Abstract
Chlamydia trachomatis is an intracellular bacterium responsible for ocular, respiratory, and sexually transmitted diseases. The genome contains a nine-member polymorphic membrane protein (Pmp) family unique to members of the order Chlamydiales. Genomic and molecular analyses were performed for the entire pmp gene family for the 18 reference serological variants (serovars) and genovariant Ja to identify specific gene and protein regions that differentiate chlamydial disease groups. The mean genetic distance among all serovars varied from 0.1% for pmpA to 7.0% for pmpF. Lymphogranuloma venereum (LGV) serovars were the most closely related for the pmp genes and were also the most divergent, compared to ocular and non-LGV urogenital disease groups. Phylogenetic reconstructions showed that for six of nine pmp genes (not pmpA, pmpD, or pmpE), the serovars clustered based on tissue tropism. The most globally successful serovars, E and F, clustered distantly from the urogenital group for five pmp genes. These pmp genes may confer a biologic advantage that may facilitate infection and transmission for E and F. Surprisingly, serovar Da clustered with the ocular group from pmpE to pmpI, which are located together in the chromosome, providing statistically significant evidence for intergenomic recombination and acquisition of a genetic composition that could hypothetically expand the host cell range of serovar Da. We also identified distinct domains for pmpE, pmpF, and pmpH where substitutions were concentrated and associated with a specific disease group. Thus, our data suggest a possible structural or functional role that may vary among pmp genes in promoting antigenic polymorphisms and/or diverse adhesions-receptors that may be involved in immune evasion and differential tissue tropism.
Chlamydia trachomatis is the most common sexually transmitted bacterial pathogen worldwide (5, 51). Genital infections caused by this obligate intracellular pathogen may lead to significant long-term complications such as ectopic pregnancy, tubal factor infertility, and chronic pelvic pain (7), as well as facilitate the transmission of human immunodeficiency virus (14). The differential immunoreactivity of mono- and polyclonal antibodies directed against epitopes on the major outer membrane protein has identified 18 distinct serotypes. Serotyping correlates well with genotyping, which is based on ompA, the gene that encodes major outer membrane protein (10, 48, 49). Serotyping groups chlamydial strains into three classes: B class (serovars B, D, E, L1, L2, and L2a), C class (A, C, H, I, Ia, J, Ja, K, and L3), and intermediate class (F and G). However, these classes do not correlate with tissue tropism or biologic properties of the organism, although individual serotypes have a propensity for certain tissues and diseases. For example, serovars A, B, Ba, and C are associated with trachoma, a chronic ocular disease, while serovars Ba, D to K, Da, and Ia are responsible for urogenital infections. However, these serovars may also cause conjunctivitis and infant pneumonia. The lymphogranuloma venereum (LGV) serovars, L1 to L3 and L2a, cause more invasive urogenital diseases (7, 13).
Seven chlamydial genomes have been sequenced over the last 7 years, revealing a polymorphic membrane protein (Pmp) family unique to Chlamydiales that likely plays an important role in chlamydial biology and disease pathogenesis (46). There are 17 pmp genes identified for Chlamydophila caviae (formerly Chlamydia psittaci; strain GPIC) (36), 21 for Chlamydia pneumoniae (strain CWL029) (21), and 9 for Chlamydia trachomatis (serovar D/UW-3 and mouse pneumonitis strain Nigg) (35, 42). The pmp genes are predicted to be localized in the outer membrane, due to the presence of a C-terminal phenylalanine, a cleavable peptide leader sequence (15, 18, 39, 44), and the fact that all Pmps have been detected by proteomics as membrane constituents, at least for serovar L2 (40, 41, 45). The C. trachomatis Pmps account for 3.15% of the organism's coding capacity (18) and are immunogenic for humans (16, 17, 20, 23). By protein structure analysis, they are predicted to be autotransporters (19, 47) that mediate the translocation of the N terminus to the bacterial surface (19, 47). They also contain multiple GGAI motifs, which have been associated in other organisms with adhesion to the host cell (18).
We recently performed statistical modeling and phylogenetic analyses of 18 reference serovars and genovariant Ja for pmpC (15) and included the partial sequences of pmpE, pmpH, and pmpI for 15 serovars that were available from another study (43). pmpC and pmpH were found to group serovars according to cell tropism properties, including a distinct clade for the most prevalent serovars, E and F. pmpC also revealed remnants of two putative insertion sequence (IS)-like elements differentially present in some serovars, suggesting a role in genome plasticity (15). Because of these findings, the goal of the present study was to sequence the remaining five pmp genes (pmpA, -B, -D, -F, and -G) for all 18 C. trachomatis serovars and genovariant Ja to completion and provide a detailed genomics and molecular analysis, including phylogenetics of the entire pmp family. We also identified specific protein regions that differentiate disease groups, as these may be responsible for significant tissue tropism properties of the organism.
MATERIALS AND METHODS
Cell culture and genotyping of C. trachomatis reference serovars.
The C. trachomatis reference strains (18 serovars and genovariant Ja) were identical to those used in our previous pmp gene studies (15, 16) to permit comparisons with previous data: A/SA-1, B/TW-5, Ba/Apache-2, C/TW-3, D/UW-3, Da/TW-448, E/Bour, F/IC-Cal3, G/UW-57, H/UW-4, I/UW-12, Ia/IU-4168, J/UW-36, Ja/UW-92, K/UW-31, L1/440, L2/434, L2a/TW-396, and L3/404. HeLa cells were grown in T-150-cm2 flasks as previously described (11, 12) and were inoculated with the respective serovar in SPG (0.25 M sucrose, 10 mM Na2HPO4, 5 mM l-glutamic acid, 10-μg/ml gentamicin, 100-μg/ml vancomycin, 25-U/ml nystatin [pH 7.4]) at room temperature for 2 h on an orbital shaker. Flasks were incubated in minimal essential medium (Gibco-Invitrogen Corporation, Carlsbad, CA) supplemented with 0.5-μg/ml cycloheximide and 10% fetal bovine serum (HyClone, Logan, Utah) at 37°C in 5% CO2. Chlamydiae were harvested at 48 to 72 h postinoculation and purified by discontinuous density centrifugation in Renografin (4), resuspended in SPG, and stored at −80°C.
Serovar confirmation of each reference strain was indirectly performed by ompA sequencing and further nucleotide comparison with available GenBank data as previously described (15). Briefly, DNA was extracted and amplified using 1× OptiBuffer (Bioline, London, United Kingdom), 2.8 mM MgCl2, 200 μM deoxynucleoside triphosphates (Promega, Madison, WI), 25 pmol of each primer, 1.5 U Bio-X-Act Proof-reading DNA Polymerase (Bioline), and 5 μl of strain DNA, in a final volume of 25 μl. PCR was run in a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA) with the following thermocycling profile: 1 min at 95°C; 1 min at 50°C; 30 cycles, each consisting of 1 min at 70°C, 30 s at 95°C, and 30 s at 50°C; and a final elongation step of 10 min at 70°C. The amplicons were gel purified and sequenced as previously described (15) using the BigDye Terminator Cycle Sequencing kit, version 1.1, according to the manufacturer's instructions (Applied Biosystems) in an ABI 3700 automated sequencer (Applied Biosystems). The sequences were compared with available ompA chlamydial sequences from GenBank using MegAlign software (DNASTAR, Madison, WI) to identity and confirm each ompA genotype.
Amplification and sequencing of pmpA, pmpB, pmpD, pmpF, and pmpG for each serovar.
PCR primers for pmpA, pmpB, pmpD, pmpF, and pmpG (Table 1) were designed with Primer Select software (DNASTAR) based on already available GenBank sequences of the respective pmp genes and intergenomic regions for serovar D (UW-3; GenBank no. AE001273). Due to the large size of pmpB (5,256 bp) and pmpD (4,596 bp), two PCR primer pairs (Table 1) were designed for each to generate two overlapping amplicons for the entire gene. PCR reagents and thermocycling profiles were the same as described for ompA, except for the primers (Table 1), annealing temperature, and extension time: 57°C and 4 min for PmpA-1/PmpA-2, 57°C and 3 min 20 s for PmpB-1/PmpB-2 and PmpB-3/PmpB-4, 56°C and 2 min for PmpD-5/PmpD-6, and 56°C and 4 min for PmpD-9/PmpD-10, PmpF-1/PmpF-2, and PmpG-5/PmpG-6. Amplicons were visualized in ethidium bromide-stained 0.8% agarose gels, purified, and sequenced as described above for ompA.
TABLE 1.
Oligonucleotide primers used for PCR and sequencing
| Gene | Primer | Primer sequence (5′ to 3′) | Gene locationa | Amplicon size (bp) |
|---|---|---|---|---|
| pmpA | PmpA-1b | TGCCCTCGCCAAATGTGGAACT | (−)406-(−)385c | 3,552 |
| PmpA-2b | CCGCAGTAGCTGACAGCCATTTCAT | (+)163-(+)139d | ||
| A-3e | CTGTTTATAGTTCCCGTTCA | (−)56-(−)37d | ||
| A-5e | ATCCTTTGTAGATCAACGTG | 600-619 | ||
| A-7e | ATCCAACTTGCAGCGAACAA | 1270-1289 | ||
| A-6e | AAGTGATGTGTTCTGGTAAG | 2308-2289 | ||
| A-4e | TACTAGGAAGATACAGAGGT | (+)50-(+)31d | ||
| pmpB | PmpB-1b | CTGCGGAGGCTATGTTGGCTTCT | (−)163-(−)141c | 2,914 |
| PmpB-2b | TCCTCCCGCTATTTGCCCTGTT | 2751-2729 | ||
| PmpB-3b | AATCCCTCCCAATCATCGTTGAAC | 2595-2618 | 2,911 | |
| PmpB-4b | TCTTGGATCGAGCTCGCCTCAG | (+)251-(+)229d | ||
| B-5e | CCTTAACTCTGTCTGAGATA | 380-400 | ||
| B-9e | TTCAATCTTCTAAACAGAGT | 1000-1020 | ||
| B-6e | CAGAGATATTCAGTTGGTCT | 2209-2189 | ||
| B-7e | GATTGAGTCTTTAGGATCTG | 3188-3208 | ||
| B-10e | TATCTAACCAAAACATAGCT | 3761-3781 | ||
| B-8e | TAAGATCCATAGAGATACGA | 4814-4794 | ||
| pmpD | PmpD-5b | ACGCGTTGTTGGACCGATTAT | (−)67-(−)47c | 1,817 |
| PmpD-6b | ATACTACTGCAGCGTTGTGGAGAA | 1750-1727 | ||
| PmpD-9b | GAATTTAGGCGCGATTTCGTTCTC | 1353-1376 | 3,563 | |
| PmpD-10b | TTTGCCATTTCGATATCTTTCGTGA | (+)318-(+)294d | ||
| D-4e | CGCCTCCTTGCGAAGCAGACT | 1117-1097 | ||
| D-11e | TTCAGAGATAATCGAGGGAG | 2044-2063 | ||
| D-13e | GAATGCTACGGAAGGACATG | 2820-2839 | ||
| D-12e | CGACATAAGGATTGAAATGC | 4183-4164 | ||
| pmpE | PmpE-2b | AACGGTTCTTGGGTCACCACAAAT | (−)173-(−)149c | 3,090 |
| PmpE-1b | GGCCGGAGCCCCCTCTATTAC | (+)22-(+)2d | ||
| E-4e | AGCGAGAATCAGTCTTGTTT | 574-593 | ||
| E-3e | GTTTGAATACAGATGTTGTCC | 623-603 | ||
| E-5e | CTTCAATAAGGAAGCTGATC | 1257-1276 | ||
| E-6e | AAACTCAAGATCCAGAACCA | 1850-1869 | ||
| E-7e | ATCAACGCATATACTGACAG | 2466-2485 | ||
| pmpF | PmpF-2b | CTTATCTTCAGCGCATTCGTCCTTC | (−)91-(−)67c | 3,664 |
| PmpF-1b | GAATGGCTCCGCCTTCTCTTATTTT | (+)468-(+)444d | ||
| F-7e | GCATTCGTCCTTCTCTCTTT | (−)80-(−)60c | ||
| F-4e | TGACAGGGAATCGAACTCT | 513-543 | ||
| F-6e | AAATTATATCCATACAGGGA | 1092-1112 | ||
| F-8e | TTCACTAGTTTTTTTCTGCT | 2041-2021 | ||
| F-5e | GAGTGAAATTTCCCTTTAGA | 2630-2610 | ||
| F-3e | ATTCTAGAAGGAACCTCTCT | (+)76-(+)56d | ||
| pmpG | PmpG-5b,f | AGGCCGTGAATGAG(G/A)TAGTAACCATA | (−)480-(−)454d | 3,704 |
| PmpG-6b | AAAGCGCAGTCTCCATTCAAGTAAAC | (+)182-(+)156d | ||
| G4e | CGAATAGGAACACAAACAAG | (+)90-(+)70d | ||
| G9e | ATGAGGTTTTCATATGCTGG | 2524-2505 | ||
| G9-Be | ATTCCCAAACCCGTAGCTA | 2505-2487 | ||
| G10e | CTAGCCAATCATACCTATCA | 1900-1881 | ||
| G11e | ATCAGCAGATAAACTGAGCT | 1254-1235 | ||
| G12e | GCAGTATTTTCTTGGAAGAC | 638-619 | ||
| pmpH | PmpH-3b | ACCGCGATGCTTTAGGTCAGG | (−)819-(−)799c | 3,982 |
| PmpH-4b | TTTCTATACCATCCGCCCTCTCCT | (+)114-(+)90d | ||
| H-5e | TTGCGGAGAAAAGGGAATGA | 351-370 | ||
| H-2e | CTTCGCCTGCTCCGGAAATACTC | 409-387 | ||
| H-6e | GAACACAGCATACGATGATT | 825-844 | ||
| H-7e | TAATCAGTCCGGTAATGGAT | 1428-1447 | ||
| H-8e | TCTGCAGATTGGACGAAAC | 1954-1972 | ||
| H-9e | CATTCGAAATGTCATTGGCA | 2527-2546 | ||
| pmpI | PmpI-14b | GACTATTCCCGTTGCCTCCTACTC | (−)312-(−)288c | 3,181 |
| PmpI-13b | GCTGGTTTGATGTTGGACTATTGG | (+)234-(+)210d | ||
| I-2e | AACAAACCAACCCAAAACTAAAAT | (−)273-(−)250c | ||
| I-7e | CGCAATAATGGTAGCATGAG | 391-410 | ||
| I-11e | CGGGGCTATTTATGCCAAGCACAT | 798-821 | ||
| I-12e | AAAGCGTAACTATCCACGC | 1433-1451 |
Primer locations are based on the sequence of strain D for the respective pmp gene (GenBank no. AE001273).
PCR primers that were used also for automated sequencing.
(−), region upstream from the start codon.
(+), region downstream from the stop codon.
Primers that were used only for automated sequencing.
The degeneration (G/A) of this primer was due to the polymorphism presented by different chlamydial serovars.
The sequences were compared with the corresponding pmp of serovar D/UW-3 using MegAlign (DNASTAR). Nucleotide changes were confirmed by reamplifying the original genomic DNA and by resequencing the new amplicons so that mismatches that may have occurred during the previous PCR could be detected. We found this strategy to be more accurate than exclusively sequencing both DNA strands of each pmp derived from a single PCR-sequencing set.
Amplification and sequencing of pmpE, pmpH, and pmpI for serovars Da, Ia, and L2a and genovariant Ja.
Partial sequences of pmpE, pmpH, and pmpI were previously available for 15 serovars (excluding Da, Ia, and L2a and genovariant Ja) (43). These sequences did not include ∼80 bases at the N terminus and ∼20 bases at the C terminus for pmpE and pmpI. We had previously sequenced the N terminus of pmpH for the 15 serovars (15). All serovars, including Da, Ia, and L2a, and genovariant Ja were sequenced to completion for these three pmp genes as described above. In addition, prior to the GenBank release of the partial pmpI sequence, our laboratory had sequenced pmpI for all 18 serovars and Ja. Therefore, our sequences for pmpI were used in this study.
PCR reagents and thermocycling profiles were the same as described above except for primers (Table 1), annealing temperature, and extension time: 57°C and 3 min 30 s for PmpE-1/PmpE-2, 57°C and 4 min 30 s for PmpH-3/PmpH-4, and 56°C and 3 min 30 s for PmpI-13/PmpI-14. The sequencing strategy was identical to that described above.
Genomic analysis of the pmp family for all known serovars and genovariant Ja.
The genomic analysis of all pmp genes was performed using Molecular Evolutionary Genetics Analysis software, version 2.1 (MEGA; Institute of Molecular Evolutionary Genetics, Pennsylvania State University; http://www.megasoftware.net) as previously described (15). Pairwise comparisons were performed between individual serovar sequences, between group means where sequences were clustered according to disease properties (ocular, genital, and LGV), and between serovar sequences within each of these groups. Briefly, mean genetic distances and pairwise comparisons were based on the number of nucleotide differences including both transitions and transversions where only valid sites were compared (gaps were excluded). The genetic distance evaluation was also done using the p-distance method that calibrates the obtained differences relative to the total number of sites under comparison that varied from 2,637 for pmpI to 5,283 for pmpC. A similar strategy was applied to the respective proteins where distances were evaluated based on all amino acid changes and respective protein lengths. For the protein analysis, the total number of valid sites under comparison varied from 878 for PmpI to 1,758 for PmpC.
To study the molecular evolution of the pmp family, a bootstrap test of phylogeny was conducted where neighbor-joining trees (38) were created using the Kimura two-parameter model (22), which takes into account transitional and transversional substitution rates while assuming that the four nucleotide frequencies are the same and that rates of substitution do not vary among sites (30). Phylogenetic reconstructions were also done at the protein level using the gamma distance model, which takes into account the inequality of the substitution rates among sites (30).
Another molecular evolution study was performed where each member of the pmp family was studied by the Nei-Gojobori method (29), where the ratio of nonsynonymous (dN) to synonymous (dS) substitutions was calculated. The p-distance option was used to normalize the nonsynonymous and synonymous differences against the number of potential nonsynonymous and synonymous sites (the latter is much smaller than the former, as substitutions occurring in the first two bases of each codon usually yield a residue substitution). For short genetic distances (e.g., on the order of 10% or less) as for our data, p-distances are nearly indistinguishable from other genetic distances. Also, considering the different protein regions that fit the autotransporter model proposed for some Pmps (see the introduction), this method was also applied separately to the 3′ end of each pmp (corresponding to the 40-kDa C-terminal domain) and to the 5′ half (defined by us to involve all conserved GGAI motifs) to evaluate possible evolutionary differences between these two major protein domains. Significant differences in mean dN and dS substitutions were determined by comparing 95% confidence intervals.
For all bioinformatics tests, a standard error (SE) computation was performed by bootstrap analysis with 1,000 replications.
A search for specific pmp regions that phylogenetically define tropism differences among serovars was also conducted: pmp genes with phylogenetic reconstructions showing serovars belonging to a specific disease group clustered in an evolutionary branch that was distant from the other serovars were subjected to a SimPlot software analysis (http://sray.med.som.jhmi.edu/SCRoftware/simplot/) (24). In SimPlot, serovars were grouped according to disease properties, followed by nucleotide pairwise comparison between the mean of the three groups using the Jukes-Cantor (one-parameter) correction for superimposed substitutions (gaps were included), yielding a graphic representation of nucleotide changes relative to their gene positions.
Analysis of protein features of the pmp family.
Based on either presumptive (15, 18, 42) or experimentally demonstrated (28, 40, 41, 45) membrane location of Pmps for C. trachomatis (in most studies, only one serovar was used), several features that are characteristic of membrane proteins were studied for the entire Pmp family for all 18 serovars and genovariant Ja, namely, the presence of a cleavable peptidase signal, the presence of a C-terminal phenylalanine, and the distribution of tryptophan residues. Briefly, the first was evaluated using SignalIP 3.0 server software (1, 31) for prediction of cleavage sites and a signal peptide based on a combination of several artificial neural networks and hidden Markov models (32). The second and the third were determined by using EditSeq software (DNASTAR).
A search for cysteine residues was also performed, since they appeared to be nonrandomly distributed for some Pmps previously studied (15); in addition, disulfide bonds can hinder autotransporter function (19). The distribution of the tetrapeptide GGAI motif, which is characteristic of the Pmp family, throughout the protein was also examined. These two analyses were performed with EditSeq software (DNASTAR).
Nucleotide sequence accession numbers.
The sequences of C. trachomatis pmp genes determined in this study were submitted to GenBank under accession numbers AY884090 through AY884108 (for pmpA), AY884109 through AY884127 (for pmpB), AY299408 through AY299426 (for pmpD), AY967735 through AY967738 (for pmpE), AY887644 through AY887662 (for pmpF), AY967739 through AY967757 (for pmpG), AY967759 through AY967761 (for pmpH), and AY299427 through AY299445 (for pmpI).
RESULTS
Genomic analysis of the pmp family.
The sequences from all pmp genes for serovar D/UW-3 analyzed in this study were identical to the corresponding pmp sequences for serovar D/UW-3 deposited at GenBank (GenBank no. AE001273).
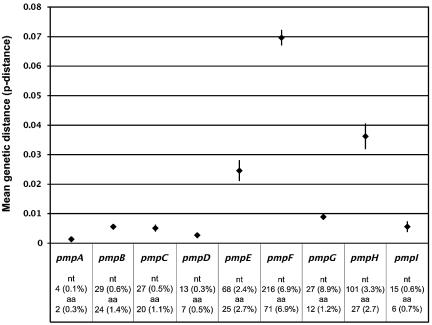
The mean genetic distance between all C. trachomatis serovars (Table 2) varied from the least polymorphic at 4.0 (SE, 1.2) (0.1%) nucleotides (nt) and 2.4 (SE, 0.9) (0.2%) amino acid differences for pmpA, to the most polymorphic at 215.5 (SE, 8.1) (7.0%) nucleotide and 71.4 (SE, 4.9) (6.9%) amino acid differences for pmpF (Fig. 1). The analysis of the genetic distance within each disease group showed that the LGV serovars were the most closely related for all pmp genes (except for pmpB and pmpE) with, in general, ≤0.1% nucleotide differences; four Pmps (PmpA, PmpD, PmpF, and PmpI) had 100% similarity among LGV serovars (Table 2). The urogenital group was the most heterogeneous, in particular for pmpF, which showed 3.3% nucleotide and amino acid differences among urogenital serovars.
TABLE 2.
Average number of replacements observed and percent genetic distances of the nine pmp genes of C. trachomatis for all 18 serovars and the genovariant Jaa
| Gene | Mean | Within-group means (SE; % change)
|
Between-group means (SE; % change)
|
||||
|---|---|---|---|---|---|---|---|
| Ocular | Genital | LGV | Ocular/genital | Ocular/LGV | Genital/LGV | ||
| pmpA | 4.0 (1.1; 0.1) | 2.8 (1.2; 0.1) | 3.5 (1.2; 0.1) | 0.5 (0.5; <0.1) | 4.2 (1.1; 0.1) | 6.0 (2.0; 0.2) | 4.3 (1.5; 0.2) |
| 2.4 (0.9; 0.2) | 2.8 (1.2; 0.3) | 2.0 (0.9; 0.2) | 0.0 (0.0; 0.0) | 3.1 (1.0; 0.3) | 3.8 (1.6; 0.4) | 1.9 (1.0; 0.1) | |
| pmpB | 29.2 (3.1; 0.6) | 1.2 (0.8; <0.1) | 14.8 (2.4; 0.3) | 16.3 (3.0; 0.3) | 13.4 (2.3; 0.3) | 59.8 (6.9; 1.2) | 57.4 (6.5; 1.1) |
| 24.1 (2.7; 1.4) | 1.2 (0.8; <0.1) | 10.4 (2.0; 0.6) | 12.2 (2.5; 0.7) | 10.5 (2.2; 0.6) | 52.5 (6.9; 3.0) | 49.1 (6.1; 2.8) | |
| pmpC | 26.8 (3.2; 0.5) | 4.5 (1.5; <0.1) | 20.7 (3.2; 0.4) | 5.3 (1.6; 0.1) | 21.5 (3.6; 0.4) | 41.8 (6.5; 0.8) | 40.3 (5.2; 0.8) |
| 20.0 (2.8; 1.1) | 3.0 (1.2; 0.2) | 15.0 (2.7; 0.9) | 4.2 (1.5; 0.2) | 15.3 (2.7; 0.9) | 32.8 (5.7; 1.9) | 30.7 (4.8; 1.8) | |
| pmpD | 12.7 (1.9; 0.3) | 7.0 (1.7; 0.2) | 10.7 (1.7; 0.2) | 1.0 (0.6; <0.1) | 12.0 (2.0; 0.3) | 15.5 (3.4; 0.3) | 17.2 (3.1; 0.4) |
| 7.4 (1.7; 0.5) | 5.3 (1.6; 0.4) | 6.0 (1.5; 0.4) | 0.0 (0.0; 0.0) | 7.2 (1.6; 0.5) | 11.0 (3.2; 0.7) | 9.5 (2.8; 0.6) | |
| pmpE | 67.6 (4.9; 2.4) | 4.3 (1.4; 0.2) | 49.2 (4.4; 1.8) | 4.5 (1.4; 0.2) | 108.4 (8.1; 3.9) | 116.0 (9.4; 4.2) | 49.5 (4.9; 1.8) |
| 24.5 (2.7; 2.7) | 2.7 (1.1; 0.3) | 15.8 (2.2; 1.7) | 1.5 (0.8; 0.2) | 39.5 (5.1; 4.3) | 46.0 (6.2; 5.0) | 18.6 (2.9; 2.0) | |
| pmpF | 215.5 (8.1; 7.0) | 3.0 (1.4; 0.1) | 102.1 (5.7; 3.3) | 0.0 (0.0; 0.0) | 237.4 (11.9; 7.7) | 418.0 (18.5; 13.5) | 320.1 (14.6; 10.3) |
| 71.4 (4.9; 6.9) | 1.8 (1.1; 0.2) | 34.3 (3.4; 3.3) | 0.0 (0.0; 0.0) | 74.7 (7.2; 7.3) | 140.5 (11.0; 13.7) | 108.6 (8.8; 10.6) | |
| pmpG | 27.0 (2.9; 0.9) | 3.5 (1.4; 0.1) | 9.1 (1.9; 0.3) | 2.0 (1.0; <0.1) | 14.2 (3.1; 0.5) | 59.8 (6.9; 2.0) | 57.1 (6.6; 1.9) |
| 12.0 (2.1; 1.2) | 2.8 (1.2; 0.3) | 4.0 (1.2; 0.4) | 2.0 (1.0; 0.2) | 6.6 (1.9; 0.7) | 25.8 (5.0; 2.6) | 24.9 (4.8; 2.5) | |
| pmpH | 100.6 (6.0; 3.3) | 1.5 (0.8; <0.1) | 30.9 (2.4; 1.0) | 2.7 (1.3; <0.1) | 131.6 (10.3; 4.4) | 189.1 (12.4; 6.3) | 170.2 (11.1; 5.7) |
| 26.8 (3.02; 2.7) | 0.0 (0.0; 0.0) | 8.3 (1.3; 0.8) | 1.3 (0.9; 0.1) | 33.6 (5.1; 3.4) | 59.3 (7.1; 6.0) | 43.9 (5.9; 4.4) | |
| pmpI | 14.7 (2.4; 0.6) | 5.7 (1.9; 0.2) | 9.2 (1.9; 0.4) | 0.5 (0.5; <0.1) | 14.4 (3.0; 0.6) | 21.8 (4.6; 0.8) | 22.3 (4.2; 0.9) |
| 6.3 (1.5; 0.7) | 1.0 (0.7; 0.1) | 4.8 (1.3; 0.6) | 0.0 (0.0; 0.0) | 8.0 (2.3; 0.9) | 7.5 (2.5; 0.9) | 7.5 (2.2; 0.9) | |
Upper-row data are nucleotide substitutions; lower-row data are amino acid substitutions.
FIG. 1.
Mean genetic distance within each of the nine C. trachomatis pmp genes based on the average p-distance between all possible pairs of sequences (different serovars) for the same pmp gene. Minimum and maximum values represent lower and upper limits of the 95% confidence interval of the estimate (vertical bar), while values plotted at the horizontal level represent the mean estimate. Absolute and percent values for the nucleotide and amino acid mean genetic distance for each pmp gene are also presented.
The genetic distance between disease groups was highly divergent for the LGV group for almost all pmp genes, where with the exception of pmpE and pmpF, the distance between this group and the urogenital and ocular groups was quite similar (Table 2). The most striking differences were for pmpF with high mean differences for the ocular-urogenital groups as follows: mean nucleotide difference, 237.4 (SE, 11.9) (7.7%); mean amino acid difference, 74.7 (SE, 7.2) (7.3%). For the urogenital-LGV group, the results were as follows: mean nucleotide difference, 320.1 (SE, 14.6) (10.3%); mean amino acid difference, 108.6 (SE, 8.8) (10.6%). For the ocular-LGV group, the results were as follows: mean nucleotide difference, 418.0 (SE, 18.5) (13.5%); mean amino acid difference, 140.5 (SE, 11.0) (13.7%). pmpF also had the highest distance between all disease groups, which was also confirmed by phylogenetic analysis (see below).
All genes except pmpA, pmpC, and pmpI revealed small deletion events for some serovars, varying from 1 for pmpD and pmpG to 21 codon deletions for pmpH (Table 3). Furthermore, pmpB had an insertion of 60 bp corresponding to a 20-amino-acid sequence at nt position 2427 for all serovars except for serovar Ia. This insertion included an imperfect direct target repeat (DTR) of 8 bp in the C terminus (2480CT[T/C]CAGCA2487) also present in the region preceding the insertion. As extensively discussed previously for pmpC, where we discovered putative DTRs for two IS-like elements (15), we think that this insertion might reflect a remnant fragment of an IS-like element, where the DTRs were created by duplication of the target sequence upon insertion of the IS element. A BLAST search revealed no homology with any gene or gene segments.
TABLE 3.
Deletion events for pmp genes among C. trachomatis reference serovars
| Gene | Deletion event
|
||
|---|---|---|---|
| Position (nt)a | No. of codons | Serovar(s) | |
| pmpA | —b | — | |
| pmpBc | 1157 | 2 | A-D, G-K |
| 1157 | 1 | LGV | |
| 1964 | 1 | Da, E, F | |
| 3836 | 3 | All but LGV | |
| pmpCc | — | — | |
| pmpD | 3124 | 1 | E, F, LGV |
| 432 | 1 | All but D, E, F, G, H, Ia | |
| 1122 | 2 | A-C, Da | |
| pmpE | 1237 | 1 | A-C, Da, I, J, Ja, K |
| 1486 | 4 | A-C, Da | |
| 1607 | 4 | A-C, Da | |
| 1291 | 1 | LGV | |
| pmpF | 2112 | 1 | H, I, J, Ja |
| 2130 | 1 | LGV | |
| pmpG | 1045 | 1 | L2 |
| 488 | 2 | I, J, Ja, K | |
| 483 | 12 | LGV | |
| 699 | 2 | LGV | |
| pmpH | 859 | 1 | L1, L3 |
| 878 | 1 | A-C, Da | |
| 1662 | 1 | A-C, Da | |
| 1872 | 2 | A-C, Da | |
| pmpI | — | — | |
The position is based on the relative pmp gene sequence of serovar D (UW-3) (GenBank no. AE001360).
—, no deletion event noted.
The remnant fragments of the putative IS-like elements present in pmpB (this paper) and pmpC (15) were not included in this table.
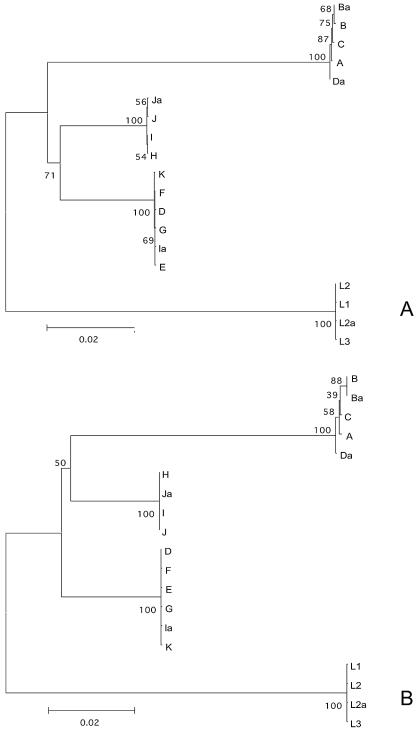
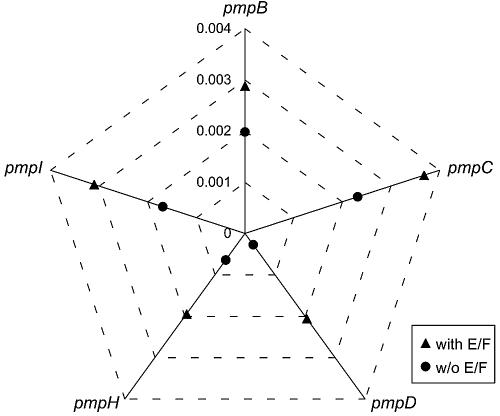
Phylogenetic reconstructions showed that, for six of nine pmp genes, serovar groupings were based on biologic properties related to tissue tropism. This was especially evident for pmpF (Fig. 2) and pmpH and to a lesser extent for pmpB, pmpC, pmpG, and pmpI (see Fig. S1 to S5 in the supplemental material). Serovars E and F clustered in a phylogenetically distant branch from the urogenital group for pmpD (Fig. 3) and pmpI, and to a lesser extent for pmpH. However, serovar Da was also included in the serovar E and F clade for pmpB and pmpC. The analysis of the genetic heterogeneity within this group revealed that the urogenital serovars became considerably genetically closer when serovars E and F were removed from the group (Fig. 4).
FIG. 2.
Phylogenetic reconstruction of the nucleotide (A) and amino acid (B) sequences showing the evolutionary history of pmpF by neighbor-joining tree topologies, based on distance estimates using a Kimura two-parameter model for substitution events (nucleotides) and the gamma distance model (amino acids). These reconstructions are based on pmpF sequences of the 18 serovars and the genovariant Ja of C. trachomatis. Branch lengths are proportional to distances between serovars. The values at the nodes are the bootstrap confidence levels representing the percentage of 1,000 bootstrap replicates for which the serovar to the right was separated from the others.
FIG. 3.
Phylogenetic reconstruction of the nucleotide (A) and amino acid (B) sequences showing the evolutionary history of pmpD by neighbor-joining tree topologies, based on distance estimates using a Kimura two-parameter model for substitution events (nucleotides) and the gamma distance model (amino acids). These reconstructions are based on pmpD sequences of the 18 serovars and the genovariant Ja of C. trachomatis. Branch lengths are proportional to distances between serovars. The values at the nodes are the bootstrap confidence levels representing the percentage of 1,000 bootstrap replicates for which the serovar to the right was separated from the others.
FIG. 4.
Mean genetic distance (p-distance) within the C. trachomatis genital group (11 serovars) with (▴) and without (•) serovars E and F, for pmpB to -D and pmpH to -I.
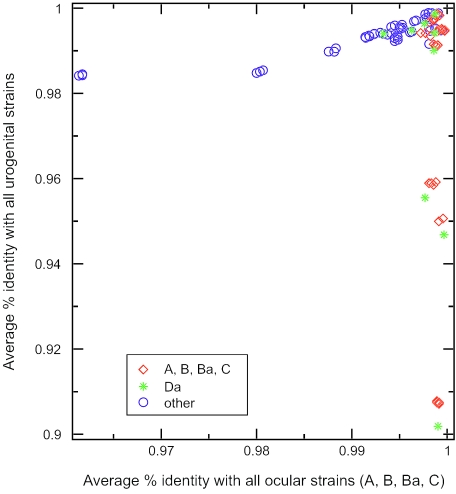
The phylogenetic analysis of serovar Da showed that it clustered with the ocular instead of the urogenital group for pmpE (see Fig. S6 in the supplemental material), pmpF, pmpG, pmpH, and pmpI; these five pmp genes occur in the second half of the chromosome. The most striking example was for pmpF, where serovar Da was genetically distant from the mean of the urogenital group by 258 nt, where there were only 3-nt differences from the mean of the ocular group (corresponding to 80 and 2 amino acids, respectively) (Fig. 2). We also took the arithmetic mean of the distances to all ocular, LGV, and urogenital serovars, which included serovar Da, and graphed 100* (1 p-distance) for both axes (Fig. 5). The distance from a serovar sequence to itself was not included in the average. Here, serovar Da clustered with the ocular group for the same pmp genes (pmpE through pmpI). Bootstrap support of ≥99% for nucleotide trees grouping serovar Da with ocular strains in pmpE through pmpI but with urogenital strains for pmpB and pmpC implied a P value of 0.01 × 0.01 = 0.0001 and that both groupings were not spurious. For third-codon position trees (not shown), which largely rule out any effects due to protein evolutionary pressure, the bootstrap support for these groupings was always at least 95%, implying a P value of 0.0025.
FIG. 5.
Similarity of pmp sequences to those from other ocular and urogenital strains. For the more variable pmp genes, the ocular strains (red) are well separated from the urogenital ones (blue asterisks). The exception is the urogenital serovar Da (green asterisks), which spans the two clusters, providing evidence for recombination. The lower-right Da points represent pmp genes pmpE, -F, and -H, each near the corresponding pmp genes from ocular strains. The upper-left Da points represent pmp genes pmpB and pmpC. Urogenital sequences from the more variable pmp genes (i.e., pmpE, -F, and -H) are not shown because they fall far off the scale to the left. Note that the percent identity is expressed as the fraction identity.
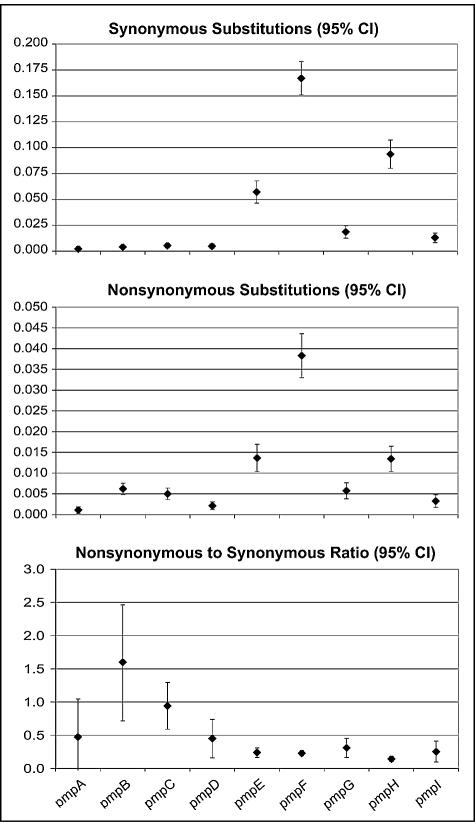
The analysis of the molecular evolution of the pmp genes based on the ratio of nonsynonymous to synonymous substitutions showed dN/dS values ranging from 0.14 (SE, 0.02) for pmpH to 1.59 (SE, 0.45) for pmpB (Fig. 6). The most polymorphic genes among all C. trachomatis serovars (pmpE, pmpF, and pmpH) had the highest number of nonsynonymous and synonymous substitutions resulting in the lowest dN/dS ratio. Also, the dN/dS ratio calculated separately for the 3′ end (corresponding to the 40-kDa C-terminal domain) and to the 5′ half (corresponding to the N terminus containing all conserved GGAI motifs) showed a remarkable difference only for pmpE, where the N terminus appeared to be less evolutionarily conserved than the C terminus (1.21 [SE, 0.61] and 0.22 [SE, 0.05], respectively).
FIG. 6.
Top and middle graphs show mean synonymous and nonsynonymous mutation substitutions, respectively, for each of the nine pmp genes, based on the method of Nei-Gojobori (p-distance model). Minimum and maximum values represent lower and upper limits of the 95% confidence interval of the estimate (vertical bar), while values plotted at horizontal level (diamonds) represent the mean estimates. The graph on the bottom shows the nonsynonymous-to-synonymous mutation ratio for each pmp gene, based on the mean estimates shown in the prior two graphs.
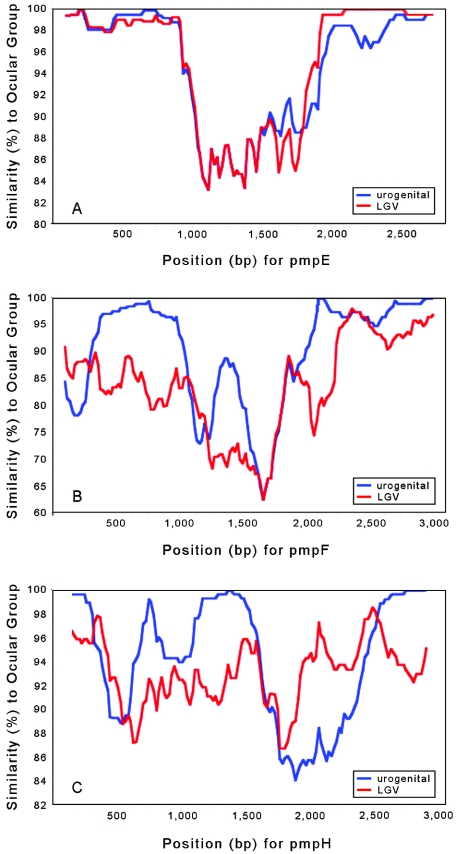
SimPlot analysis was performed to search for specific pmp regions that phylogenetically define tissue tropism differences among serovars. SimPlot revealed distinct gene domains for pmpE, pmpF, and pmpH where nucleotide substitutions were concentrated and associated with a specific disease group instead of being randomly distributed (Fig. 7). The most notable example was for pmpE, where there was segregation of the ocular group distinct from urogenital and LGV groups in the mid-region of the gene (from nt ∼1050 to 1900) (Fig. 7A). For pmpF (Fig. 7B), several main gene regions could be observed: one where the LGV group separated from the ocular and other urogenital serovars (from nt ∼300 to 1000), two regions where there was a clear separation of all groups (from nt ∼1250 to 1500 and ∼nt 2050 to 2250), and another two regions where the segregation was seen for the ocular group (from nt ∼1000 to 1200 and ∼nt 1600 to 2000). For pmpH (Fig. 7C), the ocular group was distinct from the other groups at nt 350 to 650 and nt 1600 to 1900; the LGV group had lower similarity to the other groups from nt 1100 to 1550 and from nt 2500 until the end of the gene. A clear separation of the three disease groups was found at nt 1900 to 2350. For PmpE, PmpF, and PmpH, these clusters of point mutations corresponded to clusters of nonconservative amino acid changes in the protein alignment (data not shown).
FIG. 7.
SimPlot graphs for pmpE (A), pmpF (B), and pmpH (C) of nucleotide similarity (using the Kimura two-parameter method) between the three disease groups, where each group represents the mean genetic distance of the respective serovars. Genital and LGV groups are compared to the ocular group. Ocular group, serovars A/SA-1, B/TW-5, Ba/Apache-2, and C/TW-3; urogenital group, urogenital group of serovars as described in Materials and Methods.
Analysis of protein features of the pmp family.
As expected for membrane proteins, all nine Pmps (except for PmpA) for the 18 serovars and genovariant Ja contained a cleavable peptidase signal at the N-terminal domain and a C-terminal phenylalanine. The distribution of tryptophan residues was also conserved among the serovars and was almost exclusively clustered at the C-terminal half of all Pmps (see Fig. S7 in the supplemental data).
The number and distribution of the conserved cysteine residues varied from nine to 26 cysteines for PmpA and PmpD, respectively (see Fig. S7 in the supplemental data). The distribution of the GGAI motifs that were conserved for all serovars varied from 2 for PmpH to 11 for PmpD, respectively, and were almost exclusively located in the N-terminal half of the protein (see Fig. S7 in the supplemental data). With few exceptions, they seemed to be grouped in clusters. Furthermore, for PmpD, PmpE, PmpF, PmpG, PmpH, and PmpI, the distribution of these motifs seemed nonrandom, as ∼60% of them are adjacent or within 10 nt of the cysteine residues.
DISCUSSION
The small genome of C. trachomatis (∼1.1 Mbp, about 25% of Escherichia coli's genome) is thought to have undergone progressive genome reduction from a larger precursor organism (33), while maintaining the ability to infect different tissue types and cause human diseases of major public health significance. It is thus presumed that this intracellular pathogen retains only the most important genes for survival and propagation. Consequently, the presence of a nine-member polymorphic membrane protein family that is unique to Chlamydiales and that represents 3.2% of the coding capacity of the genome (18) suggests that these proteins likely play a role in diversifying exposed antigens for escape from host immune surveillance (33). Our results support this theory.
We found extensive polymorphisms among serovars for many of the pmp genes, the most notable of which was pmpF, where the mean genetic distance among all C. trachomatis serovars was 215.5 nt (7.0% change) (Table 2). Considering the variability in pmp and Pmp sequences, we expected to find an evolutionary divergence for serovars with different cellular tropism. In support of this and in contrast to other chlamydial membrane proteins that have been studied, six of the nine pmp genes presented phylogenetic reconstructions where serovars were clustered according to disease properties. Further, genetic distances were significant in distinguishing the ocular, urogenital, and LGV disease groups for pmpF and pmpH (Table 2).
Unexpectedly, the urogenital serovar Da showed an evolutionary trend similar to the ocular serovars for the pmp cluster from pmpE to pmpI. However, the high similarity with the ocular group within this large genomic region (∼15,000 bp) and the remarkable genetic distance between the ocular and the urogenital groups for pmpE to pmpI (Table 2; Fig. 5) point to a recombination event instead of the accumulation of point mutations throughout evolution. Also, the phylogenetic analysis based on third-codon position trees that rule out any effects due to evolutionary pressure support these groupings (P = 0.0025). Thus, assuming that a partial arsenal of ocular Pmps can be effective, it appears that serovar Da has acquired a genetic composition that could hypothetically expand its host cell range. We know that urogenital serovars can cause a self-limited conjunctivitis (9) and we previously described ompA mosaics of B/D and Ba/D, the former of which was isolated from a patient with trachoma in Tunisia (8), while the latter was isolated from a patient with a sexually transmitted disease in the United States (26). While these occurrences do not directly support host cell expansion, they do support recombination among strains with different cell appetences. These collective data lend further support to the plasticity of the genome and the evidence for adaptive responses to environmental or host pressures.
We previously described two putative IS-like elements for pmpC (15). As for pmpC, the IS-like element identified for pmpB might reflect the prior presence of a complete insertion with a nearly precise excision phenomenon, leaving the imperfect downstream DTR and remnants of the IS at the site of the insertion (3, 6, 25). The differential presence among serovars of remnants of this putative IS-like element for pmpB (as for pmpC) may indicate either a differential insertion event among the serovars or a selective excision upon insertion. A complete deletion of the IS-like element may also have occurred for some serovars, a phenomenon that involves the two DTRs and DNA replication slippage (2, 37). It has been suggested that DTRs may lead to sequence deletions, as observed in this paper, and genome reductive evolution of intracellular bacteria (37), as has been documented for C. trachomatis (35). Considering the evidence for a high frequency of deletions occurring between two direct repeats in close proximity (2, 34), we would predict the complete deletion of these IS remnants during pmp evolution.
Phylogenetic analyses revealed that the pmp genes have evolved quite heterogeneously. pmpB showed the highest dN/dS ratio (1.59 [SE, 0.45]) (Table 2), indicating an evolutionary trend toward protein modification, which may occur via host immune pressure or horizontal gene transfer. In contrast, the most polymorphic pmp genes, pmpF and pmpH, encode the most evolutionary conserved proteins (with dN/dS values of 0.23 [SE, 0.02] and 0.14 [SE, 0.02], respectively) (Table 2). PmpF and PmpH may therefore play an important functional role where the occurrence of amino acid substitutions may be disadvantageous, while not being especially exposed to immune pressure. On the other hand, the phylogenetic clustering of serovars by disease properties and the remarkable genetic distance between serovars from different disease groups (Table 2) for pmpF and pmpH suggest an evolutionary adaptation of serovars to a specific tissue. Thus, we speculate that the biologic role of PmpF and PmpH may be associated with adhesion and entry into the host cell, where amino acid substitutions could be disadvantageous, by altering essential structural and functional characteristics. This is supported by the recent finding that antibodies specific to C. pneumoniae Pmp21 blocked chlamydial infectivity in the epithelial cells, which suggests a role for pmp genes in the adhesion process (50).
We also found that for five of the nine pmp genes (pmpB, -C, -D, -H, and -I), serovars E and F diverged separately from the other urogenital serovars and were closely related in a distant clade. This is interesting because these two serovars are the most prevalent among sexually transmitted diseases worldwide. The findings are distinct from ompA, where serovars E and F are evolutionarily closer to other serovars (27). The influence of these serovars on the heterogeneity of the urogenital group is illustrated in Fig. 4, where the exclusion of serovars E and F from this group for the five pmp genes dramatically decreased the mean genetic distance within the urogenital disease group. It is possible that these pmp genes confer a biological advantage for serovars E and F over the other serovars that may facilitate infectivity and transmission. Globally, as the majority of genes among the pmp family present phylogenetic reconstructions that largely segregate serovars according to disease properties, it indicates that Pmps may be involved in modulation of tissue tropism by either structural or functional constraints. While there is still a lack of data on the conformation and structure of Pmps, we have previously shown diverse immunoreactivity of patient sera to recombinant PmpC and heterogeneous transcriptome profiles for pmpC for serovars representing the three disease groups (16).
We also examined the distribution of polymorphisms in pmp genes that might segregate at least one disease group. For pmpE, pmpF, and pmpH, it was possible to observe a nonrandom distribution of point mutations with clustering in specific regions that clearly distinguished disease groups. pmpE contained one large gene region that distinguished the ocular serovars from all others; five other discrete regions were observed for pmpF and pmpH that differentiated at least one disease group (Fig. 7). For all these regions, clusters of point mutations also corresponded to smaller clusters of residue substitutions, which suggests that, if Pmps are hypothetically involved in tropism for chlamydial strains, these specific peptide segments on Pmps would certainly be responsible for those biological differences, either by the modification of the protein conformation structure or by alteration of its function. Unfortunately, the lack of similarity between Pmps and other proteins for which the three-dimensional structure has already been determined makes any prediction of hypothetical changes in protein folding due to these specific regions highly speculative.
Only recently has the translation and membrane localization of all Pmps been demonstrated by proteomics for C. trachomatis (serovar L2) (41), although previous studies had detected some Pmps (also for L2) (40, 45). The membrane location of Pmps was supported by our results, as demonstrated by a cleavable peptidase signal at the N-terminal domain (except for PmpA), a C-terminal phenylalanine, and tryptophan residues that were almost exclusively clustered at the C terminus for all serovars and Ja (see Fig. S6 in the supplemental material). Also, some Pmps, including PmpA, had high numbers of cysteine residues (PmpD; 26 residues), which are usually involved in the intra- and intermolecular interactions of membrane proteins through disulfide bonds. Although PmpA did not contain a signal peptide, the lack of one can be overcome by the presence of an anchor sequence (which is less well defined than the signal peptide) in a protein region that spans the C-terminal domain of the membrane and initiates translocation. Further, PmpA was the only Pmp reported by Skipp et al. (41) to localize to the reticulate body and not the elementary body, suggesting that it may be an important reticulate body membrane constituent but not under the influence of immune modulation.
The GGAI motifs were generally conserved among all serovars, were almost exclusively located in the N-terminal half of the protein, and in general were grouped in clusters (see Fig. S7 in the supplemental material). For PmpD, -E, -F, -G, -H, and -I, they were close or adjacent to cysteine residues, indicating a nonrandom distribution. In the autotransporter model proposed for chlamydial Pmps (19, 47), the β-barrel structure of the C-terminal domain forms a pore in the membrane, mediating the translocation of the N terminus to the outer membrane and becoming exposed in bacterial surface. This theoretical model was experimentally demonstrated for C. pneumoniae Pmp21 (ortholog of C. trachomatis PmpD) (50). In view of this, the N-terminal location of the GGAI motifs, in close proximity to cysteine residues which are responsible for the highly disulfide cross-linked proteins of the outer membrane complex of C. trachomatis, suggests both a relevant biological role for the GGAI motifs and a hypothetical cooperative action with the cysteine residues for structural or functional constraints of the passenger domain. Considering this model, we have also separately studied the molecular evolution of both the N-terminal (passenger) domain and the ∼40 kDa C-terminal domain for each Pmp; curiously, PmpE (but none of the other Pmps) showed a remarkable difference of dN/dS between those two domains (1.21 [SE, 0.61] and 0.22 [SE, 0.05], respectively). Accordingly, it seems that the N-passenger domain of PmpE has been undergoing amino acid changes throughout evolution, which supports the above-described theoretical model in which this domain is exposed in the bacterial surface and subjected to immune pressure. In contrast, the C-terminal region shows clear evolutionary conservation, which supports conserved function without immune pressure for this domain.
The comparative genetic, genomic, and molecular analyses performed in this study suggest a possible structural or functional role that may vary among Pmps in promoting antigenic polymorphisms on the bacterial surface and/or promoting a set of different receptors that may be involved in host cell adhesion and differential tissue tropism. Additional genetic and genome sequencing, especially of clinical isolates, will be required to tease out the effects of each genetic event described above.
Supplementary Material
Acknowledgments
This work was supported by grants from the Fundação Para a Ciência e Tecnologia (62%) and FEDER (38%) (POCTI/39822/MGI/2001) (to M.J.B.), the Comissão de Fomento da Investigação em Cuidados de Saúde (no. 187/01) (to J.P.G.), and the Public Health Service (grants AI39499 and AI59647) from the National Institutes of Health (to D.D.).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 2.Bi, X., and L. F. Liu. 1996. A replicational model for DNA recombination between direct repeats. J. Mol. Biol. 256:849-858. [DOI] [PubMed] [Google Scholar]
- 3.Blot, M. 1994. Transposable elements and adaptation of host bacteria. Genetica 93:5-12. [DOI] [PubMed] [Google Scholar]
- 4.Caldwell, H. D., J. Kromhout, and J. Schachter. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 31:1161-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2002. Sexually Transmitted Disease Surveillance. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, Atlanta, Ga.
- 6.Chalmers, R., and M. Blot. 1999. Insertion sequences and transposons, p. 151-168. In R. L. Charlebois (ed.), Organization of the prokaryotic genome. American Society for Microbiology, Washington, D.C.
- 7.Dean, D. 1997. Chlamydia trachomatis sexually transmitted diseases, p. 473-490. In D. H. Connor, D. A. Schwartz, and F. W. Chandler (ed.), Pathology of infectious diseases. Appleton and Lange Publishers, Stamford, CT.
- 8.Dean, D. 1994. Molecular characterization of new Chlamydia trachomatis serological variants from a trachoma endemic region of Africa, p. 259-262. In J. Orfila, G. I. Byrne, M. A. Chernesky, J. T. Grayston, R. B. Jones, G. L. Ridgway, R. Saikku, J. Schachter, W. E. Stamm, and R. S. Stephens (ed.), Chlamydial infections. Societa Editrice Esculapio, Bologna, Italy.
- 9.Dean, D. 2002. Pathogenesis of chlamydial ocular infections, p. 1-22. In W. Tasman and E. A. Jaeger (ed.), Duane's foundations of clinical ophthalmology, vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 10.Dean, D., M. Patton, and R. S. Stephens. 1991. Direct sequence evaluation of the major outer membrane protein gene variant regions of Chlamydia trachomatis subtypes D′, I′, and L2′. Infect. Immun. 59:1579-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dean, D., and V. C. Powers. 2001. Persistent Chlamydia trachomatis infections resist apoptotic stimuli. Infect. Immun. 69:2442-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dean, D., R. Suchland, and W. Stamm. 2000. Evidence for long-term cervical persistence of Chlamydia trachomatis by omp1 genotyping. J. Infect. Dis. 182:909-916. [DOI] [PubMed] [Google Scholar]
- 13.Fields, P. I., and R. C. Barnes. 1992. The genus Chlamydia, p. 3691-3709. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes. Springer-Verlag, New York, N.Y.
- 14.Fleming, D. T., and J. N. Wasserheit. 1999. From epidemiological synergy to public health policy and practice: the contribution of other sexually transmitted diseases to sexual transmission of HIV infection. Sex. Transm. Infect. 75:3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomes, J. P., W. J. Bruno, M. J. Borrego, and D. Dean. 2004. Recombination in the genome of Chlamydia trachomatis involving the polymorphic membrane protein C gene relative to ompA and evidence for horizontal gene transfer. J. Bacteriol. 186:4295-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomes, J. P., R. C. Hsia, S. Mead, M. J. Borrego, and D. Dean. 2005. Immunoreactivity and differential developmental expression of known and putative Chlamydia trachomatis membrane proteins for biologically variant serovars representing distinct disease groups. Microbes Infect. 7:410-420. [DOI] [PubMed] [Google Scholar]
- 17.Goodall, J. C., G. Yeo, M. Huang, R. Raggiaschi, and J. S. Gaston. 2001. Identification of Chlamydia trachomatis antigens recognized by human CD4+ T lymphocytes by screening an expression library. Eur. J. Immunol. 31:1513-1522. [DOI] [PubMed] [Google Scholar]
- 18.Grimwood, J., and R. S. Stephens. 1999. Computational analysis of the polymorphic membrane protein superfamily of Chlamydia trachomatis and Chlamydia pneumoniae. Microb. Comp. Genomics 4:187-201. [DOI] [PubMed] [Google Scholar]
- 19.Henderson, I. R., and A. C. Lam. 2001. Polymorphic proteins of Chlamydia spp.—autotransporters beyond the Proteobacteria. Trends Microbiol. 9:573-578. [DOI] [PubMed] [Google Scholar]
- 20.Hsia, R. C., I. Ahmed, B. Batteiger, O. Sekkides, G. Ridgway, and P. M. Bavoil. 2000. Presented at the Fourth Meeting of the European Society for Chlamydia Research, Helsinki, Finland, 20-23 August 2000.
- 21.Kalman, S., W. Mitchell, R. Marathe, C. Lammel, J. Fan, R. W. Hyman, L. Olinger, J. Grimwood, R. W. Davis, and R. S. Stephens. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21:385-389. [DOI] [PubMed] [Google Scholar]
- 22.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 23.Lindquist, E. A., and R. S. Stephens. 1998. Transcriptional activity of a sequence variable protein family in Chlamydia trachomatis, p. 259-262. In R. S. Stephens, G. I. Byrne, G. Christiansen, I. N. Clarke, J. T. Grayston, R. G. Rank, G. L. Ridgway, P. Saikku, J. Schachter, and W. E. Stamm (ed.), Chlamydial infections. Proceedings of the Ninth International Symposium on Human Chlamydial Infection. International Chlamydia Symposium, Napa, Calif.
- 24.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Millman, K., C. M. Black, R. E. Johnson, W. E. Stamm, R. B. Jones, E. W. Hook, D. H. Martin, G. Bolan, S. Tavaré, and D. Dean. 2004. Population-based genetic and evolutionary analysis of Chlamydia trachomatis urogenital strain variation in the United States. J. Bacteriol. 186:2457-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Millman, K. L., S. Tavaré, and D. Dean. 2001. Recombination in the ompA gene but not the omcB gene of Chlamydia contributes to serovar-specific differences in tissue tropism, immune surveillance, and persistence of the organism. J. Bacteriol. 183:5997-6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mygind, P. H., G. Christiansen, P. Roepstorff, and S. Birkelund. 2000. Membrane proteins PmpG and PmpH are major constituents of Chlamydia trachomatis L2 outer membrane complex. FEMS Microbiol. Lett. 186:163-169. [DOI] [PubMed] [Google Scholar]
- 29.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 30.Nei, M., and S. Kumar. 2000. Molecular evolution and phylogenetics. Oxford University Press, New York, N.Y.
- 31.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen, H., and A. Krogh. 1998. Prediction of signal peptides and signal anchors by a hidden Markov model. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:122-130. [PubMed] [Google Scholar]
- 33.Palmer, G. H. 2002. The highest priority: what microbial genomes are telling us about immunity. Vet. Immunol. Immunopathol. 85:1-8. [DOI] [PubMed] [Google Scholar]
- 34.Pierce, J. C., D. Kong, and W. Masker. 1991. The effect of the length of direct repeats and the presence of palindromes on deletion between directly repeated DNA sequences in bacteriophage T7. Nucleic Acids Res. 19:3901-3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Read, T. D., G. S. Myers, R. C. Brunham, W. C. Nelson, I. T. Paulsen, J. Heidelberg, E. Holtzapple, H. Khouri, N. B. Federova, H. A. Carty, L. A. Umayam, D. H. Haft, J. Peterson, M. J. Beanan, O. White, S. L. Salzberg, R. C. Hsia, G. McClarty, R. G. Rank, P. M. Bavoil, and C. M. Fraser. 2003. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 31:2134-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rocha, E. P. 2003. An appraisal of the potential for illegitimate recombination in bacterial genomes and its consequences: from duplications to genome reduction. Genome Res. 13:1123-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 39.Schiffer, M., C. H. Chang, and F. J. Stevens. 1992. The functions of tryptophan residues in membrane proteins. Protein Eng. 5:213-214. [DOI] [PubMed] [Google Scholar]
- 40.Shaw, A. C., K. Gevaert, H. Demol, B. Hoorelbeke, J. Vandekerckhove, M. R. Larsen, P. Roepstorff, A. Holm, G. Christiansen, and S. Birkelund. 2002. Comparative proteome analysis of Chlamydia trachomatis serovar A, D and L2. Proteomics 2:164-186. [DOI] [PubMed] [Google Scholar]
- 41.Skipp, P., J. Robinson, C. D. O'Connor, and I. N. Clarke. 2005. Shotgun proteomic analysis of Chlamydia trachomatis. Proteomics 5:1558-1573. [DOI] [PubMed] [Google Scholar]
- 42.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, and L. Olinger. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 43.Stothard, D. R., G. A. Toth, and B. E. Batteiger. 2003. Polymorphic membrane protein H has evolved in parallel with the three disease-causing groups of Chlamydia trachomatis. Infect. Immun. 71:1200-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Struyve, M., M. Moons, and J. Tommassen. 1991. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J. Mol. Biol. 218:141-148. [DOI] [PubMed] [Google Scholar]
- 45.Tanzer, R. J., and T. P. Hatch. 2001. Characterization of outer membrane proteins in Chlamydia trachomatis LGV serovar L2. J. Bacteriol. 183:2686-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vandahl, B. B., S. Birkelund, and G. Christiansen. 2004. Genome and proteome analysis of Chlamydia. Proteomics 4:2831-2842. [DOI] [PubMed] [Google Scholar]
- 47.Vandahl, B. B., S. Birkelund, H. Demol, B. Hoorelbeke, G. Christiansen, J. Vandekerckhove, and K. Gevaert. 2001. Proteome analysis of the Chlamydia pneumoniae elementary body. Electrophoresis 22:1204-1223. [DOI] [PubMed] [Google Scholar]
- 48.Wang, S. P., and J. T. Grayston. 1991. Three new serovars of Chlamydia trachomatis: Da, Ia, and L2a. J. Infect. Dis. 163:403-405. [DOI] [PubMed] [Google Scholar]
- 49.Wang, S. P., C. C. Kuo, R. C. Barnes, R. S. Stephens, and J. T. Grayston. 1985. Immunotyping of Chlamydia trachomatis with monoclonal antibodies. J. Infect. Dis. 152:791-800. [DOI] [PubMed] [Google Scholar]
- 50.Wehrl, W., V. Brinkmann, P. R. Jungblut, T. F. Meyer, and A. J. Szczepek. 2004. From the inside out—processing of the chlamydial autotransporter PmpD and its role in bacterial adhesion and activation of human host cells. Mol. Microbiol. 51:319-334. [DOI] [PubMed] [Google Scholar]
- 51.Wilson, J. S., E. Honey, A. Templeton, J. Paavonen, P. A. Mardh, and B. Stray-Pedersen. 2002. A systematic review of the prevalence of Chlamydia trachomatis among European women. Hum. Reprod. Update 8:385-394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.