetracycline resistance. The recombination was mediated by λ-Red (plasmid pKD46) (8), resulting in the loss of the tetRA element and replacement with the coding sequences of fliA or flgM from A. aeolicus by selection on tetracycline-sensitive medium (28). (B) Isolation of A. aeolicus fliA (σ28) and flgM mutants using error-prone PCR and λ-Red-mediated replacement of the tetRA element. Replacement of the tetRA element were performed as described for panel A, except the donor PCR products of coding sequences from fliA (σ28) or flgM were generated by error-prone PCR (as indicated by the sunburst symbol).
Regulation of σ28 activity by the cognate negative regulator FlgM is species specific.
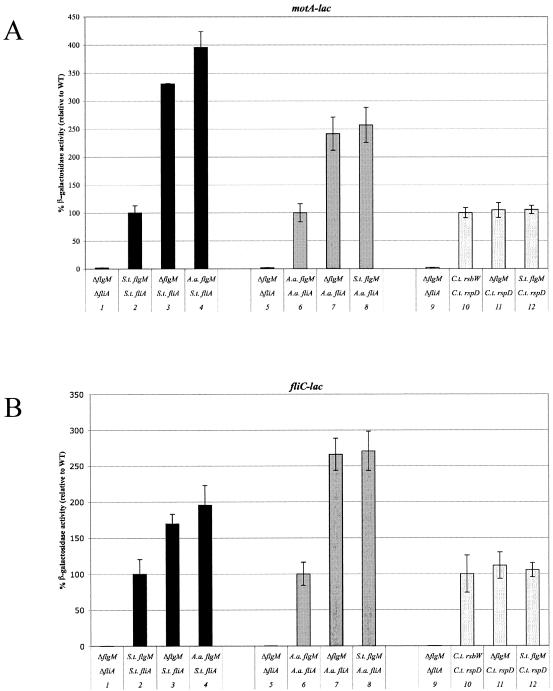
To investigate the interactions of cognate sigma factor and anti-sigma factor activities between species, hybrid S. enterica serovar Typhimurium strains were constructed by use of λ-Red (Fig. 3A; also see Materials and Methods). The fliA+ gene (σ28) of S. enterica serovar Typhimurium was replaced with the σ28 homologs from A. aeolicus and C. trachomatis, fliA+ and rspD+, respectively. Further, the negative regulatory gene flgM+ of S. enterica serovar Typhimurium was replaced with A. aeolicus flgM+. We assayed σ28-mediated gene expression in the hybrid strains using lac gene transcriptional fusion reporters to two σ28-dependent promoters, motA and fliC. The motA and fliC genes encode vital components of the flagellum; MotA is required for flagellar rotation, and FliC is the main structural component of the flagellar filament (25).
Relative β-galactosidase levels of motA-lac and fliC-lac expression in WT S. enterica serovar Typhimurium increased 3- and 1.7-fold, respectively, in the absence of the S. enterica serovar Typhimurium FlgM (Fig. 4, compare lanes 2 and 3 in both panels). Likewise, in S. enterica serovar Typhimurium strains with A. aeolicus σ28, in the absence of A. aeolicus FlgM, motA-lac and fliC-lac expression was increased around 2.5-fold (Fig. 4, compare lanes 6 and 7). This is in support of structure studies on A. aeolicus σ28/FlgM interaction which suggest that FlgM binding to σ28 inhibits its interaction with core RNAP (41). Interestingly, in the presence of S. enterica serovar Typhimurium FlgM, A. aeolicus σ28-mediated levels of motA-lac and fliC-lac expression were similar to that of an S. enterica serovar Typhimurium flgM deletion strain, indicating the absence of negative regulation on A. aeolicus σ28 activity (Fig. 4, compare lanes 7 and 8). In S. enterica serovar Typhimurium σ28 strains, the relative levels of β-galactosidase activity in the S. enterica serovar Typhimurium flgM deletion strain were similar to those of strains containing A. aeolicus FlgM (Fig. 4, compare lanes 3 and 4), suggesting that A. aeolicus FlgM has no effect on S. enterica serovar Typhimurium σ28 activity. These findings show inhibition of A. aeolicus σ28 activity by its cognate negative regulator FlgM but not by S. enterica serovar Typhimurium FlgM. Similarly, S. enterica serovar Typhimurium σ28 activity is inhibited by its cognate negative regulator FlgM but not by A. aeolicus FlgM. Collectively, these studies indicate that the anti-σ28 FlgM factors interactionswith σ28, and their negative regulation on σ28 activities are species specific.
FIG. 4.
β-Galactosidase activities of lac gene transcriptional fusion reporters to σ28-dependent genes motA and fliC in S. enterica serovar Typhimurium. The σ28 coding sequences were replaced by λ-Red recombination with coding sequences from either σ28 from A. aeolicus (fliA) or C. trachomatis (rspD). The flgM coding sequence was replaced with A. aeolicus flgM. C. trachomatis RsbW, putative negative regulator of RspD, was encoded in trans by plasmid pJK627. (A) MotA-lac β-galactosidase activities were compared relative to those of the following isogenic WT σ28 strains (WT = 100%): TH6238, black bars; TH8196, gray bars; TH9033, white bars. Lane 1, ΔfliA, ΔflgM (TH8197); lane 2, S. enterica serovar Typhimurium (S.t.) fliA, S. enterica serovar Typhimurium flgM (TH6238); lane 3, S. enterica serovar Typhimurium fliA, ΔflgM (TH8194); lane 4, S. enterica serovar Typhimurium fliA, A. aeolicus (A.a.) flgM (TH8120); lane 5, ΔfliA, ΔflgM (TH8197); lane 6, A. aeolicus fliA, A. aeolicus flgM (TH8122); lane 7, A. aeolicus fliA, ΔflgM (TH8196); lane 8, A. aeolicus fliA, S. enterica serovar Typhimurium flgM (TH8059,); lane 9, ΔfliA, ΔflgM (TH8197); lane 10, C. trachomatis (C.t.) rspD, C. trachomatis rsbW (TH9031); lane 11, C. trachomatis rspD, ΔflgM (TH9032); lane 12, C. trachomatis rspD, S. enterica serovar Typhimurium flgM (TH9033). (B) FliC-lac β-galactosidase activities were compared relative to those of the following isogenic WT σ28 strains (WT = 100%): TH4098, black bars; TH8125, gray bars; TH9040, white bars. Lane 1, ΔfliA, ΔflgM (TH8201); lane 2, S. enterica serovar Typhimurium fliA, S. enterica serovar Typhimurium flgM (TH4098); lane 3, S. enterica serovar Typhimurium fliA, ΔflgM (TH8198); lane 4, S. enterica serovar Typhimurium fliA, A. aeolicus flgM (TH8123); lane 5, ΔfliA, ΔflgM (TH8201); lane 6, A. aeolicus fliA, A. aeolicus flgM (TH8125); lane 7, A. aeolicus fliA, ΔflgM (TH8200); lane 8, A. aeolicus fliA, S. enterica serovar Typhimurium flgM (TH8061); lane 9, ΔfliA, ΔflgM (TH8201); lane 10, C. trachomatis rspD, C trachomatis. rsbW (TH9040); lane 11, C. trachomatis rspD, ΔflgM (TH9041); lane 12, C. trachomatis rspD, S. enterica serovar Typhimurium flgM (TH9042).
Hybrid strains of S. enterica serovar Typhimurium containing C. trachomatis rspD+ (C. trachomatis σ28) were constructed in motA-lac and fliC-lac backgrounds. C. trachomatis RsbW, the putative negative regulator of RspD (39), was encoded in trans by plasmid pJK627. However, there was no difference in the relative β-galactosidase levels in the presence of RsbW or in its absence (Fig. 4, lanes 10 and 11, respectively). S. enterica serovar Typhimurium FlgM also did not negatively regulate C. trachomatis σ28 activity (Fig. 4, lanes 12). C. trachomatis RsbW is 36% identical and 58% similar to homolog RsbW in Bacillus subtilis. In B. subtilis, anti-sigma factor RsbW and anti-anti-sigma factor RsbV, as well as other regulatory proteins, regulate σB activity but differ mechanistically from anti-σ28 FlgM homologs (16). The lack of anti-σ28 activity by C. trachomatis RsbW in S. enterica serovar Typhimurium suggests that additional factors for C. trachomatis σ28 inhibition may be required or that σ28 inhibition may be independent of RsbW.
Isolation of fliA mutants defective in A. aeolicus σ28/FlgM interactions.
We sought to isolate mutants defective in A. aeolicus σ28/FlgM interactions using a genetic screen. S. enterica serovar Typhimurium strain TH8125 contains A. aeolicus fliA+ and A. aeolicus flgM+ genes as well as a σ28-dependent fliC-lac reporter construct. TH8125 is white (Lac−) on MacConkey lactose (Mac-lac) medium, and the same strain deleted for A. aeolicus flgM is red (Lac+). Therefore, a phenotypic screen was used to identify mutants of TH8125 defective in A. aeolicus σ28/FlgM interactions (red on Mac-lac). Error-prone PCR was used to mutagenize A. aeolicus fliA (σ28) and A. aeolicus flgM genes. The mutagenized gene products were electroporated into S. enterica serovar Typhimurium strain TH8125 that had a tetRA insertion in either A. aeolicus fliA or A. aeolicus flgM. Subsequent recombination of fliA and flgM was achieved by use of λ-Red (Fig. 3B). The loss of tetracycline resistance, indicative of replacement with the mutagenized gene products, was selected on tetracycline-sensitive medium (28). The transformants were subsequently screened on Mac-lac medium, and red (Lac+) colonies were sequenced for putative mutations in either A. aeolicus fliA or A. aeolicus flgM.
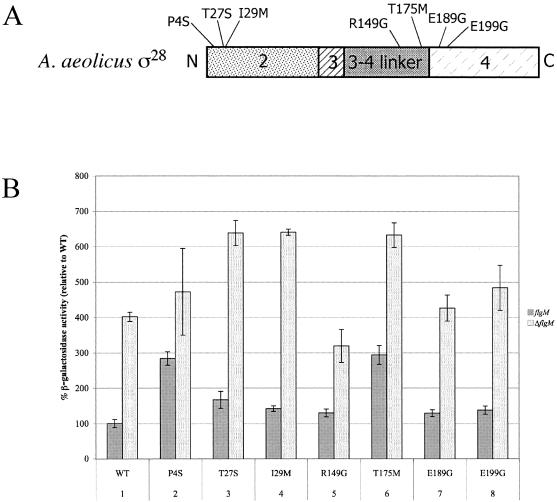
In the A. aeolicus fliA mutagenesis screen, 300 Tcs colonies were isolated, of which 28 were Lac+ on Mac-lac medium. Twelve Lac+ mutants were sequenced, and seven contained missense mutations in A. aeolicus fliA (Fig. 5A and Table 2).Since the remaining five had multiple mutations in A. aeolicus fliA and are difficult to interpret, they were excluded from the study. β-Galactosidase assay results for A. aeolicus σ28 mutants compared to those for WT (TH8125) demonstrated an increase in activity in the presence of A. aeolicus FlgM (Fig. 5B). The relative activity of A. aeolicus σ28 mutants ranged from 1.3- to 2.9-fold higher than that for WT (TH8125) (Table 2).
FIG. 5.
(A) Summary of mutations isolated in A. aeolicus fliA. Domain regions correspond to regions shown in Fig. 1. Mutations resulting in single amino acid changes are listed in Table 2. (B) β-Galactosidase activities of fliC-lac in hybrid S. enterica serovar Typhimurium strains with A. aeolicus fliA mutants isolated defective for σ28/FlgM interactions. β-Galactosidase activities were compared to those of TH8125. Dark gray bars indicate the presence of FlgM; light gray bars indicate the absence of FlgM. Lane 1, WT (TH8125, TH8200); lane 2, P4S (TH8612, TH9607); lane 3, T27S (TH8613, TH9608); lane 4, I29M (TH8614, TH9609); lane 5, R149G (TH8615, TH9610); lane 6, T175M (TH8616, TH9611); lane 7, E189G (TH8617, TH9612); lane 8, E199G (TH8618, TH9647).
TABLE 2.
Summary of mutants isolated in A. aeolicus fliA
| Strain | Mutation relative to the start codon | Comments | Change in % of codon frequency from WT to mutant | Relative activity (P value) compared toa:
|
|
|---|---|---|---|---|---|
| TH8125 in the presence of FlgMb | TH8200 in the absence of FlgMc | ||||
| TH8125 | WT | 1 | |||
| TH8200 | ΔflgM | 1 | |||
| TH8612 | C→T +10 bp | CCT→TCT P4S, domain σ2 | 7.8 to 8.5 | 2.8 (<0.001) | 1.2 (>0.05) |
| TH8613 | A→T +79 bp | ACA→TCA T27S, domain σ2 | 4.7 to 5.9 | 1.7 (0.012) | 1.6 (<0.001) |
| TH8614 | A→G +87 bp | ATA→ATG, 129M, domain σ2 | 4.2 to 23.6 | 1.4 (0.006) | 1.6 (0.003) |
| TH8615 | A→G +445 bp | AGA→GGA, R149G, domain σ3-4 linker | 2.2 to 6.3 | 1.3 (0.032) | 0.8 (0.042) |
| TH8616 | C→T +524 bp | ACG→ATG, T175M, domain σ3-4 linker | 17.1 to 23.6 | 2.9 (<0.001) | 1.6 (0.013) |
| TH8617 | A→G +566 bp | GAA→GGA, E189G, domain σ4 | 55 to 14.5 | 1.3 (0.030) | 1.1 (>0.05) |
| TH8618 | A→G +596 bp | GAA→GGA, E199G, domain σ4 | 55 to 14.5 | 1.4 (0.016) | 1.2 (>0.05) |
Relative β-galactosidase activities were calculated from Fig. 5B.
P values were determined using two-tailed t tests on β-galactosidase activities compared to TH8125.
P values were determined using two-tailed t tests on β-galactosidase activities compared to TH8200.
To examine whether the mutants were more active due to increased activity of σ28, β-galactosidase assays were performed in the absence of A. aeolicus FlgM (TH8200) (Fig. 5B). The relative activity of A. aeolicus σ28 mutants compared to WT (TH8200) ranged from 0.8- to 1.6-fold higher (Table 2). Mutants T27S, I29M, and T175M had more σ28 activity than WT (TH8200) in the absence A. aeolicus FlgM, suggesting that their ability to bypass FlgM inhibition was due to increased σ28 activity and that they are not defective for FlgM interaction. Mutants I29M and T175M had changes in codon frequency that may have resulted in increased levels of σ28 expression (Table 2). The codon frequency change of mutant T27S was nominal (Table 2), suggesting that an increased σ28 activity is not due to increased translation. The increased activity could be attributed to increased stability, as was found for the H14D mutants of S. enterica serovar Typhimurium σ28 that were able to bypass FlgM inhibition (4).
Mutation R149G had more σ28 activity than WT (TH8125) in the presence of FlgM but had less σ28 activity in the absence of FlgM (TH8200) (Fig. 5B and Table 2). This class of mutant was also found in the S. enterica serovar Typhimurium σ28 defective for FlgM interaction (4). Although they had more σ28 activity in the presence of FlgM, they were implied to have less σ28 activity in the absence of FlgM, due to amino acid substitutions that increased the turnover of σ28 (4).
Mutants P4S, E189G, and E199G have σ28 activity levels similar to that of WT (TH8200) in the absence A. aeolicus FlgM and more σ28 activity than WT (TH8125) in the presence of A. aeolicus FlgM, suggesting they are defective for FlgM interaction (Fig. 5B and Table 2). Residue E189 is buried within the σ28 structure, where there are extensive contacts with the other interacting domains (41). This mutation may be involved in stabilizing the σ28 structure in a manner that renders it resistant to FlgM inhibition through allosteric effects. In contrast, P4 lies at the N terminus of σ28, is not buried within the σ28, and appears too distant to make contacts with FlgM (41). Nonetheless, the P4S mutation could involve a conformational change to alter inhibition by FlgM or modify σ28 protein stability or interaction with RNAP.
The E199G mutation found in A. aeolicus σ28 corresponds to location E203 of S. enterica serovar Typhimurium (4). An S. enterica serovar Typhimurium σ28 E203D mutant exhibited 10-fold less affinity to S. enterica serovar Typhimurium FlgM and was the most attenuated for the destabilizing of the S. enterica serovar Typhimurium σ28-RNAP holoenzyme by S. enterica serovar Typhimurium FlgM (4). By analogy to S. enterica serovar Typhimurium σ28, the A. aeolicus σ28 E199 position is an important contact for FlgM in inhibition of σ28 interaction, with RNAP likely to be involved in the “stripping” of σ28 from the σ28-RNAP holoenzyme complex by FlgM (4, 41). The E199G mutation is located in the conserved region 4 of the σ70 family of transcription factors (31). Interestingly, many mutations isolated in S. enterica serovar Typhimurium defective in σ28/FlgM interactions mapped to σ28 region 4 (4, 20). This domain is also a target for transcriptional regulation in many other systems (12). Anti-sigma factors Rsd, SpoIIAB, and FlgM bind to region 4 of the sigma factors to inhibit its interaction with RNAP (12).
Previous structural studies showed that when FlgM is bound to σ28, it could occlude the interaction of this domain with the β-flap of RNAP that is important for proper recognition of the −35 promoter element (22, 30, 41, 45). Further, studies suggest FlgM helix H3′ interacts with region 4 (where E199 of A. aeolicus σ28 is located) on the σ28-RNAP holoenzyme complex, and either by a conformational change in the interaction with FlgM or through enzyme “breathing,” region 4 is released for binding by helix H4′ of FlgM. This inhibits region 4 interaction with the RNAP β-flap and the subsequent dissociation of σ28 from RNAP (4, 41). E199 in σ28 lies within the face of the helix in s4, where it interacts with H3′ helix of FlgM (41). Isolation of E199G in A. aeolicus σ28 that is resistant to FlgM inhibition supports this model.
Isolation of flgM mutants defective in A. aeolicus σ28/FlgM interactions.
Three hundred Tcs clones were isolated from the A. aeolicus flgM mutagenesis screen, of which 25 were Lac+ on Mac-lac indicator medium. Twelve Lac+ colonies were sequenced, and eight had either single base pair changes or deletions in FlgM (Fig. 6 and Table 3). The remaining four had multiple mutations, and they were excluded from the study. All strains with A. aeolicus flgM mutations were assayed for σ28 activity by use of β-galactosidase assays to compare the levels of fliC-lac expression with a strain harboring wild-type A. aeolicus flgM (Table 3).
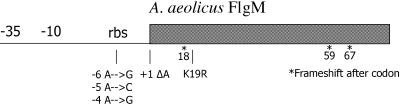
FIG. 6.
Summary of mutations isolated in A. aeolicus flgM. rbs, ribosome binding site.
TABLE 3.
Summary of mutations isolated in A. aeolicus flgM
| Strain | Mutation relative to the start codon | Commentsa | Change in % of codon frequency from WT to mutant | % fliC-lac β-galactosidase activity relative to WT | Relative activityb |
|---|---|---|---|---|---|
| TH8125 | WT | 100 | 1 | ||
| TH8200 | ΔflgM | 395 | 4.0 | ||
| TH8604 | A→G −6 bp | RBS | 116 | 1.2 | |
| TH8605 | A→C −5 bp | RBS | 316 | 3.2 | |
| TH8606 | A→G −4 bp | RBS | 95 | 0.95 | |
| TH8607 | A→G +1 bp | ATG→GTG M1V | 26.3 to 26.4 | 168 | 1.7 |
| TH8608 | A→G +56 bp | AAG→AGG K19R, rare codon | 11.7 to 1.2 | 116 | 1.2 |
| TH8609 | ΔA +55-56 bp | Frameshift after codon 18 | 395 | 4.0 | |
| TH8610 | ΔA +177 bp | Frameshift after codon 59 | 374 | 3.7 | |
| TH8611 | ΔA +199 bp | Frameshift after codon 67 | 437 | 4.4 |
RBS, ribosome binding site.
Relative activity calculated from β-galactosidase activities in column 5.
Of the mutations characterized, several were found in the ribosome binding site, one altered in the start codon, and another introduced a rare codon; all mutations presumably lowered the levels of FlgM, thereby enabling increased σ28 activity (Fig. 6 and Table 3). Three mutations resulted in frameshifts in FlgM after codon positions 18, 59, and 67. Frameshift mutation after codon 67 suggests that the last 21 amino acids in FlgM are important for inhibition of σ28 activity. Previous studies of FlgM show the C terminus is important for σ28 interaction (9, 41). Alternatively, the resulting frameshift mutation could destabilize FlgM protein; therefore, additional biochemical methods are needed to support the in vivo findings.
We found a bias in favor of A. aeolicus flgM null mutant isolation, because the screen was based on the loss of anti-σ28 activity. Previous isolations of FlgM mutants of S. enterica serovar Typhimurium defective in interaction with S. enterica serovar Typhimurium σ28 indicated a bias against flgM null alleles. This is because S. enterica serovar Typhimurium strains carrying flgM null alleles and remaining fliA+ are sick, and excess σ28 activity is lethal in S. enterica serovar Typhimurium. The FlgM mutants obtained were predominantly single amino acid substitution mutants that retained some anti-σ28 activity (5, 9). Since it was easy to isolate flgM null alleles of A. aeolicus in a strain expressing A. aeolicus σ28, we conclude that excess A. aeolicus σ28 activity, unlike excess S. enterica serovar Typhimurium σ28 activity, does not inhibit cell growth.
DISCUSSION
We have demonstrated the use of S. enterica serovar Typhimurium as an in vivo heterologous system to assess the sigma factor activities of homologs from other genetically intractable species, such as A. aeolicus and C. trachomatis. We demonstrate that the λ-Red technology provides a simple method to replace S. enterica serovar Typhimurium genes with functional homologues from bacterial species that are normally genetically intractable. Chromosomal replacement of both the A. aeolicus and C. trachomatis fliA+ genes complemented an S. enterica serovar Typhimurium fliA mutant for motility and production of FliC flagellin. While complementation by A. aeolicus fliA+ was not as robust as that with C. trachomatis fliA+, these assays were done without the aid of a rare-codon tRNA plasmid that was required to obtain large amounts of A. aeolicus FliA from expression in E. coli for crystallization (41). Indeed, by not using a rare-codon tRNA plasmid in these studies, the sensitivity of our genetic selection allowed for the isolation of a class of A. aeolicus fliA mutants with increased efficiency of translation. In addition, we isolated mutations in A. aeolicus of the class expected to be defective for σ28/FlgM interactions: the increase in class 3 transcription was only in the presence of FlgM and not in the absence of FlgM.
We found that FlgM inhibition of S. enterica serovar Typhimurium and A. aeolicus σ28-dependent class 3 transcription was species specific. A. aeolicus FlgM would inhibit only A. aeolicus σ28 activity and not S. enterica serovar Typhimurium σ28 activity; S. enterica serovar Typhimurium FlgM would inhibit only S. enterica serovar Typhimurium σ28 activity and not A. aeolicus σ28 activity. We also tested the possibility that RsbW was a negative regulator of C. trachomatis σ28, as had been suggested previously (39). However, we found that RsbW did not inhibit C. trachomatis σ28 in S. enterica serovar Typhimurium. Either RsbW does not inhibit C. trachomatis σ28 activity or other factors are required for such an activity to be observed.
The ability to replace the S. enterica serovar Typhimurium fliA and flgM genes with homologous genes of genetically intractable organisms allowed us to use standard Salmonella genetic methods to study interactions of sigma and anti-sigma factors from bacterial species that are otherwise difficult to analyze genetically. It also provided an additional benefit in that plasmid artifacts are avoided when such genes are expressed from the normal S. enterica serovar Typhimurium chromosomal location. We also demonstrate the ability to use λ-Red recombination to replace any gene to be targeted for mutagenesis with the tetRA element. The tetRA element is an excellent tool for targeted mutagenesis because of the ability to select for inheritance of (Tcr) and replacement of (Tcs) the tetRA element. This general strategy could be employed with any bacterial species where the λ-Red system can be expressed and is functional, and a selectable/counterselectable marker(s), such as tetRA or sacB linked to a drug-resistance gene, can be used to perform targeted mutagenesis.
Acknowledgments
We thank Karl O. Stetter, Thomas P. Hatch, and You-xun Zhang for the kind gifts of A. aeolicus genomic DNA, pET28a-rsbW C. trachomatis L2, and pLF28, respectively. We also thank Meg Chadsey for construction of pMC147. We are grateful to Lakshmi Rajagopal, Amy Siegesmund, and members of the Hughes laboratory for critical reading of the manuscript.
This work was supported by funding from PSH grant GM56141 from the National Institutes of Health.
REFERENCES
- 1.Aldridge, P., and K. T. Hughes. 2002. Regulation of flagellar assembly. Curr. Opin. Microbiol. 5:160-165. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein, H. J. 2000. Recent changes to RasMol, recombining the variants. Trends Biochem. Sci. 25:453-455. [DOI] [PubMed] [Google Scholar]
- 3.Blair, D. F. 1995. How bacteria sense and swim. Annu. Rev. Microbiol. 49:489-522. [DOI] [PubMed] [Google Scholar]
- 4.Chadsey, M. S., and K. T. Hughes. 2001. A multipartite interaction between Salmonella transcription factor sigma28 and its anti-sigma factor FlgM: implications for sigma28 holoenzyme destabilization through stepwise binding. J. Mol. Biol. 306:915-929. [DOI] [PubMed] [Google Scholar]
- 5.Chadsey, M. S., J. E. Karlinsey, and K. T. Hughes. 1998. The flagellar anti-sigma factor FlgM actively dissociates Salmonella typhimurium sigma28 RNA polymerase holoenzyme. Genes Dev. 12:3123-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clamp, M., J. Cuff, S. M. Searle, and G. J. Barton. 2004. The Jalview Java alignment editor. Bioinformatics 20:426-427. [DOI] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daughdrill, G. W., M. S. Chadsey, J. E. Karlinsey, K. T. Hughes, and F. W. Dahlquist. 1997. The C-terminal half of the anti-sigma factor, FlgM, becomes structured when bound to its target, sigma 28. Nat. Struct. Biol. 4:285-291. [DOI] [PubMed] [Google Scholar]
- 10.Davis, R. W., D. Botstein, J. R. Roth, and Cold Spring Harbor N.Y. Laboratory of Quantitative Biology. 1980. Advanced bacterial genetics: a manual for genetic engineering. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 11.Deckert, G., P. V. Warren, T. Gaasterland, W. G. Young, A. L. Lenox, D. E. Graham, R. Overbeek, M. A. Snead, M. Keller, M. Aujay, R. Huber, R. A. Feldman, J. M. Short, G. J. Olsen, and R. V. Swanson. 1998. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature 392:353-358. [DOI] [PubMed] [Google Scholar]
- 12.Dove, S. L., S. A. Darst, and A. Hochschild. 2003. Region 4 of sigma as a target for transcription regulation. Mol. Microbiol. 48:863-874. [DOI] [PubMed] [Google Scholar]
- 13.Gillen, K. L., and K. T. Hughes. 1991. Negative regulatory loci coupling flagellin synthesis to flagellar assembly in Salmonella typhimurium. J. Bacteriol. 173:2301-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes, K. T., K. L. Gillen, M. J. Semon, and J. E. Karlinsey. 1993. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science 262:1277-1280. [DOI] [PubMed] [Google Scholar]
- 16.Hughes, K. T., and K. Mathee. 1998. The anti-sigma factors. Annu. Rev. Microbiol. 52:231-286. [DOI] [PubMed] [Google Scholar]
- 17.Josenhans, C., and S. Suerbaum. 2002. The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291:605-614. [DOI] [PubMed] [Google Scholar]
- 18.Karlinsey, J. E., S. Tanaka, V. Bettenworth, S. Yamaguchi, W. Boos, S. I. Aizawa, and K. T. Hughes. 2000. Completion of the hook-basal body complex of the Salmonella typhimurium flagellum is coupled to FlgM secretion and fliC transcription. Mol. Microbiol. 37:1220-1231. [DOI] [PubMed] [Google Scholar]
- 19.Kirov, S. M. 2003. Bacteria that express lateral flagella enable dissection of the multifunctional roles of flagella in pathogenesis. FEMS Microbiol. Lett. 224:151-159. [DOI] [PubMed] [Google Scholar]
- 20.Kutsukake, K., S. Iyoda, K. Ohnishi, and T. Iino. 1994. Genetic and molecular analyses of the interaction between the flagellum-specific sigma and anti-sigma factors in Salmonella typhimurium. EMBO J. 13:4568-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kutsukake, K., Y. Ohya, and T. Iino. 1990. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J. Bacteriol. 172:741-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuznedelov, K., L. Minakhin, A. Niedziela-Majka, S. L. Dove, D. Rogulja, B. E. Nickels, A. Hochschild, T. Heyduk, and K. Severinov. 2002. A role for interaction of the RNA polymerase flap domain with the sigma subunit in promoter recognition. Science 295:855-857. [DOI] [PubMed] [Google Scholar]
- 23.Laskowski, R. A., V. V. Chistyakov, and J. M. Thornton. 2005. PDBsum more: new summaries and analyses of the known 3D structures of proteins and nucleic acids. Nucleic Acids Res. 33:D266-D268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lonetto, M., M. Gribskov, and C. A. Gross. 1992. The sigma 70 family: sequence conservation and evolutionary relationships. J. Bacteriol. 174:3843-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macnab, R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 26.Macnab, R. M. 1999. The bacterial flagellum: reversible rotary propellor and type III export apparatus. J. Bacteriol. 181:7149-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maloy, S. R. 1990. Experimental techniques in bacterial genetics. Jones and Bartlett, Boston, Mass.
- 28.Maloy, S. R., and W. D. Nunn. 1981. Selection for loss of tetracycline resistance by Escherichia coli. J. Bacteriol. 145:1110-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsudaira, P. 1987. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem. 262:10035-10038. [PubMed] [Google Scholar]
- 30.Murakami, K. S., S. Masuda, E. A. Campbell, O. Muzzin, and S. A. Darst. 2002. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science 296:1285-1290. [DOI] [PubMed] [Google Scholar]
- 31.Paget, M. S., and J. D. Helmann. 2003. The sigma70 family of sigma factors. Genome Biol. 4:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poteete, A. R. 2001. What makes the bacteriophage lambda Red system useful for genetic engineering: molecular mechanism and biological function. FEMS Microbiol. Lett. 201:9-14. [DOI] [PubMed] [Google Scholar]
- 33.Ramos, H. C., M. Rumbo, and J. C. Sirard. 2004. Bacterial flagellins: mediators of pathogenicity and host immune responses in mucosa. Trends Microbiol. 12:509-517. [DOI] [PubMed] [Google Scholar]
- 34.Rappleye, C. A., and J. R. Roth. 1997. A Tn10 derivative (T-POP) for isolation of insertions with conditional (tetracycline-dependent) phenotypes. J. Bacteriol. 179:5827-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., and W. R. David. 2001. Molecular cloning: a laboratory manual, 3rd. ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Sayle, R. A., and E. J. Milner-White. 1995. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 20:374. [DOI] [PubMed] [Google Scholar]
- 37.Schachter, J. 1999. Infection and disease epidemiology, p. 139-170. In R. S. Stephens (ed.), Chlamydia: intracellular biology, pathogenesis, and immunity. ASM Press, Washington, D.C.
- 38.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 39.Shen, L., M. Li, and Y. X. Zhang. 2004. Chlamydia trachomatis sigma28 recognizes the fliC promoter of Escherichia coli and responds to heat shock in chlamydiae. Microbiology 150:205-215. [DOI] [PubMed] [Google Scholar]
- 40.Sobolev, V., A. Sorokine, J. Prilusky, E. E. Abola, and M. Edelman. 1999. Automated analysis of interatomic contacts in proteins. Bioinformatics 15:327-332. [DOI] [PubMed] [Google Scholar]
- 41.Sorenson, M. K., S. S. Ray, and S. A. Darst. 2004. Crystal structure of the flagellar sigma/anti-sigma complex sigma(28)/FlgM reveals an intact sigma factor in an inactive conformation. Mol. Cell 14:127-138. [DOI] [PubMed] [Google Scholar]
- 42.Starnbach, M. N., and S. Lory. 1992. The fliA (rpoF) gene of Pseudomonas aeruginosa encodes an alternative sigma factor required for flagellin synthesis. Mol. Microbiol. 6:459-469. [DOI] [PubMed] [Google Scholar]
- 43.Stephens, R. S. (ed.). 1999. Chlamydia: intracellular biology, pathogenesis, and immunity. ASM Press, Washington, D.C.
- 44.Studholme, D. J., and M. Buck. 2000. The alternative sigma factor sigma(28) of the extreme thermophile Aquifex aeolicus restores motility to an Escherichia coli fliA mutant. FEMS Microbiol. Lett. 191:103-107. [DOI] [PubMed] [Google Scholar]
- 45.Vassylyev, D. G., S. Sekine, O. Laptenko, J. Lee, M. N. Vassylyeva, S. Borukhov, and S. Yokoyama. 2002. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature 417:712-719. [DOI] [PubMed] [Google Scholar]
- 46.Wadhams, G. H., and J. P. Armitage. 2004. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 5:1024-1037. [DOI] [PubMed] [Google Scholar]
- 47.Way, J. C., M. A. Davis, D. Morisato, D. E. Roberts, and N. Kleckner. 1984. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene 32:369-379. [DOI] [PubMed] [Google Scholar]
- 48.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]